Abstract
Specific chromosomal abnormalities are increasingly recognised to be associated with particular tumour subtypes. These cytogenetic abnormalities define the sites of specific genes, the alteration of which is implicated in the neoplastic process. We used comparative genomic hybridisation (CGH) to examine DNA from different breast and ovarian cancer cell lines for variations in DNA sequence copy number compared with the same normal control. We also compared different sources of the MCF7 breast line by both CGH and cDNA expression arrays. Some of the differences between the subcultures were extensive and involved large regions of the chromosome. Differences between the four subcultures were observed for gains of 2q, 5p, 5q, 6q, 7p, 7q, 9q, 10p, 11q, 13q, 14q, 16q, 18p and 20p, and losses of 4q, 5p, 5q, 6q, 7q, 8p, 11p, 11q, 12q, 13q, 15q, 19p, 19q, 20p, 21q, 22q and Xp. However, few variations were found between two subcultures examined, 5 months apart, from the same initial source. The RNA arrays also demonstrated considerable variation between the three different subcultures, with only 43% of genes expressed at the same levels in all three. Moreover, the patterns of the expressed genes did not always reflect our observed CGH aberrations. These results demonstrate extensive genomic instability and variation in RNA expression during subculture and provide supportive data for evidence that cell lines do evolve in culture, thereby weakening the direct relevance of such cultures as models of human cancer. This work also reinforces the concern that comparisons of published analyses of cultures of the same name may be dangerous.
Keywords: CGH, expression arrays, cell culture, breast cancer, ovarian cancer
Long-term culture in vitro has represented the most commonly used experimental model of cancer for the past 40 years (Goldin et al, 1979, pp. 165–245; Boyd, 1989, pp. 1–12; Monks et al, 1991). Even though the focus of the NCI screen has changed in recent years to become more tissue specific and molecularly targeted, heavy reliance is still placed on well-characterised long-term cell culture lines. This is of concern since phenotypic diversity has been demonstrated between established cell lines and the primary tumour from which they were cultured (Band et al, 1990; Lukashov and Goudsmit, 1995; Sen et al, 1995). Moreover, differences in biological properties have been demonstrated between sublines of both haematopoietic and breast cell lines (Osborne et al, 1987; Band et al, 1990; Crepin et al, 1990; Lukashov and Goudsmit, 1995). However, long-term subcultures have not been examined for variation in expression profiling and genomic (chromosomal) abnormalities.
In this study, we report considerable variation between different cultures of the MCF7 cell line and lesser variation between different cultures of three ovarian cell lines. The extent of differences was more pronounced for a particular cell line interlaboratory than within the same laboratory at different time points. These results provide supportive data for evidence that cell lines do evolve in culture, thereby weakening the direct relevance of such cultures as models of human cancer. This work also reinforces the concern that comparisons of published analyses of cultures of the same name may be dangerous.
In order to identify new drug resistance genes, we have applied the technique of comparative genomic hybridisation (CGH) (Kallioniemi et al, 1992) to screen for chromosomal abnormalities specific to the acquisition of resistance in long-term culture cell lines (Leyland-Jones et al, 1999). We have demonstrated the validity of this approach by identifying high-level amplifications of genes known to be associated with specific resistance mechanisms (Hiorns et al, 1999). CGH ratios were determined for a series of breast and ovarian carcinoma cell lines. We noted considerable variation in the CGH patterns between two cultures of the same cell line obtained from different laboratories. This observation prompted us to investigate further cell lines. For four of the cell lines (one breast and three ovarian), we made a comparison of different sources of the tumour cell line (against the same normal control). For one breast (MCF7) and one ovarian (CH1) cell line, we made a comparison of the aberrations present in cultures from the same laboratory 5 months apart. For one cell line (MCF7), we obtained cultures that had been independently maintained in different geographical locations over a number of generations. In order to compare the expression levels of certain genes with their genomic representation, we used cDNA arrays (Botwell, 1999) to examine different patterns of RNA expression between these different cultures.
Among the methods available for comprehensive analysis of RNA expression, cDNA microarrays were chosen for their ability to detect static information on specific gene expression and dynamic information on the levels of gene expression in a single experiment. From the commercially available microarrays, we chose the Atlas™ cancer arrays (Clontech, Palo Alto, USA) since the cDNAs on the filters are known genes preselected for their involvement in cancer and grouped according to their involvement in specific processes. The sensitivity of this filter-based, 32P-probe-labelled methodology is known to be limited to high and medium abundance genes.
MATERIALS AND METHODS
Cell lines
These cell lines were deliberately chosen to represent realistic diversity in the scientific community. In particular, for the MCF7, the McGill lines came from an NIH source; the two differently timed aliquots were carefully controlled in the senior author's laboratory. The Nottingham and ICR cell lines were both obtained from ATCC and were obviously grown in different laboratories, but under conditions suitable for collaboration.
