Abstract
Worldwide variation in the distribution of BRCA1 and BRCA2 mutations is well recognised, and for the Belgian population no comprehensive studies about BRCA1/2 mutation spectra or frequencies have been published. We screened the complete coding region of both genes in 451 individuals from 349 Belgian families referred to a family cancer clinic and identified 49 families with a BRCA1 and 26 families with a BRCA2 mutation. Six major recurrent mutations (BRCA1 IVS5+3A>G, 2478–2479insG, E1221X and BRCA2 IVS6+1G>A, 6503-6504delTT, 9132delC) accounted for nearly 60% of all mutations identified. Besides 75 true pathogenic mutations, we identified several variants of unknown clinical significance. In combination with a family history, an early average age of female breast cancer diagnosis (P<0.001), and the presence of a relative with ovarian cancer (P<0.0001) or multiple primary breast cancers (P=0.002), increased the chance for finding a mutation. Male breast cancer was indicative of a BRCA2 mutation segregating in the family (P=0.002). Mutations in the 5′-end of BRCA1 and BRCA2 were associated with a significantly increased risk for ovarian cancer relative to the central portion of the gene. Our study suggests a role for additional breast cancer susceptibility genes in the Belgian population, since mutation detection ratios were low in high-risk breast cancer-only families as compared to breast–ovarian cancer families. Given the large proportion of recurring mutations, molecular testing can now be organised in a more cost-effective way. Our data allow optimisation of genetic counselling and disease prevention in Belgian breast/ovarian cancer families.
Keywords: hereditary breast cancer, BRCA1, BRCA2, mutation analysis, HBOC
Since the mapping and the cloning of two genes that confer susceptibility to both breast and ovarian cancer, BRCA1 and BRCA2 (Miki et al, 1994; Wooster et al, 1995; Tavtigian et al, 1996), it became possible to offer genetic testing to families with a predisposition for breast and/or ovarian cancer. Consequently, individuals at risk can now be identified as candidates for surveillance programmes. A large number of distinct mutations in the BRCA1 and BRCA2 genes have been reported worldwide, but population-specific variation in the distribution of BRCA1/2 mutations is well recognised. In some populations or ethnic groups, founder mutations form a sufficient proportion of the total to justify the adoption of specific molecular screening strategies.
To date, no comprehensive studies in the Belgian population have been published. Only data from small series are available (Claes et al, 1999a, 1999b) or from studies in which the analysis was restricted to a few BRCA1/2 exons or to BRCA1 only (Peelen et al, 1997; Sibille-Hoang et al, 1998; Goelen et al, 1999). We performed mutation screening of the complete coding region of BRCA1 and BRCA2 in 349 unrelated Belgian families referred to our family cancer clinic and report here the nature and distribution of the mutations identified. We found phenotypic differences between families in whom a disease-causing mutation was identified vs BRCA1/2 mutation-negative families. We also evaluated in our cohort of patients if we could find an association between mutation site and relative risk of breast or ovarian cancer.
MATERIALS AND METHODS
Study population
Breast and/or ovarian cancer-prone families visiting the Centre for Medical Genetics at the Ghent University Hospital were selected for molecular testing of the BRCA1/2 genes if fulfilling one of the following inclusion criteria:
families with at least three first-degree relatives(*) with breast and/or ovarian cancer;
families with at least two first- and/or second-degree relatives(*) with breast and/or ovarian cancer before an average age of 50 years;
sporadic patients diagnosed with breast or ovarian cancer before the age of 38 years;
sporadic patients diagnosed with multiple primary breast cancers or concomitant breast and ovarian cancer and all tumours occurred before an average age of 50 years; and
sporadic patients with male breast cancer
(*) in case of male gene transmission, two affected females related through a male were considered to be first-degree relatives.
Counselling and genetic testing were provided by a multidisciplinary team of genetic counsellors, gynaecologists, oncologists, a psychologist and molecular biologists (De Vos et al, 1999). Before taking a blood sample, an informed consent was obtained. This study was approved by the ethics committee of the Ghent University Hospital.
In total, 58 sporadic patients (without a family history) and 291 families with a history of breast and/or ovarian cancer were analysed. To investigate familial clustering of the disease, we made a distinction between families with hereditary breast and/or ovarian cancer (HBOC) and familial breast and/or ovarian cancer (FBOC). Hereditary breast and/or ovarian cancer families are at high risk (>3 times population risk) and defined as families with at least three first-degree relatives with breast and/or ovarian cancer (or second-degree relatives in case of paternal inheritance) in at least two successive generations and at least one of them diagnosed before the age of 50 years. Families with at least two first-degree relatives (or second-degree relatives, in case of paternal inheritance) with breast and/or ovarian cancer diagnosed at young age, but not fulfilling the criteria for HBOC, are FBOC families. These families are at moderate increased risk (two to three times population risk). In total, we analysed 91 HBOC families and 200 FBOC families (Table 1 ).
Table 1. Unclassified variants in BRCA1 and BRCA2.
