Abstract
We cloned and characterized a cDNA corresponding to a cdc5+ homolog of the higher plant, Arabidopsis thaliana. The cDNA, named AtCDC5 cDNA, encodes a polypeptide of 844 amino acid residues. The amino acid sequence of N-terminal one-fourth region of the predicted protein bears significant similarity to that of Schizosaccharomyces pombe Cdc5 and Myb-related proteins. Overexpression of the AtCDC5 cDNA in S. pombe cells is able to complement the growth defective phenotype of a cdc5 temperature-sensitive mutant. These results indicate that the AtCDC5 gene is a plant counterpart of S. pombe cdc5+. This is the first report of a cdc5+-like gene in a multicellular organism. We also demonstrated that a recombinant AtCDC5 protein possesses a sequence specific DNA binding activity (CTCAGCG) and the AtCDC5 gene is expressed extensively in shoot and root meristems. In addition, we cloned a PCR fragment corresponding to the DNA binding domain of human Cdc5-like protein. These results strongly suggest that Cdc5-like protein exists in all eukaryotes and may function in cell cycle regulation.
Recent studies of the cell-cycle regulation of many eukaryotes have revealed that the basic mechanisms for the control of the cell cycle are common to all eukaryotes. Plants are not exceptions. It has been demonstrated that higher plants have genes for cyclin-dependent kinases and cyclins that resemble those of yeasts and other eukaryotes in structure and function (1–15). The eukaryotic cell cycle consists of four principal stages; G1, S, G2, and M phase. In the S phase, chromosomes are replicated, while in the M phase, the replicated chromosomes are segregated into two daughter cells. The main events controlling the cell cycle involve regulation of the onset of the S and M phase (16). In the late G1 phase, a controlling system operates to determine whether cells should commit to enter the cell cycle. Such a regulatory system has been shown to occur in a variety of eukaryotic organisms, and is called “START” for yeast, or “restriction point” for mammalian cells. Once cells pass through START or restriction point, they come out from developmental states and enter S phase, subsequently followed by mitosis. In yeast, a cyclin-dependent kinase is pivotal in the passage through START. Although the target proteins are not clearly identified, the kinase activity of Cdc28p in Saccharomyces cerevisiae and Cdc2 in Schizosaccharomyces pombe is required for START. Regulatory subunits for these kinases, namely cyclins, are also necessary for START. After passage of START, cell-cycle-coupled expression of the genes whose products are needed for the G1–S transition or the progression of S phase occurs (17, 18). In S. cerevisiae, two transcriptional factor complexes, SWI4-SWI6 complex (SBF) and DSC1, are required for the expression of these genes (19–23). In S. pombe, Cdc10 and Res1/Sct1 are required for the expression of genes which are necessary for G1–S transition or S phase (Cdc10 is also required for START) and share homology with the SWI4 and SWI6 proteins (24–26). Although multicellular organisms have similar mechanisms, they have several cyclin dependent kinases and more abundant cyclins (27). A transcriptional factor, E2F, is presumed to play a key role in the gene expression coordinate G1–S transition in mammalian cells (28).
Another important controlling mechanism occurs in the G2–M transition. This checkpoint mechanism ensure that mitosis starts after the completion of DNA synthesis (16, 29–31). Although the mechanisms for the checkpoint remain unknown, cyclin-dependent kinase and mitotic cyclins have a pivotal role in yeast cells. The level of mitotic cyclins increase in S through G2 phase, and contribute to the activation of Cdc28p/Cdc2. The elevation of mitotic cyclins is thought to be caused mainly by the accumulation of mRNA for these proteins (32, 33). It is presumed that the cell-cycle-coordinated expression of many genes, as well as mitotic cyclins, are required for the progression of the G2–M phase (28, 34, 35). However, the controlling mechanisms of the induction of these genes have not been clarified yet. Recently, Ohi et al. (36) described cdc5+, another cell-division-cycle gene from S. pombe. The cdc5+ gene is essential for the progression of the G2 phase in S. pombe. The structure of Cdc5 protein is similar to those of Myb-related proteins and its Myb-like domain bears DNA binding activity. Therefore, Cdc5 protein is presumed to be a transcriptional regulator whose function is required in G2 although its target genes are unknown. Here, we describe a cDNA encoding the Cdc5-like protein of a higher plant, Arabidopsis thaliana. This is the first demonstration that cdc5+-like genes exist in multicellular organisms.