A series of cell lines previously established from breast and ovarian carcinomas were passaged independently for a number of years in long-term culture. Cell lines MCF7(b), A2780(a), 41M(a) and CH1(a) were obtained from the laboratory of LRK; cell lines OVCAR-3, A2780(b), 41M(b) and CH1(b and c) from the laboratory of LAS; cell lines MCF7 (c and d) from the laboratory of BL-J; and cell line MCF7(a) from the laboratory of TDB. Two aliquots of the MCF7 cell line (c and d) and two of the CH1 cell line (b and c) were sampled 5 months apart, having been kept in continuous passage during the intervening period.
41 M and A2780 are ovarian carcinoma cell lines established from previously untreated patients. OVCAR-3 was established from the malignant ascites of a patient with progressive ovarian adenocarcinoma. CH1 was established from an ovarian carcinoma patient previously treated with, and resistant to, cisplatin and carboplatin. MCF7 is a breast cancer cell line established from a previously untreated patient. All cell lines were aneuploid. All cell lines were maintained as monolayers in Dulbecco's minimum essential medium supplemented with 10% foetal calf serum, 50 μg ml−1 gentamicin, 0.5 μg ml−1 hydrocortisone and 2 mM L-glutamine, in a 5% CO2 atmosphere. For the RNA expression studies, all three MCF7 cell lines were plated at the same densities, appeared to grow at equal rates and were sampled at the same time points.
CGH
DNA was extracted from harvested cells by the conventional phenol/chloroform technique. DNA (1 μg) from each cell line was labelled for 20 h with Biotin-High-Prime (Boehringer Mannheim, Lewes, UK) using their supplied protocol. ‘Normal’ control DNA (1 μg) was labelled with Digoxigenin-High-Prime (Boehringer). Unincorporated nucleotides were removed using Nick columns (Pharmacia, St Albans, UK). Equal amounts (1 μg) of each DNA were combined together with a 50-fold excess of COT1 DNA (Gibco/BRL, Paisley, UK), ethanol precipitated and resuspended in 10 μl Hybrisol VIII (Oncor, Gaithersburg, MD, USA). This probe mixture was then denatured at 75°C for 8 min, chilled on ice and warmed to 37°C, followed by hybridisation to slides containing ‘normal male’ lymphocyte metaphases (Vysis, Downers Grove, IL, USA). Hybridisation and washing conditions were described previously as (Hiorns, in press).
Slides were examined using an Axioskop microscope (Carl Zeiss, Welwyn Garden City, UK) equipped with appropriate filters. Images were collected using a CCD camera (Photometrics, Tucson, AZ, USA) and Quips™ software (Vysis). In order to control against small changes in hybridisation efficiency, at least 10 metaphases were captured for each hybridisation; the chromosomes were karyotyped and the axis defined based on the DAPI banding pattern. The DAPI image as used as a mask for the red and green images to exclude background fluorescence. The total image intensity for the masked red and green images was then independently normalised to accommodate differences in the image capture, so that the average red : green ratio for each cell was 1.0. The red : green ratios for cross-sections of each chromosome were measured perpendicular to the axis. The ratio profiles of 10 metaphases were averaged and represented graphically. Chromosomal imbalances affecting more than 30% of the cell population were identified when the ratio was greater than 1.15–1.20 for gains and less than 0.85–0.80 for losses (all imbalances were within 99% confidence limits). The heterochromatic regions consisting of clustered repetitive sequences were excluded from the analysis as were the telomeres of all chromosomes.
Expression arrays
Total RNA was extracted from cell lines in exponential growth phase, using the Atlas™ Pure RNA Isolation Kit (Clontech, Palo Alto, USA) including all optional purification steps. A measure of 1 μg of each RNA was reverse-transcribed using gene-specific primers and Moloney murine leukaemia virus in the presence of [α-32P]dATP. The cDNA probes were purified from unincorporated nucleotides using Chroma Spin-200 columns (Clontech, Palo Alto, USA) and hybridised overnight at 68°C to Atlas™ cancer arrays (Clontech, version 1 which analyses 588 human cDNA's, nine housekeeping cDNA's and negative controls). A series of high-stringency washes were performed, using the protocol supplied. The hybridisation pattern was detected using a phosphorimager. Signals greater than twice the background were considered positive. The ratio of signal intensity for the expressed genes was compared to that of the housekeeping genes using AtlasImage™ software (Clonetech). Comparisons between the cell lines were carried out on a paired basis using the same software. Differences between expression levels were considered significant according to the accepted criteria of (i) a ratio difference of more than two-fold, or (ii) an intensity difference of more than the two backgrounds combined.