| Exon/intron | Nucleotide change | Amino-acid change | BIC | No. of families | Type of family | Polarity change | Conserved in dog/mouse/rat/chicken | Pathogenic status |
|---|---|---|---|---|---|---|---|---|
| BRCA1 | ||||||||
| 2 | IVS2–14C>T | Noncoding | Y | 1 | F, brca only | / | / | UV/P (no aberrant transcript observed by cDNA analysis) Claes et al (2003) |
| 10 | IVS10+8C>T | Noncoding | N | 1 | Sporadic | / | / | UV/P (no aberrant transcript observed by cDNA analysis) |
| 11 | 2196G>A | D693N | Y | 3 | F, brca-onlya F, brovca; H, brca only | N (negatively charged → uncharged polar) | N/N/N/N | UV/P (in two of the families true pathogenic BRCA2 mutations (IVS6+1G>A, 1617–1618delAG) were segregating) |
| 11 | 3238G>A | S1040N | Y | 3 | H, brca-onlya F, brovca; F, brca only | n | N/Y/Y/N | UV/P [in two of the families pathogenic mutations were segregating (BRCA1 E1221X, BRCA2 1617–618delAG)] |
| 11 | 3298A>C | E1060A | Y | 5 | Sporadica F, brca-only; F, brovca H, brca only | N (negatively charged → positively charged) | Y/Y/Y/N | UV/P (variant in linkage disequilibrium with BRCA1 E1221X) |
| 11b | 4145A>C | S1342S | N | 1 | H, brca only | / | / | UV |
| 17 | 5112G>A | V1665M | Y | 1 | H, brca only | N | Y/Y/Y/Y | UV/P (not segregating with the disease) |
| BRCA2 | ||||||||
| 3 | 407A>G | N60S | Y | 1 | F, brca only | N | Y/N/N/N | UV |
| 4 | IVS4+33A>G | Noncoding | N | 1 | F, brovca | / | / | UV/P (patient is carrying a pathogenic mutation (BRCA1 Q1281X); no aberrant transcript observed by cDNA analysis) |
| 10 | 1022–5insT | Noncoding | N | 1 | H, brovca | / | / | UV/P (no aberrant transcript observed by cDNA analysis) Claes et al (2003) |
| 10b | 1441G>A | G405R | N | 1 | H, brca only | Y | Y/N/Y/N | UV |
| 10 | 1571G>A | R448H | Y | 1 | F, brca only | N | N/N/N/N | UV |
| 10 | 1613A>G | E462G | Y | 1 | F, brca only | Y | Y/Y/Y/Y | UV |
| 11 | 3199A>G | N991D | Y | 2 | H, brca-only F, brovca | Y | N/N/N/N | UV/P (in one of the families BRCA1 5382insC was segregating and the patient was homozygous for N991D) |
| 11 | 7052A>G | E2275G | N | 1 | F, brca only, male brca | Y | Y/N/N/Y | UV/P (variant cosegregating with BRCA2 9132delC in one of the families) |
| 14 | 7285G>C | G2353R | Y | 1 | F, brovca | Y | Y/N/N/N | UV |
| 14 | 7641A>G | T2471T | N | 1 | H, brca only | / | / | UV (identified in two sisters with brca) |
| 15 | 7691G>A | R2488K | Y | 2 | H, brca only | N | Y/Y/Y/Y | UV/P (in one family not segregating with the disease; variant in linkage disequilibrium with 2166C>T (S646S)) |
| 15 | 7830G>C | A2534A | N | 1 | Sporadic | / | / | UV/P |
| 17 | 8172C>T | S2648S | N | 1 | Sporadic | / | / | UV/P |
| 18 | 8395C>G | D2723H | Y | 1 | H, brca only | N (negatively charged → positively charged) | Y/Y/Y/Y | UV |
| 18 | 8471G>A | G2748D | Y | 1 | F, brca only; | Y | Y/Y/Y/Y | UV |
| 20 | 8795A>C | E2856A | Y | 3 | F, brca-only; H, brca-only sporadic | Y | Y/Y/Y/Y | UV (not segregating with the disease in one of the families) |
| 22 | 9078G>T | K2950N | Y | 1 | F, brca only | N (positively charged → uncharged polar) | Y/Y/Y/Y | UV (identified in two sisters with brca, one of them carrying a truncating BRCA1 mutation) |
| 23 | 9266C>T | T3013I | Y | 1 | F, brca only | Y | N/N/N/Y | UV |
| 25 | 9520T>C | Y3098H | Y | 1 | F, brca only | N (positively charged → uncharged polar) | N/N/N/Y | UV |
| 25 | IVS25+9A>C | Noncoding | Y | 1 | H, brovca | / | / | UV/P (identified in patient carrying a BRCA1 mutation; no aberrant transcript observed at cDNA level) Claes et al (2003) |
| 27 | 10338G>A | R3370R | Y | 1 | H, brca only | / | / | UV/P (identified in a patient carrying BRCA2 Y42C) |
| 27 | 10462A>G | I3412V | Y | 1 | F, brca only | N | N/N/N/N | UV/P (not segregating with the disease in the family) |
H=hereditary breast cancer; F=familial breast cancer; brca=breast cancer; ovca=ovarian cancer; Brca-only=site-specific breast cancer family; brovca=breast cancer family with at least one case of ovarian cancer; UV=unclassified variant; P=polymorphism; Y=yes; N=no.
In this family two UVs were identified (BRCA1 S1040N and D693N) and one pathogenic mutation (BRCA2 1617–1618delAG).
Two genetic variants identified in the same patient.
When feasible, we investigated multiple affected family members in order to exclude the presence of a phenocopy. In total, 451 individuals from 349 families were selected for genetic testing (i.e. on average 1.29 individuals per family). For most patients, clinical files and pathological records were retrieved and re-evaluated. In 22 families, no affected relative was available for testing for various reasons (11 breast cancer-only families (10 FBOC and one HBOC) and 11 breast–ovarian cancer families (nine FBOC and two HBOC)), and in these families, 28 asymptomatic first-degree relatives of breast/ovarian cancer patients were analysed. Furthermore, two asymptomatic women were analysed whose mothers had died of a breast cancer diagnosed before the age of 35 years, but without a further family history.
Mutation detection
We screened the complete coding region of BRCA1 and BRCA2 in all families. For the first 85 families, the following strategy was used: in the first instance, BRCA1 exon 11 and BRCA2 exons 10 and 11 were analysed with the protein truncation test (PTT) (Hogervorst et al, 1995; Friedman et al, 1997). If no mutation was found, all other exons and splice sites of BRCA1 were investigated by heteroduplex analysis (HA) as described (Claes et al, 1999a). In 68 families in whom no mutations were identified, HA for all remaining coding exons of BRCA2 was performed.
For 203 families, we applied another strategy. BRCA1 exon 11 and BRCA2 exon 11 were investigated by PTT and all remaining coding exons and splice sites with denaturing gradient gel electrophoresis (DGGE) (van der Hout et al, 1999), a more sensitive technique than HA. The PCR set-up was automated using a robot (RoboAmp 4200, MWG Biotech, Ebersberg, Germany).