MATERIALS AND METHODS
Plant Materials.
A. thaliana (Columbia ecotype) was used in this study.
Complementation Analysis.
S. pombe KGY372 (cdc5-120, leu1-32, ura4-D18, h−) and KGY450 (cdc5+/cdc5::ura4+, ura4-D18/ura4-D18. leu1-32/leu1-32, ade6-M210/ade6-M216, h+/h−) were used for the complementation assay (36). Recombinant plasmids in which the AtCDC5 cDNA was inserted in pREP1 (37) were constructed. The chimeric plasmid DNAs were introduced into the cells of these strains as described elsewhere (38). In the case of KGY372, transformed colonies were streaked on an EMM2-agar (thiamin + or −) plate and incubated at 36°C or 26°C for 2–3 days. In the case of KGY450, transformed cells were induced sporulation. Tetrads were dissected under a microscope, then germinated and cultured on a yeast extract/peptone/dextrose-agar plate.
In Situ Hybridization.
The AtCDC5 cDNA was introduced in the EcoRI site of pGEM4z (Promega). For the antisense probe, this plasmid DNA was linearized with HincII and the RNA probe was synthesized with the SP6 polymerase. For the sense probe, the plasmid DNA was linearized with BglII and the RNA probe was synthesized with the T7 polymerase. RNA probes were prepared by in vitro transcription with digoxigenin-dUTP (Boehringer Mannheim) according to manufacturer’s instructions. Both RNA probes, ≈1.4 kb, were hydrolyzed in 60 mM Na2CO3, 40 mM NaHCO3 at 60°C for 40 min. Whole mount in situ hybridization was performed as described by de Almeida Engler et al. (39) using 3–5 day old Arabidopsis seedlings.
Production of Glutathione S-Transferase (GST) Fusion Protein.
The DNA fragment corresponding to amino acids 1–144 of AtCDC5 was inserted in an expression vector, pGEX4T-2 (Pharmacia). Cells of the Escherichia coli strain JM109 (40), carrying this recombinant plasmid were grown in Luria-Bertani broth at 37°C. The methods for the production and the isolation of the GST fusion protein were described (41).
Random Binding Site Selection.
Two random oligodeoxynucleotides were used. The 68-mer contains 26 random bases; 5′-(left arm; AAGCTTGGTACCGTCGACATC)-(N)26-(right arm; CTAGTACTCGAGCTCAAGCTT) and the 46-mer contains 14 random bases; 5′-(left arm; GAGAAGCTTGTCTAGG)-(N)14-(right arm; GGAACTCGGATCCTGG)-3′. Double-stranded probes were synthesized by the Klenow fragment with [32P]dCTP using primers corresponding to right arms. Gel mobility shift-assay was performed as described (42). DNA fragments that were recognized by the GST-AtCDC5 protein were purified with nitrocellulose membranes as DNA–protein complex, or purified by the gel mobility shift as a shifted band. The recognized DNA fragments were amplified by PCR with primers corresponding to arm sequences. Obtained DNA fragments were subjected to next round of the selection, and purified by 8–10 times of the selection procedure. The recognized DNA fragments were cut out at the restriction sites from the arm sequence, and cloned with pGEM4z or pSKII plasmids for sequence analysis.
Zoo Blot and Northern Blot Hybridization Analysis.
Total DNA from various organisms was digested with EcoRI restriction endonuclease, fractionated on a 0.7% agarose gel, and then transferred to a nylon membrane (Hybond-N; Amersham). Low stringency hybridization conditions were described elsewhere (43). Total RNAs (30 μg) isolated from various organs were used in the Northern blot analysis. The methods for Northern blotting and hybridization conditions were described previously (43).
Conditions for the PCR.
The two oligodeoxynucleotides were synthesized as primers for the amplification of the cDNA fragment corresponding to the putative DNA binding domain of a human Cdc5-like protein. Primer 1 (5′-GGNGGNGTNTGGAARAAY-3′) was designed to correspond to the amino acid sequence (GGVWKN) found in the N terminal of the AtCDC5 protein. Primer 2 (5′-GTATTAGCCAAGCGGGC-3′) was designed to correspond to the nucleotide sequence found in expressed sequence tag clones (T59208 and T59167). The PCRs were performed as follows. The reaction mixtures contained 10 pg/μl human cDNA, 0.2 mM of each dNTP, 0.1 pmol/μl of both primers and 0.025 unit/μl of EX-Taq DNA polymerase in 1× buffer provided by manufacturer (Takara Shuzo, Kyoto). The mixtures were subjected to 94°C for 90 sec, 54°C for 2 min, and 72°C for 3 min for 45 cycles.