RESULTS
Breast cancer cell line CGH results: overall agreement with literature
The genomic imbalances identified in the MCF7 cell line are in broad agreement with the frequently observed changes in the literature, including both those reported for primary human breast carcinomas (Nishizaki et al, 1997; Tirkkonen et al, 1998) and cell lines (Kallioniemi et al, 1994; Jones et al, 2000). The most frequently observed changes in our four different MCF7 subcultures included consistent gains of 1q, 3p, 3q, 8q, 12q, 15q, 17q and 20q together with persistent losses of 9p, 17p, 18q and X. In particular, we observed the gains 3p14 and 14q reported by Kallioniemi et al (1994) but not observed by Jones et al (2000). However, the gain of 14q was only observed in the cell lines from one of our three laboratories (subcultures c and d).
Striking differences between four MCF7 subcultures
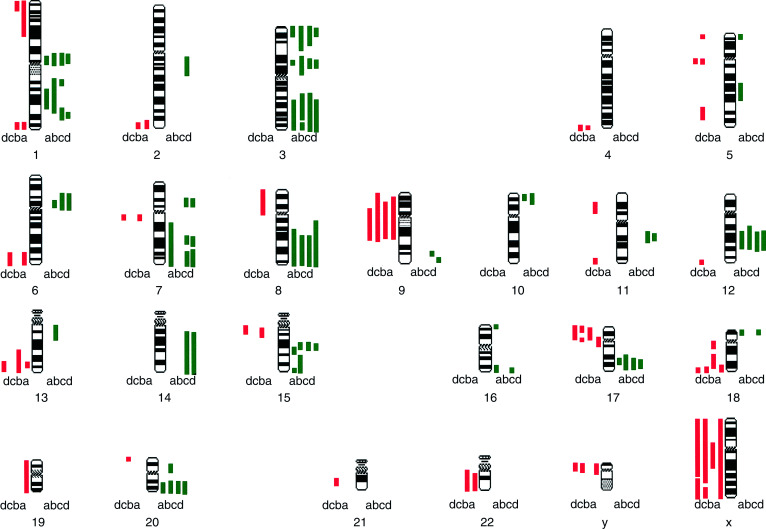
We note surprising variation in the chromosomal aberration patterns between the four different MCF7 subcultures (illustrated in Figure 1). The following differences between the four subcultures were observed: gains of 2q, 5p, 5q, 6q, 7p, 7q, 9q, 10p, 11q, 13q, 14q, 16q, 18p and 20p; and losses of 4q, 5p, 5q, 6q, 7q, 8p, 11p, 11q, 12q, 13q, 15q, 19p, 19q, 20p, 21q, 22q and X. Some of the differences between the subcultures were extensive involving large regions of the chromosome. Many of the changes between our cell lines are similar to those observed by Jones et al (2000). It is reassuring to note fewer variations between the two subcultures from the same laboratory (c and d) which were examined over a period of 5 months in continuous passage. In all, 15 differences were noted between subcultures (c) and (d) in contrast to 22 differences between (a) and (b), 30 differences between (a) and (c), and 34 differences between (b) and (c).
Figure 1.
Idiogram of the human karyotype showing regions of amplification (green) and deletion (red) in DNA from the MCF7 cell line from different sources: (a) University of Nottingham; (b) Institute of Cancer Research (Sutton); (c) McGill University (Montreal) April 1996; and (d) McGill University (Montreal) August 1996.
Fewer CGH differences between ovarian cell line subcultures
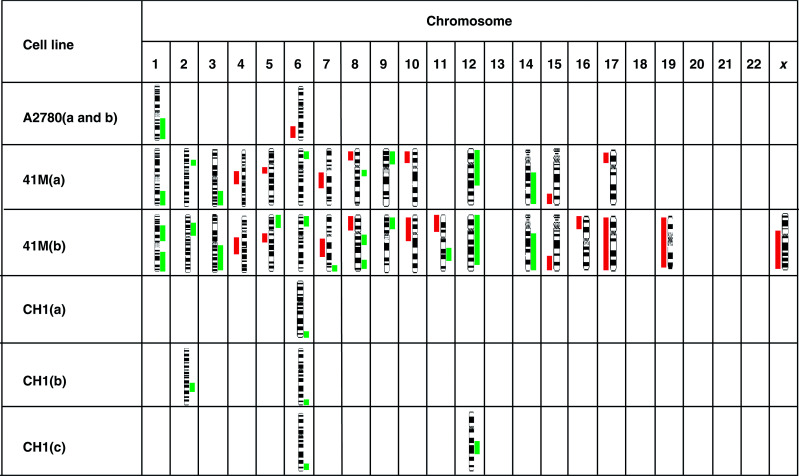
The genomic aberrations identified by CGH in the ovarian cell lines studied (A2780, CH1 and 41M) are listed in Table 1 . The ovarian cancer cell lines showed a wide variation in the number and location of genomic imbalances. In contrast to the MCF7 breast line, each ovarian cancer cell line showed a much smaller degree of variation both between different laboratories and with time.