As PTT only allows the detection of truncating mutations, direct sequencing of BRCA1 exon 11 and BRCA2 exon 11 was performed for the last 62 families using the dye terminator chemistry on the ABI3100. All remaining coding exons and splice sites were investigated with DGGE.
When a mobility shift was observed on HA, PTT or DGGE gels, cycle sequencing was performed using dye primer chemistry on the ALF Express (Amersham Biosciences, Buckinghamshire, England) or dye terminator chemistry on the ABI377 or ABI3100 (Applied Biosystems, Foster City, USA) according to the manufacturer's instructions.
Furthermore, a specifically designed PCR assay was used to screen all families for four recurrent BRCA1 rearrangements (deletion exon 22, deletion exon 13, duplication of exon 13, deletion exons 8–9) (Petrij-Bosch et al, 1997; Puget et al, 1999; Rohlfs et al, 2000). Recently, multiplex ligation-dependent probe amplification (MLPA), a sensitive and comprehensive high-throughput test to detect single or multiple exon deletions and amplifications in the BRCA1 gene, has been made commercially available by MRC-Holland. We analysed with MLPA 117 families in whom no mutation was identified by conventional PCR-based techniques (all 55 remaining HBOC, 52 FBOC and 10 sporadic patients).
All mutations were reconfirmed on a second blood sample from the index case before offering counselling and access to genetic testing for at-risk adult family members.
Statistical analysis
The SPSS version 11.01 statistical analysis program was used for all analyses. The analyses were used to describe the specific characteristics (phenotypes) of the families. Characteristics included the following: at least one family member diagnosed with ovarian cancer, with bilateral breast cancer/multiple ipsilateral primary breast cancers, with male breast cancer or with both breast and ovarian cancer. These cancer diagnosis descriptors were analysed as dichotomous variables (presence vs absence) with the two-sided χ2 test and Fisher's exact test. Independent samples T-test was used to evaluate continuous variables, such as the mean age at diagnosis of breast cancer and ovarian cancer and average number of breast/ovarian cancers, between families carrying each genotype (BRCA1, BRCA2, either mutation, neither mutation). All statistical tests were two-sided. P-values less than 0.05 were considered to be statistically significant.
RESULTS
BRCA1 and BRCA2 mutations
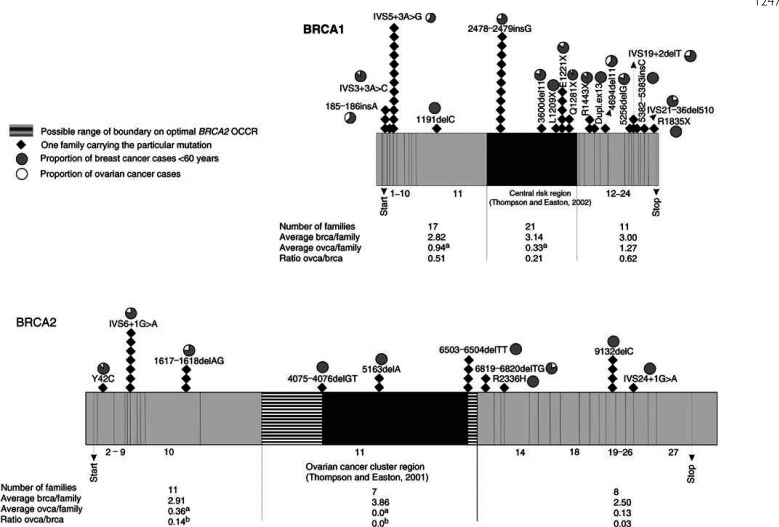
In total, 49 (18 distinct) BRCA1 mutations and 26 (10 distinct) BRCA2 mutations were identified (Figure 1). The vast majority of the mutations are predicted to lead to a premature stop codon (39 frameshifts, 15 nonsense mutations and 24 splice site disruptions). Nontruncating amino-acid substitutions occurring in only a limited number of families were considered as unclassified variants (Table 1). For the statistical analyses, these families were considered as BRCA1/2 mutation negative. Only BRCA2 Y42C was thought to be a pathogenic mutation. Y42 is a highly conserved amino acid and Y to C is a radical amino-acid change, compromising in vivo the interaction between BRCA2 and replication protein A (Wong et al, 2003).
Figure 1.
Diagrammatic representation of BRCA1 and BRCA2 exons, showing 49 BRCA1 and 26 BRCA2 germline mutations identified in Belgian breast/ovarian cancer families selected for genetic testing. A correlation with the phenotype in each mutation-positive family was made based on the ratio of ovarian cancer cases to breast cancer cases. The central regions in both genes, proposed to be associated with an increased ovarian : breast cancer ratio by Thompson and Easton (2001, 2002), are marked. Furthermore, the distribution of breast (brca) and ovarian cancers (ovca) in families according to the site of BRCA1 and BRCA2 mutations are indicated. (a) Differences in risk of ovarian cancer were statistically significant for BRCA1 5′ vs central portion (P=0.027) and BRCA2 5′ vs central portion (P=0.038). (b) Differences in relative risks of ovarian vs breast cancer were statistically significant for BRCA2 5′ vs central portion (P=0.017).
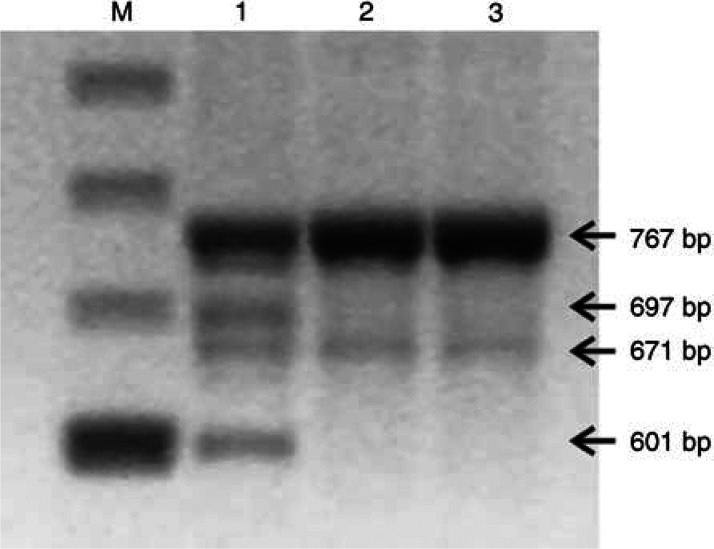
All splice site mutations were studied at the RNA level (Claes et al, 2002; Claes et al, 2003). RT–PCR analysis for BRCA2 R2336H in the last codon of exon 13 was not yet described and revealed the wild-type allele and three smaller transcripts, representing a complete loss of exon 13, loss of exon 12 and loss of exons 12 and 13. Loss of exon 12 was also observed in transcripts from normal individuals (Figure 2).