RESULTS
Structure of the cDNA for a cdc5+ Homolog of Arabidopsis.
In the course of isolating genes related to the Arabidopsis ABI3 gene that encodes a transcriptional regulator, we cloned a novel cDNA from normally grown Arabidopsis rosette plants. The determination of the nucleotide sequence revealed that this cDNA consists of 2807 bp and possesses one putative ORF. Because the postulated start codon is preceded by an in-frame stop codon (data not shown) and Northern blot analysis using this cDNA as a probe gave a single band of mRNA of ≈3.1 kb (see Fig. 5B), this cDNA appears to encode a full-length polypeptide. The predicted protein consists of 844 amino acids with a calculated molecular mass of 95.8 kDa, and contains abundant hydrophilic amino acids (≈40%), such as ABI3, although there was no sequence similarity between them (Fig. 1). The N-terminal region, however, is similar to those of Myb-related proteins and, in particular, to that of S. pombe Cdc5 with greater than 85% identity (Fig. 2 A and C). In addition, the structure of the succeeding region of the Myb-related domain, amino acids 120–230, is also closely related to that of S. pombe Cdc5 (Fig. 2A). These structural characteristics of the predicted protein indicated that this cDNA may encode a plant Cdc5-related protein. Thus, we designated the gene corresponding to this cDNA as the AtCDC5 gene.
Figure 5.
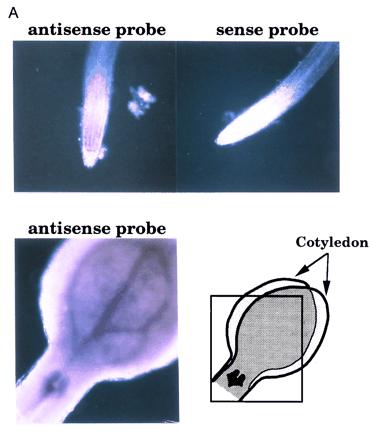
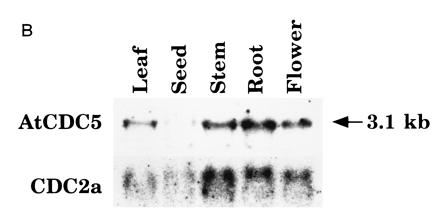
The expression pattern of AtCDC5. (A) In situ analysis of AtCDC5. Using antisense and sense RNA probes, whole mount in situ hybridization was performed. Microscopic views of root tips and a vegetative shoot apex are shown. Note that high background signals in cotyledons was due to their overlapping as indicated. (B) Northern blot analysis of AtCDC5 and CDC2a. RNAs from leaves, developing siliques, shoots, roots, and flowers were hybridized with the AtCDC5 cDNA probe or the CDC2a cDNA probe (3).
Figure 1.

Deduced amino acid sequence of AtCDC5 cDNA. The putative Myb-like DNA binding domain is underlined.
Figure 2.