Table 1. Amplifications (green) and deletions (red) of genomic sequences identified in ovarian tumours and the ovarian tumour-derived cell lines: Ovcar, A2780, 41M and CH1.
Similarities between MCF7 subculture expression patterns
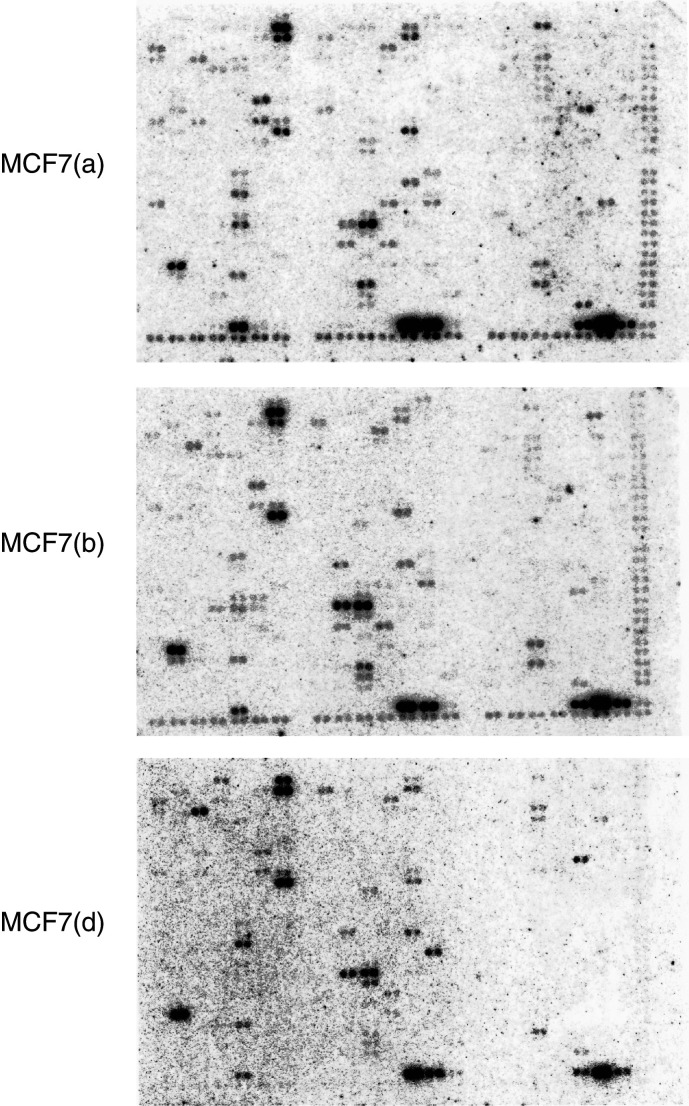
Similar patterns of RNA expression were observed for all three of the MCF7 subcultures (a, b and d) both with respect to specific genes and the levels of gene expression (see Figure 2). In all, 116 genes were expressed at levels greater than 0.01% in one or more of the cultures; this number is similar to that reported by Sehgal et al (1998) in GMTT glioblastoma cells but less than that reported by Xie et al (1998) in HL-60 leukaemia cells. A total of 52 genes showed the same relative levels of expression in all three cultures.
Figure 2.
Clonetech ‘Cancer Array’ filters hybridised with cDNA from MCF7 sublines from different sources: (a) University of Nottingham; (b) Institute of Cancer Research (Sutton); and (d) McGill University (Montreal). Each hybridisation was to a new replicate filter; (none were reprobed).
Sixty-four genes demonstrated variability in levels of expression between the different subcultures
The chromosomal location and relative expression levels for these genes are listed in Table 2 . All 64 genes either showed a ratio difference greater than two-fold or an intensity difference between two subcultures greater than the two background intensities combined (with the vast majority of differences greater than six-fold).
Table 2. Comparative expression levels of the 45% of genes differentially expressed between at least two of the cell lines.