Figure 2.
RT–PCR was performed with primers spanning exons 11–15 of the BRCA2 gene (nucleotides 6948–7714; GenBank accession number NM_000059) on RNA extracted from lymphocytes as described by Claes et al (2003). RT–PCR on RNA from the patient carrying BRCA2 R2336H (lane 1) showed a full-length fragment of 767 bp and three faster migrating bands (697 bp: skipping of out of frame exon 13 (stop 2345); 671 bp: skipping of in-frame exon 12 (deletion of 32 amino acids); 601 bp: skipping of out of frame exons 12 and 13 (stop 2311)). In healthy control persons (lanes 2 and 3), besides the full-length fragment, a band (671 bp) representing the skipping of in-frame exon 12, was also observed. M, marker.
The large majority of the mutations (>80%) were recurrent. BRCA1 IVS5+3A>G and BRCA2 IVS6+1G>A, respectively, represent the most frequent mutations in BRCA1 and BRCA2. BRCA1 IVS5+3A>G is a Belgian founder mutation (Claes et al, 1999b). For BRCA2 IVS6+1G>A haplotypes could not be established since only one carrier per family was available for study. A founder effect is very likely since this mutation has not yet been reported in other populations, and the donor site of BRCA2 exon 6 does not represent a mutational hotspot region given the limited number of mutations reported in the BIC database in this region. The recurrence of the other mutations was also due to founder effects (data not shown).
In some populations, large intragenic deletions/duplications constitute a substantial fraction of mutations (Hogervorst et al, 2003; Montagna et al, 2003). We investigated the prevalence of single or multi-exon deletions/duplications in our Belgian study population. Therefore, we analysed with MLPA 117 families in whom no mutation was identified by conventional PCR-based techniques (all 55 remaining uninformative HBOC families, 52 FBOC families and 10 sporadic patients). In none of them a genomic rearrangement was detected.
Clinical characteristics indicative of a germline mutation
In patients with a family history of breast/ovarian cancer, significantly more mutations were identified compared to sporadic patients (71 mutations in 291 unrelated families (24.4%) and four mutations in 58 sporadic patients (6.9%); P=0.003). In breast–ovarian cancer families, mutation frequencies were significantly higher than in breast cancer-only families: 44.8% (30/67) vs 18.3% (41/224) (P<0.0001) (Table 2 ). The highest mutation detection ratio was obtained in breast–ovarian cancer families fulfilling the criteria for hereditary disease (75%=21/28), decreasing to 23.1% (9/39) in families with familial breast and ovarian cancer (P<0.0001). In breast cancer-only families, mutation frequencies were 23.8% (15/63) and 16.1% (26/161) in families with, respectively, hereditary and familial breast cancer (P=0.185). Especially, BRCA1 mutations conferred an increased risk for ovarian cancer: on average, there were nearly three times as many ovarian cancers in BRCA1 families than in BRCA2 families (0.89 vs 0.32; P=0.091). The average number of breast cancer cases was comparable (3.70 in BRCA1 and 3.72 in BRCA2 families; P=0.965). The ratio of ovarian to breast cancers was on average 0.41 : 1 for BRCA1 families and 0.07 : 1 for BRCA2 families (P=0.015).
Table 2. Overview of all HBOC and FBOC families investigated and mutation detection frequencies.
|
FBOC |
HBOC |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
#mutations found (%) |
#mutations found (%) |
||||||||
| Phenotypes | N | BRCA1 | BRCA2 | BRCA | N | BRCA1 | BRCA2 | BRCA | |
| Breast–ovarian cancer | No male breast cancer patient | 39 | 9 (23.1%) | 0 | 9 (23.1%) | 25 | 13 (52.0%) | 6 (24.0%) | 19 (76.0%) |
| families | At least one male breast cancer patient | 0 | 0 | 0 | 0 | 3 | 0 | 2 (66.7%) | 2 (66.7%) |
| Total | 39 | 9 (23.1%) | 0 | 9 (23.1%) | 28 | 13 (46.4%) | 8 (28.6%) | 21 (75.0%) | |
| Breast cancer-only | No male breast cancer patient | 155 | 13 (8.4%) | 11 (7.1%) | 24 (15.5%) | 60 | 11 (18.3%) | 3 (5.0%) | 14 (23.3%) |
| families | At least one male breast cancer patient | 6 | 0 | 2 (33.0%) | 2 (33.0%) | 3 | 0 | 1 (33.3%) | 1 (33.0%) |
| Total | 161 | 13 (8.1%) | 13 (8.1%) | 26 (16.1%) | 63 | 11 (17.5%) | 4 (6.3%) | 15 (23.8%) | |
| Total | 200 | 22 (11.0%) | 13 (6.5%) | 35 (17.5%) | 91 | 24 (26.4%) | 12 (13.2%) | 36 (39.6%) | |
FBOC=familial breast and/or ovarian cancer; HBOC=hereditary breast and/or ovarian cancer.
A male breast cancer case was indicative of a BRCA2 mutation segregating in the family (P=0.002) (Table 1). Furthermore, a relative with multiple primary breast cancers also increased the chance for finding a BRCA mutation: in 26 of 66 families (39.4%) with at least one relative diagnosed with multiple primary breast cancers, a mutation was identified compared to 45 mutations in 225 families (20%) without such a phenotype (P=0.002).