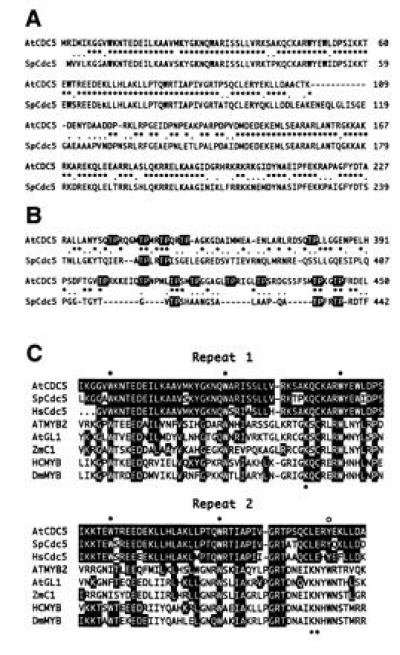
The structure of AtCDC5 compared with other Myb-related proteins. (A) Comparison of the amino acid sequence of the N-terminal region of AtCDC5 with that of S. pombe Cdc5. Identical amino acid residues are indicated with asterisks and conservative amino acid substitutions are indicated with dots. (B) The structure of the threonine-proline rich regions of AtCDC5 and Cdc5 proteins. Thr-Pro sequences are indicated with reversed characters. (C) The predicted amino acid sequence of the Myb-like repeats of AtCDC5 is compared with S. pombe Cdc5 (SpCdc5), putative human Cdc5 protein (HsCdc5), Arabidopsis ATMYB2 (42), Arabidopsis GL1 (AtGL1; ref. 44), Zea mays C1 (ZmC1; ref. 45), human c-MYB (HCMYB; ref. 46), Drosophila MYB (DmMYB; ref. 47). The amino acid residues that are identical in AtCDC5 and another Myb-related protein are indicated with reversed characters. Closed circles and open circles indicate conserved tryptophan residues. Asterisks represent the amino acid residues that are presumed to be involved in the target recognition of human c-Myb protein. Note that the first five amino acid sequences of human Cdc5 protein corresponds to primer 1.
In contrast to the high similarity in the N-terminal region, few amino acid residues are identical in the C-terminal two-thirds of AtCDC5 and yeast Cdc5 proteins. However, numerous Thr-Pro di-amino acid sequences are found in the middle regions of both proteins (Fig. 2B). Some of them are followed by Arg/Lys or Xaa-Arg/Lys residues and fit with the consensus of the target amino acid sequence of protein serine/threonine kinases.
AtCDC5 cDNA Can Rescue the S. pombe Temperature-Sensitive cdc5 Mutant.
Based on the significant structural similarity between S. pombe Cdc5 protein and Arabidopsis AtCDC5 protein, the functional similarity between them can be easily deduced. We performed complementation analyses of AtCDC5 using the S. pombe cdc5-120 mutant and a cdc5 disrupted mutant (36). The cdc5+ gene is essential for the growth of S. pombe. The cdc5-120 mutant can grow at a permissive temperature (26°C), but not at a restrictive temperature (36°C). The cells that have been transformed with a plasmid DNA carrying the cdc5+ gene were able to grow at the restrictive temperature. The AtCDC5 cDNA linked with a thiamin-inducible nmt1 promoter was introduced into the S. pombe cells of cdc5-120 mutant. When the AtCDC5 cDNA was expressed in the cells of cdc5-120, the cells could grow at the restrictive temperature although the growth rate was slightly reduced in comparison with the cells carrying the S. pombe cdc5+ gene (Fig. 3). S. pombe cells with the disrupted cdc5 gene do not grow at all (36), however, the introduction of the cdc5+ gene suppresses this phenotype. By contrast, overexpression of the AtCDC5 cDNA that was controlled by a thiamin-inducible nmt1 promoter or a constitutive active adh promoter did not complement the cdc5 null mutation (data not shown). These results suggest that the AtCDC5 protein shares partial functional similarity with S. pombe Cdc5 protein (see Discussion).
Figure 3.
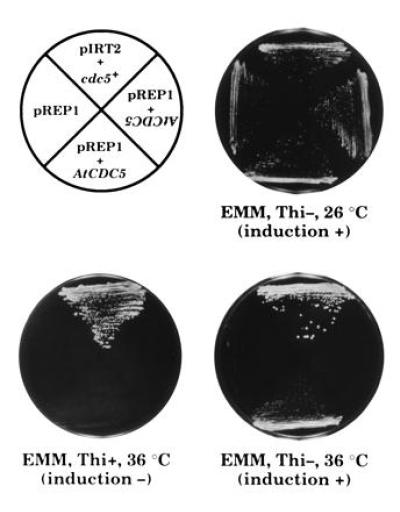
Complementation of the cdc5-120 temperature-sensitive mutation by AtCDC5 cDNA. KGY372 (cdc5-120) was transformed with recombinant plasmid DNAs, streaked on EMM-agar (3 mM thiamin + or −) plates and incubated at the permissive temperature (26°C) or nonpermissive temperature (36°C). “pIRT2+cdc5+” indicates that the cells were transformed with the chimeric plasmids DNA that consist of pIRT2 and the cdc5+ gene. “pREP1+AtCDC5,” “pREP1+AtCDC5 (inverted characters),” and “pREP1” indicates the cells transformed with the chimeric pREP1 plasmid carrying the AtCDC5 cDNA in appropriate direction, the cells transformed with the chimeric pREP1 plasmid carrying the AtCDC5 cDNA in reverse direction and the cells transformed with pREP1 vector only, respectively.