| Chromosome location | Gene name | MCF7 (a)a | MCF7 (b)a | MCF7 (d)a |
|---|---|---|---|---|
| 1p13.21 | rhoC | 3.87 | 6.48 | 3.69 |
| 1p13.3 | Glutathione-S-transferase (GST) | 7.40 | 3.57 | 3.47 |
| 2p10–12 | rhoB | 15.29 | 10.63 | 3.61 |
| 2q33–34 | IGFBP2 | — | 6.43 | — |
| 3p21.3 | rhoA H12 | 14.92 | 7.02 | 7.23 |
| 3p21.3 | Laminin receptor | 18.21 | 15.00 | 9.11 |
| 3p21.3 | Semaphorin | — | 3.54 | — |
| 4q12 | PDGF-associated protein | 7.49 | — | — |
| 5q12 | CyclinB1 (CCNB1) | 3.61 | — | — |
| 5q31.3 | Early growth response protein (EGR1) | 8.21 | 7.48 | — |
| 5q31 | Interleukin-13 precursor | 3.70 | 3.49 | — |
| 5q31 | Alpha 1 catenin | 3.69 | — | — |
| 6p21.2 | CDKN1,WAF1 | 6.62 | 6.45 | 9.06 |
| 8p12 | T-plasminogen activator | 5.98 | 4.11 | — |
| 9p12 | Bcl-2 binding athanogene-1 (BAG-1) | 3.95 | — | — |
| 9q22 | Ninjurin-1 | 8.20 | 5.58 | 5.45 |
| 9q34.3 | NOTCH1 | 3.82 | — | 11.22 |
| 10q24 | Cytohesin-1 | 4.26 | 4.02 | 2.63 |
| 11p13 | CD59 | 5.24 | — | — |
| 11p15.4–15.5 | rhoG | 3.57 | — | 3.42 |
| 11q13 | Fau | 12.61 | — | 16.58 |
| 12p13 | CD9 | 11.94 | 5.48 | — |
| 12p13 | CD27BP /SIVA | 13.97 | 4.93 | 8.81 |
| 12q13 | Cytokeratin 8 | 17.17 | 15.00 | 9.10 |
| 12q13 | Cytokeratin 18 | 18.21 | 15.00 | 9.10 |
| 12q14 | CDK4 | 6.32 | 3.00 | 3.75 |
| 14q24.3 | c-fos | 3.35 | 3.46 | — |
| 16p11.2 | MAP kinase 1 | 3.95 | — | 2.28 |
| 16p13.3 | Netrin-2 | — | 6.97 | — |
| 16p13.3 | Nm23-H4 | 3.70 | 5.53 | 8.03 |
| 17q21–22 | Cytokeratin 10 | 11.06 | 3.42 | 4.46 |
| 17q21–22 | Cytokeratin 19 | 18.17 | 13.85 | 9.11 |
| 17q21 | Desmoplakin III | 6.57 | 6.39 | 13.33 |
| 17q21.3 | NDP kinase A (NM23-H1) | 4.69 | 4.93 | 3.05 |
| 17q21.3 | NDP kinase B (NM23-H2) | 18.21 | 15.00 | 11.59 |
| 17q21.32 | Integrin beta 3 | — | 3.79 | — |
| 17q24–25 | EGFRBP-GRB2 | 3.49 | — | — |
| 17q25.3 | Rho GDP dissociation inhibitor 1 | 8.53 | 5.99 | 10.10 |
| 19p13.2 | UV excision repair protein RAD23A | 5.65 | — | 14.71 |
| 19p13.3 | CDC34 | — | 3.06 | 4.78 |
| 19q13 | Apoptosis regulator bax | 4.29 | 4.15 | 5.14 |
| 19 | Macrophage migration inhibitory factor (MIF) | 12.31 | 3.93 | 11.62 |
| 20p13 | CDC25B | 4.29 | — | 2.56 |
| 21q22.1 | Cytosolic superoxide dismutase (SOD1) | 13.73 | 3.21 | 7.53 |
| 22q11 | DVL1 | 3.90 | — | — |
| Xp11.23 | PCTK1 | — | — | 2.84 |
| Xp11.23–3 | EPA/TIMP-1 | — | 6.95 | 4.26 |
| Xq25 | Hepatoma-derived growth factor (HDGF) | 10.00 | 6.93 | 19.53 |
| CD147 antigen | 7.13 | 14.41 | 13.39 | |
| Zyxin-related protein ZRP-1 | 10.12 | — | 8.08 | |
| Cytokeratin 7 | 5.30 | 5.60 | 5.38 | |
| Cytokeratin 16 | 5.24 | 4.47 | — | |
| rhoHP1 | 5.84 | 7.10 | — | |
| P21-rac1, ras-like protein TC25 | 5.85 | — | — | |
| PLK1 | 4.80 | — | — | |
| PDCD2 | 4.14 | — | — | |
| CASP2 | 3.22 | — | — | |
| FAST kinase | 3.94 | — | 3.66 | |
| CDK5 | 4.02 | — | 2.50 | |
| c-myc-binding protein MM-1 | 13.06 | 6.85 | 4.56 | |
| Cytokeratin 5 | — | — | 2.80 | |
| PIG12 | — | — | 4.12 | |
| Integrin-beta 8 precursor | — | — | 2.30 | |
| IGFBP5 | — | — | 3.49 |
Ratio of signal intensity to background. MCF7 sources: (a) University of Nottingham, (b) Institute of Cancer Research (Sutton), and (d) McGill University (Montreal).