The mean age of diagnosis (±standard deviation) of the first female breast cancer was 45.87 (±12.80) years in mutation carriers and 49.01 (±12.35) years for persons without known mutations (P=0.001). On average, the age of onset was younger in BRCA1 compared to BRCA2 families; however, differences were not statistically significant (44.82±12.39 vs 48.00±13.42 years; P=0.086). For the mean age of diagnosis of ovarian or male breast cancer, no significant differences were obtained between mutation-positive and mutation-negative families (data not shown).
In total, four mutations were identified in 58 sporadic patients fulfilling our inclusion criteria (Table 3 ). Three mutations (BRCA1 IVS5+3A>G, 2626–2627delAA and BRCA2 6503–6504delTT) were found in 41 sporadic patients with early-onset breast cancer and one mutation (BRCA1 E1221X) in two sporadic patients diagnosed with both breast and ovarian cancer. No mutations were found in six sporadic patients with multiple primary breast cancers, neither in five sporadic ovarian cancer patients with early-onset disease nor in four sporadic males with breast cancer.
Table 3. Phenotypical features of sporadic patients investigated (N=58).
|
BRCA1 mutation-positive patients |
BRCA2 mutation-positive patients |
Mutation positive patients |
||||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| 41 female breast cancer patients (mean Dx 33.5, median Dx 33, range 26–52) | 2 | 4.9 | 1 | 2.4 | 3 | 7.3 |
| Six patients with multiple primary breast cancers (bilateral or multifocal) (mean Dx first breast cancer 39.8, median Dx 39.5, range 31–50) | 0 | 0 | 0 | 0 | 0 | 0 |
| Five ovarian cancer patients (mean Dx 35.4, median Dx35, range 25–52) | 0 | 0 | 0 | 0 | 0 | 0 |
| Two patients with both primary breast and ovarian cancer (mean Dx breast cancer 52.5 (range 50–55); mean Dx ovarian cancer 50 (range 47–53) | 1 | 50 | 0 | 0 | 1 | 50 |
| Four male breast cancer patients (mean Dx 49.3, median Dx 58.5, range 12–68) | 0 | 0 | 0 | 0 | 0 | 0 |
| 58 sporadic patients | 3 | 5.2 | 1 | 1.7 | 4 | 6.9 |
Dx=age at diagnosis (years).
Variation in cancer risk by mutation position
In our series of families, we found an increased ovarian to breast cancer ratio in the 5′-end of both genes. Figure 1 suggests that, for mutations 5′ of BRCA1 exon 11, this may result from both an increase in ovarian cancer risk and a reduction in breast cancer risk. Differences in breast cancer risk were not statistically significant. Also, for mutations in the 5′ region of BRCA2, a statistically significant increased incidence of ovarian cancer was observed relative to the central portion of the gene. However, our observations are based on a limited number of distinct mutations. For BRCA2, an ‘Ovarian Cancer Cluster Region’ (OCCR) in the middle third of the gene was proposed by Gayther et al (1997). Surprisingly, in our study no ovarian cancer cases were recorded in seven families bearing three distinct mutations in the BRCA2 OCCR.
DISCUSSION
We identified 75 disease-causing mutations in 349 Belgian breast/ovarian cancer families selected for genetic testing. The prevalence of BRCA1 mutations was approximately twice the prevalence of BRCA2 mutations. Most strikingly, six mutations (BRCA1 IVS5+3A>G, 2478–2479insG, E1221X, and BRCA2 IVS6+1G>A, 6503–6504delTT and 9132delC) accounted for nearly 60% of all mutations identified. BRCA1 2478–2479insG and BRCA2 IVS6+1G>A have not yet been reported in other populations. BRCA1 IVS5+3A>G is a Belgian founder mutation that has also been found in a few German, Dutch and French families (Claes et al, in preparation). BRCA1 E1221X and BRCA2 6503–6504delTT & 9132delC have been reported in several populations worldwide (BIC database). Given the proportion of all breast/ovarian cancer families in our population attributable to recurring mutations, a cost-effective stepwise molecular screening strategy of BRCA1 and BRCA2 may be applied in the future. A first-stage analysis, covering the recurrent mutations, can be offered to a substantial number of families, then, if a negative test result is obtained, more stringent risk criteria can be applied for complete analysis of the genes. Further examples of founder mutations in particular regions of Belgium may be found; the majority of the patients we analysed are living in North-West Belgium. Therefore, larger studies are required.
Despite the fact that Belgians historically have been in contact with many populations, only a limited number of founder mutations from other countries were detected. BRCA1 5382insC, an Ashkenazi Jewish founder mutation and the most frequent mutation in many European populations, was identified in only one Belgian family. Besides BRCA1 5382insC, no other Jewish mutations were detected in our patient population. The deletion of BRCA1 exon 22, the most recurrent Dutch mutation, was identified in one Belgian patient with a Dutch mother. The BRCA1 exon 13 6-kb duplication, a mutation likely to be derived from a British ancestor (The BRCA1 Exon 13 Duplication Screening group, 2000), was found once. Besides these two rearrangements, no other mutations involving one or more exons were detected with MLPA in all 55 remaining uninformative HBOC families, in 52 FBOC families and 10 sporadic patients in whom no mutations were identified with conventional PCR-based techniques. These preliminary data suggest that genomic rearrangements in BRCA1 do not have a major contribution in Belgian breast/ovarian cancer families. However, as indicated before, we mainly screened patients living in North-West Belgium; in other regions, this kind of mutations may be more prevalent.
Besides true pathogenic mutations, we identified several ‘unclassified variants’. Many of them were identified in high-risk families and occurred at amino acids that display substantial evolutionary conservation. Some of them were likely to be polymorphic for various reasons (Table 1). Most of the unclassified variants in our study were infrequent in our population and not reported before. A very high number of properly selected control individuals would be needed to search for statistically significant associations of these alleles with breast/ovarian cancer. We recently started LOH analysis for several variants to test the presumed association with the BRCA loci. We are also investigating if some of the unclassified variants affect correct splicing by disrupting functional exonic splicing enhancer sequences, as described for a BRCA2 amino-acid substitution (Fackenthal et al, 2002).