Differences in gene expression (for example, ZRP-1, fau, RAD23A) between some of the subcultures were profound. Moreover, large differences were observed in expression levels of key genes involved in multiple cellular processes – signal transduction, cytoskeletal maintenance, transport, adhesion, oncogenesis, apoptosis, etc.
Characteristics of highly expressed genes common to all subcultures
The majority of genes expressed at high levels in MCF7 cultures were either intermediate filament markers (1) or genes involved in their anchorage (2); for example:
Cytokeratin 8, cytokeratin 18 and WAF-1, which have previously been shown to be differentially expressed in MCF7 cells (in contrast to MDA-MB 231) (Kuang et al, 1998).
All three cultures had low levels of expression of rhoC and rhoG, and all but one had higher levels of expression of rhoA and rhoB. Absolute levels of expression varied widely between the three different cultures.
The other notable observation involved the two heterodimeric subunits (H1 and H2) of the metastasis inhibition factor NM23. NM23-H2 was consistently expressed at high levels, and NM23H1 was consistently expressed at low levels in all subcultures. Low levels of NM23 correlate with high metastatic potential (Steeg et al, 1988); they are believed to complex with G-proteins and affect developmental pathways.
DISCUSSION
The viability of using long-term cell lines as models of human tumours has been questioned in a number of reviews (Corbett et al, 1987; Orth et al, 1994; Sen et al, 1995). It is well known that cells are subject to selection both during initial culture and subsequent long-term propagation. Phenotypic diversity has been demonstrated between established cell lines and the primary tumour from which they were cultured (Band et al, 1990; Lukashov and Goudsmit, 1995; Sen et al, 1995). Differences in biological properties (morphology, phenotype, karyotype, growth rate, cloning efficiency and tumorigenicity) have been demonstrated between sublines of haematopoietic cells (Band et al, 1990; Lukashov and Goudsmit, 1995) and for the breast cell lines MDA-MB 231 (Crepin et al, 1990) and MCF7 (Osborne et al, 1987). Murine tumours derived from long-term cell culture passage are frequently less invasive, far less metastatic and less aggressive than the same tumour maintained by animal passage only (Corbett et al, 1987). The drug sensitivities for long-term passaged tumours in vitro can, and do, change frequently (Corbett et al, 1995; Valeriote et al, 1996). In contrast, the sensitivity pattern for a tumour passaged in vivo to a given set of agents remains relatively stable for many years and dozens of transplant generations. Microsatellite instability has been demonstrated in ovarian cell lines (Orth et al, 1994) and is further increased in resistant sublines (Anthoney et al, 1996). Recently, mutations in hMSH2-deficient tumour cell lines have been shown to accumulate in a time-dependent manner in the absence of growth (Richards et al, 1997). This situation is perhaps more representative of the microenvironment of a tumour than the rapid growth that occurs in cell culture.
Our CGH results for the four MCF7 subcultures show moderate variations in two subcultures of the same parental line within the same laboratory but striking differences between subcultures from three different laboratories. Whereas Jones et al (2000) have already demonstrated extensive variation among different MCF7 cell stocks, this is the first report that:
compares and contrasts MCF7 lines from different laboratories;
examines the same subculture taken at different time intervals; and
most importantly, compares the CGH analytical work with expression array results.
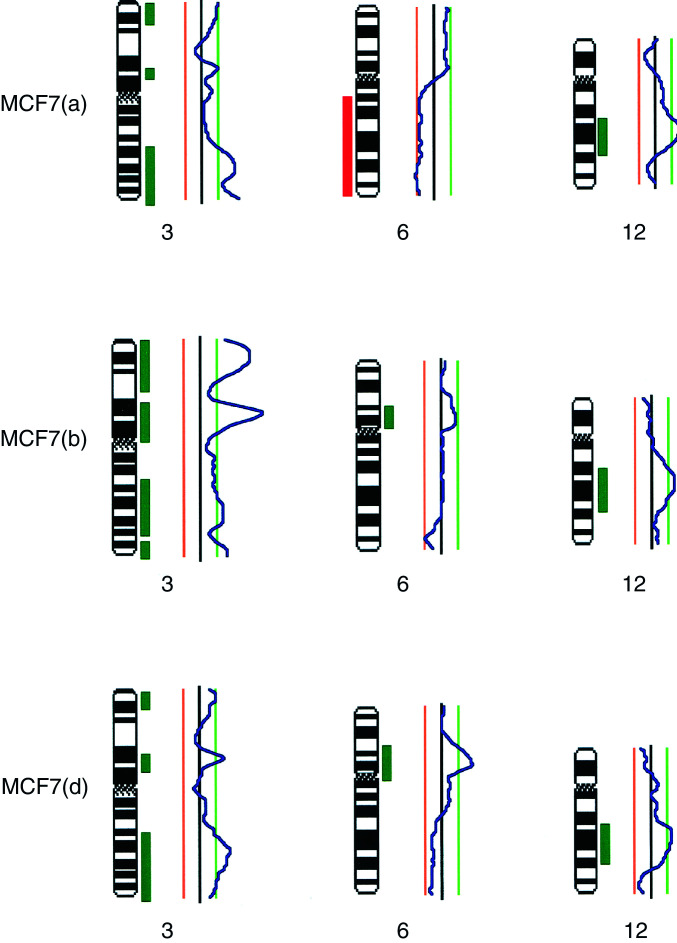
Moreover, our cDNA array results showed that the majority of expressed genes were related to intermediate filament markers and cell–cell interaction. Both CGH and expression results demonstrated a surprising level of divergence between subcultures from the different laboratories. Comparison between genomic copy number (as identified by CGH) and gene expression (as measured by arrays) is complicated by the fact that CGH does not resolve the fine structure of genome copy number changes. Thus, some lack of correlation may occur because narrow amplicons are missed in CGH or narrow deletions are missed, if part of a larger amplified region (Figure 3).
Figure 3.
CGH ratio profiles and accompanying idiograms for selected chromosomes for the three MCF7 cell lines: (a) University of Nottingham; (b) Institute of Cancer Research (Sutton); and (d) McGill University (Montreal). The ratio profiles are shown together with the threshold lines for amplification in green on the right and deletion in red on the left. The areas which exceed the threshold are also illustrated in green or red alongside the idiogram. The heterochromatic regions (which are excluded from the analysis) are shown hatched on the idiogram.
Since all three subcultures were grown in the same media at the same time, harvested in exponential phase and RNA extracted simultaneously, the differences that we observed in gene expression reflect real differences between the subcultures. Substantial variations were observed at both the genomic and RNA expression levels. Each subculture may have been subjected to different selection pressures or have reacted differently to the same selection pressures. Furthermore, overexpression can and does occur in the absence of DNA amplification and vice versa.
The combination of CGH and expression arrays may be useful in differentiating between genes that are transitorily expressed at high levels and those for which there is a long-term requirement for high-level expression. We agree with Pollack et al (1999) that genes which are both amplified and highly expressed may be more likely candidates for control of tumour initiation or progression.
These results not only question the utility of long-term culture cell lines as a basis for genomic analysis and the identification of signalling pathways, but also provide supportive data for evidence that cell lines do evolve in culture, thereby weakening the direct relevance of such cultures as models of human cancer. This work also reinforces the concern that comparisons of published analyses of cultures of the same name may be dangerous.
Acknowledgments
We thank Professors KR Harrap and A Horwich for bringing this collaboration together. This work was supported by National Cancer Institute of Canada Grant number 008030; BLJ was supported by a Haddow Fellowship awarded by the Institute of Cancer Research.
References
- Anthoney DA, McIlwrath AJ, Gallagher WM, Edlin AR, Brown R (1996) Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res 56: 1374–1381 [PubMed] [Google Scholar]
- Band V, Zajchowski C, Swisshelm K, Trask D, Kulesa V, Cohen C, Connolly J, Sager R (1990) Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res 50: 7351–7357 [PubMed] [Google Scholar]
- Botwell DDL (1999) Options available – from start to finish – for obtaining expression data by microarray. Nature Genet 21s: 25–32 [DOI] [PubMed] [Google Scholar]
- Boyd MR (1989) Status of the NCI preclinical antitumor drug discovery screen: implications for selection of new agents for clinical trial. In Cancer Principles and Practice of Oncology, Update Series, Vol. 3, DeVita VT, Hellman S, Rosenberg SA (eds). pp 1–12. New York: Academic Press [Google Scholar]
- Corbett TH, Valeriote FA, Baker LH (1987) Is the P388 murine tumor no longer adequate as a drug discovery model? Invest New Drugs 5: 3–20 [DOI] [PubMed] [Google Scholar]
- Corbett T, Valeriote F, LoRusso P, Polin L, Panchapor C, Pugh S, White K, Knight J, Demchik L, Jones J, Jones L, Lowichik N, Biernat L, Foster B, Wozniak A, Lisow L, Valdivieso M, Baker L, Leopold W, Sebolt J, Bissery MC, Mattes K, Dzubow J, Rake J, Perni R, Wentland M, Coughlin S, Shaw JM, Liversidge G, Liversidge E, Bruno J, Sarpotdar P, Moore R, Patterson G (1995) Tumour models and the discovery and secondary evaluation of solid tumor active agents. Int J Pharmacognosy 33: 102–122 [Google Scholar]
- Crepin M, Salle V, Raux H, Berger R, Hamelin R, Brouty-Boye D, Israel L (1990) Steroid hormone receptors and tumorigenicity of sublines from breast tumor metastatic MDA-MB 231 cell line. Anticancer Res 10: 1661–1666 [PubMed] [Google Scholar]
- Goldin A, Schepnz SA, Venditti JM, DeVita Jr VT (1979) Historical development and current strategy of the National Cancer Institute drug development program. In Methods in Cancer Research XVI, DeVita Jr VT, Busch H (eds). pp 165–245. New York: Academic Press [Google Scholar]