Our study permits to estimate the prevalence of BRCA1 and BRCA2 mutations in a Belgian patient population referred to a family cancer clinic. In 24.4% of the patients with a family history of breast/ovarian cancer, a mutation was identified. In families with at least one relative with ovarian cancer (P<0.0001), multiple primary breast cancers (P=0.002) or male breast cancer (P=0.002) significantly more mutations were identified compared to families without such phenotypes (Table 2). The literature is not unanimous about the predictive value of multiple primary (ipsilateral or contralateral) breast cancers for finding a mutation. Our results are in agreement with the studies of Bergthorsson et al (2001), de la Hoya et al (2002) and Ford et al (1998); however, others failed to demonstrate such a predictive value (Couch et al, 1997; Steinmann et al, 2001). Male breast cancer in combination with a family history of breast/ovarian cancer was indicative of finding a BRCA2 mutation (P=0.002), which is consistent with a recent population-based British study (Basham et al, 2002). In none of the four Belgian sporadic male breast cancer patients a mutation was identified. However, BRCA2 mutation prevalence in sporadic male breast cancer has been found to be as high as 33 and 21% in the Hungarian and Swedish population (Haraldsson et al, 1998; Csokay et al, 1999). Larger studies in Belgian patients are required before definite conclusions about mutation frequencies in this group can be drawn.
In mutation-negative families, the average age of onset of female breast cancer, but not male breast cancer, tended to be higher than in families in whom a mutation was identified. The mean age of onset for ovarian cancer was comparable in mutation-positive and mutation-negative families. These observations are in agreement with large studies on ovarian cancer patients (Risch et al, 2001; Frank et al, 2002). It has repeatedly been shown that only a small percentage of BRCA-positive ovarian cancer cases occur at ages <40 years (Boyd et al, 2000; Liede et al, 2002).
As early onset breast/ovarian cancer and the occurrence of multiple primary breast and/ovarian cancers in a single individual adds considerably to the prior probability of a mutation being present, we evaluated the prevalence of BRCA1/2 mutations in patients with these clinical characteristics and no further family history. Three recurrent mutations (BRCA1 E1221X & IVS5+3A>G and BRCA2 6503–6504delTT) and one novel mutation (BRCA1 2626–2627delAA) in 54 such patients were identified. For the recurrent mutations, a de novo event was highly unlikely. Haplotype analysis had revealed identical alleles with other Belgian patients carrying BRCA1 E1221X and IVS5+3A>G, respectively (data not shown). The patient with the BRCA2 6503–6504delTT mutation was bearing in addition two downstream polymorphisms (IVS24-16T>C and K3326X) that had been observed in all our other patients carrying the BRCA2 6503–6504delTT mutation. BRCA1 2626–2627delAA has not yet been reported in the BIC database and was identified in a sporadic patient diagnosed with breast cancer at the age of 27 years. A de novo event could not be ruled out as DNA of the parents was not (yet) available.
In only 7.5% of the sporadic patients diagnosed with breast cancer at young age, BRCA1/2 mutations were identified. We hypothesise a possible role for genetic variants in DNA double-strand break repair genes in this patient group, since a significant proportion of these patients showed elevated chromosomal radiosensitivity by in vitro assays (Baeyens et al, 2002).
In hereditary breast cancer-only families, mutation detection ratios were low (23.8%) compared to hereditary breast–ovarian cancer families (75%) (P<0.0001). Mutation frequencies did not statistically significantly differ between breast cancer-only families with and without an autosomal dominant inheritance pattern of the disease (23.8 vs 16.1%; P=0.085). As breast cancer is relatively frequent in Belgium, familial clustering of breast cancer may have occurred by chance in some families. In a few families, mutations undetectable by the techniques used may be present (e.g. regulatory mutations) or some of the genetic variants reported as being of ‘uncertain significance’ may be characterised as deleterious in the future. Nevertheless, our findings strongly suggest a role for additional breast cancer susceptibility genes.
From a clinical perspective, it would be most interesting to gain an insight into a possible relationship between mutation site and relative risk of breast or ovarian cancer. We found that in families with mutations occurring 5′ of BRCA1 exon 11 on average a significantly higher number of ovarian cancers were present than in families with mutations in the central portion of the gene (Figure 1). This trend is not consistent with the study of The Breast Cancer Linkage Consortium (BCLC) (Thompson and Easton, 2002) reporting an increased ovarian to breast cancer ratio in the central region of BRCA1 due to a lower breast cancer risk. Furthermore, they found a reduced ovarian cancer risk associated with mutations in the 3′ part of the gene, a trend that was not observed in our study.
For BRCA2, the BCLC study (Thompson and Easton, 2001) revealed an increased ovarian to breast cancer ratio for OCCR mutations, due to a reduced absolute risk of breast cancer. This observation was not confirmed in our study population. In none of our families bearing a BRCA2 OCCR mutation ovarian cancer was part of the phenotypes. Several other investigators (for instance, Frank et al, 1998; Ikeda et al, 2001; de la Hoya et al, 2002) also failed to demonstrate an increased incidence of ovarian cancer in the BRCA2 OCCR. In our study, mutations occurring 5′ of the OCCR were significantly associated with a higher ovarian cancer risk relative to the central portion of the gene.
As the data from different studies are not consistent, we think that associations between mutation position and phenotype are not sufficiently strong to influence genetic counselling and management of individual families. It is well known that large variations in cancer risks are also observed in families bearing the same mutation, suggesting the involvement of genetic and/or environmental modifiers. For counselling of affected families, it may be wiser to take into account the previous history of the family.
Acknowledgments
We thank the physicians for referring patients to our clinic, in particular, gynaecologists and oncologists from Aalsters Stedelijk Ziekenhuis, Aalst; AZ St-Jan, Brugge; AZ St-Lucas, Brugge; AZ Maria-Middelares, Gent; AZ St-Lucas, Gent; UZ Gent, Gent; AZ OLV Ter Linden, Knokke; AZ Maria's Voorzienigheid, Kortrijk; AZ H Hart, Roeselare; St-Augustinuskliniek, Veurne; AZ St.-Elisabeth, Zottegem, etc.