- Hiorns LR, Seckl MJ, Paradinas F, Sharp SY, Skelton LA, Brunstrom G, Newlands ES, Kelland LR, Leyland-Jones B (1999) A molecular cytogenetic approach to studying platinum resistance. J Inorganic Biochem 77: 95–104 [DOI] [PubMed] [Google Scholar]
- Hiorns LR. Comparative genomic hybridisation. In Cytogenetic Methods and Protocols: Methods in Molecular Biology, Swansbury GJ (ed), Totowa, NJ: Humana Press (in press) [Google Scholar]
- Jones C, Payne J, Wells D, Delhanty JD, Lakhani SR, Kortenkamp A (2000) Comparative genomic hybridization reveals extensive variation among different MCF-7 cell stocks. Cancer Genet Cytogenet 117: 153–158 [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi O-P, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D (1992) Comparative genomic hybridisation for molecular cytogenetic analysis of solid tumors. Science 258: 818–821 [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi O-P, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM (1994) Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 91: 2156–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang WW, Thompson DA, Hoch RV, Weigel RJ (1998) Differential screening and suppression subtractive hybridization identified genes differentially expressed in an estrogen receptor-positive breast carcinoma cell line. Nucleic Acids Res 15: 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyland-Jones B, Kelland LR, Harrap KR, Hiorns LR (1999) Genomic imbalances associated with acquired resistance to platinum analogues. Am J Path 155: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashov VV, Goudsmit J (1995) Increasing genotypic and phenotypic selection from the original genomic RNA populations of HIV-1 strains LA1 and MN (NM) by peripheral blood mononuclear cell culture, B-cell-line propagation and T-cell-line adaptation. AIDS 9: 1307–1311 [DOI] [PubMed] [Google Scholar]
- Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A (1991) Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 83: 757–766 [DOI] [PubMed] [Google Scholar]
- Nishizaki T, Chew K, Chu L, Isola J, Kallioniemi A, Weidner N, Waldman FM (1997) Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J Cancer 74: 513–517 [DOI] [PubMed] [Google Scholar]
- Orth K, Hung J, Gazdar A, Bowcock A, Mathis JM, Sambrook J (1994) Genetic instability in human ovarian cancer cell lines. Proc Natl Acad Sci USA 91: 9495–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK, Hobbs K, Trent JM (1987) Biological differences among MCF-7 human breast cancer cell lines from different laboratories. Breast Cancer Res Treat 9: 111–121 [DOI] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet 23: 41–46 [DOI] [PubMed] [Google Scholar]
- Richards B, Zang H, Phear G, Meuth M (1997) Conditional mutator phenotypes in hMSH2-deficient tumor cell lines. Science 277: 1523–1526 [DOI] [PubMed] [Google Scholar]
- Sehgal A, Boynton AL, Young RF, Vermeulen SS, Yonemura KS, Kohler EP, Aldape HC, Simrell CR, Murphy GP (1998) Application of the differential hybridization of Atlas human expression arrays in the identification of differentially expressed genes in human glioblastoma multiforme tumor tissue. J Surg Oncol 67: 234–241 [DOI] [PubMed] [Google Scholar]
- Sen S, Zhou H, Andersson BS, Cork A, Freireich EJ, Stass SA (1995) p53 gene mutations with chromosome 17 abnormalities in chronic myelogenous leukemia blast crisis patients persist in long-term cell lines but may be acquired in acute myeloid leukemia cells in vitro. Cancer Genet Cytogenet 82: 35–40 [DOI] [PubMed] [Google Scholar]
- Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME (1988) Evidence for a novel gene associated with low tumour metastatic potential. J Natl Cancer Inst 80: 200–204 [DOI] [PubMed] [Google Scholar]
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O-P (1998) Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer 21: 177–184 [PubMed] [Google Scholar]
- Valeriote F, Corbett T, Edelstein M, Baker L (1996) New in vitro screening model for the discovery of antileukemic anticancer agents. Cancer Invest 14: 124–141 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhao S, Xu H, McQueen T, Andreeff M (1998) Differential gene expression patterns in human myeloblastic leukemia HL-60 and multidrug resistant HL-60/dox cells analyzed by human cDNA expression array. Blood, 1600a