The study presented was supported by a research grant of the University of Ghent (GOA 12051397). B Poppe is an aspirant of the Fund for Scientific Research (FWO).
References
- Baeyens A, Thierens H, Claes K, Poppe B, Messiaen L, De Ridder L, Vral A (2002) Chromosomal radiosensitivity in breast cancer patients with a known or putative genetic predisposition. Br J Cancer 87: 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham VM, Lipscombe JM, Ward JM, Gayther SA, Ponder BA, Easton DF, Pharoah PD (2002) BRCA1 and BRCA2 mutations in a population-based study of male breast cancer. Breast Cancer Res 4: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson JT, Ejlertsen B, Olsen JH, Borg A, Nielsen KV, Barkardottir RB, Klausen S, Mouridsen HT, Winther K, Fenger K, Niebuhr A, Harboe TL, Niebuhr E (2001) BRCA1 and BRCA2 mutation status and cancer family history of Danish women affected with multifocal or bilateral breast cancer at a young age. J Med Genet 38: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, Saigo PE, Almadrones LA, Barakat RR, Brown CL, Chi DS, Curtin JP, Poynor EA, Hoskins WJ (2000) Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA 283: 2260–2265 [DOI] [PubMed] [Google Scholar]
- Claes K, Machackova E, De Vos M, Mortier G, De Paepe A, Messiaen L (1999a) Mutation analysis of the BRCA1 and BRCA2 genes results in the identification of novel and recurrent mutations in 6/16 Flemish families with breast and/or ovarian cancer but not in 12 sporadic patients with early-onset disease. Mutations in brief no. 224. Online. Hum Mutat 13: 256. [DOI] [PubMed] [Google Scholar]
- Claes K, Machackova E, De Vos M, Poppe B, De Paepe A, Messiaen L (1999b) Mutation analysis of the BRCA1 and BRCA2 genes in the Belgian patient population and identification of a Belgian founder mutation BRCA1 IVS5+3A>G. Dis Markers 15: 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes K, Poppe B, Machackova E, Coene I, Foretova L, De Paepe A, Messiaen L (2003) Differentiating pathogenic mutations from polymorphic alterations in the splice sites of BRCA1 and BRCA2. Genes Chromosomes Cancer 37: 314–320 [DOI] [PubMed] [Google Scholar]
- Claes K, Vandesompele J, Poppe B, Dahan K, Coene I, De Paepe A, Messiaen L (2002) Pathological splice mutations outside the invariant AG/GT splice sites of BRCA1 exon 5 increase alternative transcript levels in the 5′ end of the BRCA1 gene. Oncogene 21: 4171–4175 [DOI] [PubMed] [Google Scholar]
- Claes K, Poppe B, Szabo C, Thompson D, Peyrat JP, Fournier J, Coene I, Schelfhoul V, De Paepe A, Messiaen L. A common ancestor for Belgian and French patients bearing BRCA1 1VS5+3A>G. Submitted
- Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L, Ganguly A, Rebbeck T, Weber BL (1997) BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med 336: 1409–1415 [DOI] [PubMed] [Google Scholar]
- Csokay B, Udvarhelyi N, Sulyok Z, Besznyak I, Ramus S, Ponder B, Olah E (1999) High frequency of germ-line BRCA2 mutations among Hungarian male breast cancer patients without family history. Cancer Res 59: 995–998 [PubMed] [Google Scholar]
- de la Hoya M, Osorio A, Godino J, Sulleiro S, Tosar A, Perez-Segura P, Fernandez C, Rodriguez R, Diaz-Rubio E, Benitez J, Devilee P, Caldes T (2002) Association between BRCA1 and BRCA2 mutations and cancer phenotype in Spanish breast/ovarian cancer families: implications for genetic testing. Int J Cancer 97: 466–471 [DOI] [PubMed] [Google Scholar]
- De Vos M, Poppe B, Delvaux I, Mortier G, Claes K, Messiaen L, De Paepe A (1999) Genetic counselling and testing for hereditary breast and ovarian cancer: the gent(le) approach. Dis Markers 15: 191–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackenthal JD, Cartegni L, Krainer AR, Olopade OI (2002) BRCA2 T2722R is a deleterious allele that causes Exon skipping. Am J Hum Genet 71: 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Birch JM, Lindblom A, Stoppa-Lyonnet D, Bignon Y, Borg A, Hamann U, Haites N, Scott RJ, Maugard CM, Vasen H, Seitz S, Cannon-Albright LA, Schofield A, Zelada-Hedman M, Breast Cancer Linkage Consortium (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet 62: 676–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC (2002) Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol 20: 1480–1490 [DOI] [PubMed] [Google Scholar]
- Frank TS, Manley SA, Olopade OI, Cummings S, Garber JE, Bernhardt B, Antman K, Russo D, Wood ME, Mullineau L, Isaacs C, Peshkin B, Buys S, Venne V, Rowley PT, Loader S, Offit K, Robson M, Hampel H, Brener D, Winer EP, Clark S, Weber B, Strong LC, Rieger P, McClure M, Ward BE, Shattuck-Eidens D, Oliphant A, Skolnick MH, Thomas A (1998) Sequence analysis of BRCA1 and BRCA2: correlation of mutations with family history and ovarian cancer risk. J Clin Oncol 16: 2417–2425 [DOI] [PubMed] [Google Scholar]
- Friedman LS, Gayther SA, Kurosaki T, Gordon D, Noble B, Casey G, Ponder BA, Anton-Culver H (1997) Mutation analysis of BRCA1 and BRCA2 in a male breast cancer population. Am J Hum Genet 60: 313–319 [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA, Stratton MR, Easton D (1997) Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet 15: 103–105 [DOI] [PubMed] [Google Scholar]
- Goelen G, Teugels E, Bonduelle M, Neyns B, De Greve J (1999) High frequency of BRCA1/2 germline mutations in 42 Belgian families with a small number of symptomatic subjects. J Med Genet 36: 304–308 [PMC free article] [PubMed] [Google Scholar]
- Haraldsson K, Loman N, Zhang QX, Johannsson O, Olsson H, Borg A (1998) BRCA2 germ-line mutations are frequent in male breast cancer patients without a family history of the disease. Cancer Res 58: 1367–1371 [PubMed] [Google Scholar]
- Hogervorst FB, Cornelis RS, Bout M, van Vliet M, Oosterwijk JC, Olmer R, Bakker B, Klijn JG, Vasen HF, Meijers-Heijboer H, Menko FH, Cornelisse CJ, Den Dunnen JT, Devilee P, Van Ommen GJB (1995) Rapid detection of BRCA1 mutations by the protein truncation test. Nat Genet 10: 208–212 [DOI] [PubMed] [Google Scholar]
- Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH, Kluijt I, Dommering C, Verhoef S, Schouten JP, van't Veer LJ, Pals G (2003) Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res 63: 1449–1453 [PubMed] [Google Scholar]
- Ikeda N, Miyoshi Y, Yoneda K, Shiba E, Sekihara Y, Kinoshita M, Noguchi S (2001) Frequency of BRCA1 and BRCA2 germline mutations in Japanese breast cancer families. Int J Cancer 91: 83–88 [DOI] [PubMed] [Google Scholar]
- Liede A, Malik IA, Aziz Z, Rios Pd Pde L, Kwan E, Narod SA (2002) Contribution of BRCA1 and BRCA2 mutations to breast and ovarian cancer in Pakistan. Am J Hum Genet 71: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayanath P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barret JC, Lewis C, Neuhausen S, Cannon-albright L, Golgdgar D, Wiseman R, Kamb A, Skolnick MH (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71 [DOI] [PubMed] [Google Scholar]
- Montagna M, Palma MD, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E (2003) Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet 12: 1055–1061 [DOI] [PubMed] [Google Scholar]
- Peelen T, van Vliet M, Petrij-Bosch A, Mieremet R, Szabo C, van den Ouweland AMW, Hogervorst F, Brohet R, Ligtenberg MJL, Teugels E, van der Luijt R, van der Hout AH, Gille JJP, Pals G, Jedema I, Olmer R, van Leeuwen I, Newman B, Plandsoen M, van der Est M, Brink G, Hageman S, Arts PJW, Bakker MM, Willems HW, van der Looji E, Neyns B, Bonduelle M, Jansen R, Oosterwijk JC, Sijmons R, Smeets HJM, van Asperen CJ, Meijers-Heijboer H, Klijn JGM, de Greve J, King M-C, Menko FH, Brunner HG, Halley D, van Ommen G-JB, Vasen HFA, Cornelisse CJ, van’t Veer LJ, de Knijff P, Bakker E, Devilee P (1997) A high proportion of novel mutations in BRCA1 with strong founder effects among Dutch and Belgian hereditary breast and ovarian cancer families. Am J Hum Genet 60: 1041–1049 [PMC free article] [PubMed] [Google Scholar]
- Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, Hageman S, Arts PJ, Ligtenberg MJ, Meijers-Heijboer H, Klijn JG, Vasen HF, Cornelisse CJ, van’t Veer LJ, Bakker E, van Ommen GJ, Devilee P (1997) BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 17: 341–345 [DOI] [PubMed] [Google Scholar]
- Puget N, Sinilnikova OM, Stoppa-Lyonnet D, Audoynaud C, Pages S, Lynch HT, Goldgar D, Lenoir GM, Mazoyer S (1999) An Alu-mediated 6-kb duplication in the BRCA1 gene: a new founder mutation? Am J Hum Genet 64: 300–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, Jack E, Vesprini DJ, Kuperstein G, Abrahamson JL, Fan I, Wong B, Narod SA (2001) Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet 68: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs EM, Puget N, Graham ML, Weber BL, Garber JE, Skrzynia C, Halperin JL, Lenoir GM, Silverman LM, Mazoyer S (2000) An Alu-mediated 7.1 kb deletion of BRCA1 exons 8 and 9 in breast and ovarian cancer families that results in alternative splicing of exon 10. Genes Chromosomes Cancer 28: 300–307 [DOI] [PubMed] [Google Scholar]
- Sibille-Hoang C, Froment O, Joos de ter Beerst A, Lepiece V, Huberlant G, Blauwaert G, Vindevoghel A, Canon JL, Gillerot Y (1998) BRCA1 and BRCA2 mutations in Belgian families with a history of breast and/or ovarian cancer. Eur J Cancer Prev 7(Suppl 1): S3–S5 [DOI] [PubMed] [Google Scholar]
- Steinmann D, Bremer M, Rades D, Skawran B, Siebrands C, Karstens JH, Dork T (2001) Mutations of the BRCA1 and BRCA2 genes in patients with bilateral breast cancer. Br J Cancer 85: 850–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, Belanger C, Bell R, Berry S, Bogden R, Chen Q, Davis T, Dumont M, Frye C, Hattier T, Jammulapati S, Janecki T, Jiang P, Kehrer R, Leblanc JF, Mitchell JT, McArthur-Morrison J, Nguyen K, Peng Y, Samson C, Schroeder M, Snyder SC, Steele L, Stringfellow M, Stroup C, Swedlund B, Swensen J, Teng D, Thomas A, Tran T, Tranchant M, Weaver-Feldhaus J, Wong AKC, Shizuya H, Eyfjord JE, Cannon-Albright L, Labrie F, Skolnick MH, Weber B, Kamb A, Goldgar DE (1996) The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 12: 333–337 [DOI] [PubMed] [Google Scholar]
- The BRCA1 Exon 13 Duplication Screening Group (2000) The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. Am J Hum Genet 67: 207–212 [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton D (2001) Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet 68: 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton D (2002) Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomarkers Prev 11: 329–336 [PubMed] [Google Scholar]
- van der Hout AH, Mulder IM, Wu Y, van der Vlies P, Huisman M, Oosterwijk JC, Buys CHCM, Hofstra RMW (1999) A DGGE system for comprehensive mutation screening of the complete coding regions of BRCA1 and BRCA2 outside exons 11. Am J Hum Genet 65: A410 [Google Scholar]
- Wong JM, Ionescu D, Ingles CJ (2003) Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene 22: 28–33 [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792 [DOI] [PubMed] [Google Scholar]