A Recombinant AtCDC5 Protein Binds Double-Stranded DNA in a Sequence-Dependent Manner.
To analyze the DNA binding activity of the AtCDC5 protein, a GST-AtCDC5 (1–144 amino acids) fused protein was produced in E. coli cells, and partially purified on a column of glutathione-Sepharose 4B (data not shown). Using random oligonucleotide as a probe, we performed gel mobility shift assay. When the probe DNA mixture was incubated with the purified GST-AtCDC5 fused protein, a faint but clear shifted band was observed (data not shown). Next we performed random binding site selections with the GST-AtCDC5 protein. Determination of the nucleotide sequence of the selected oligonucleotides revealed that a CTCAGCG (complementary; CGCTGAG) sequence was preferentially recognized in vitro by the GST-AtCDC5 protein (Fig. 4A). We performed three independent experiments with two sets of random oligonucleotides and obtained the same sequence as the recognized sequence. Totally, about 60% of the selected DNA contained this consensus sequence. Competition DNA binding assay revealed that the GST-AtCDC5 protein specifically binds to the consensus sequence (Fig. 4B). The GST-AtCDC5 protein did not bind to a DNA fragment without the consensus sequence. These results suggest that the Myb-like domain of the AtCDC5 protein binds to DNA in a sequence-specific manner and that the recognized sequence of AtCDC5 is different from that of Myb-related protein (YAACNG).
Figure 4.
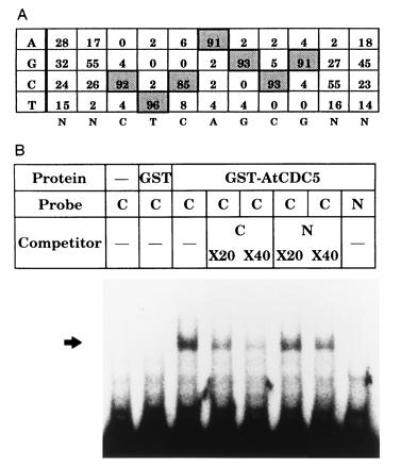
The GST-AtCDC5 fused protein possessed a sequence specific DNA binding activity. (A) Summary of the nucleotide sequence of DNA fragments recognized by the GST-AtCDC5 fused protein in random binding site selection experiments. The base constitution around the consensus sequence was indicated in percent. (B) Gel mobility shift assay. Five nanograms per lane of the GST-AtCDC5 protein was used. C, A DNA fragment with the consensus sequence (68-mer left arm-ACCAACGGGCGCTGAGCTGATGTCG-68-mer right arm); N, a DNA fragment without the consensus sequence (68-mer left arm-ACACCAAGTCTTACGCGCTGTCTCGC-68-mer right arm).
The Expression Pattern of the AtCDC5 Gene.
It has been observed that cell-division-cycle controlling genes of plants are expressed at higher levels in proliferating cells than in nonproliferating cells. To determine whether the AtCDC5 gene exhibits a similar expression pattern, we performed whole mount in situ hybridization using Arabidopsis seedlings. As shown in Fig. 5A, hybridization with an antisense probe produced significant signals in apical shoot meristems, leaf primordia, and root meristems, but hybridization with a sense probe did not. Northern blot analysis using RNAs isolated from leaves, developing siliques, shoot, roots, and flowers revealed that AtCDC5 is expressed in all tissues although the mRNA levels are different among tissues (Fig. 5B). The expression pattern of AtCDC5 was very similar to that of another cell cycle controlling gene, CDC2a. These results indicate that AtCDC5 is expressed mainly in the proliferating cells of vegetative and floral tissues, and that AtCDC5 is involved in the cell-cycle regulation.
cdc5+-Like Genes Exist in the Genome of Various Eukaryotes.
Regarding the significant similarity in structure and function between S. pombe Cdc5 and AtCDC5, we speculated that the Cdc5-like protein exists in all eukaryotes. We performed a so-called zoo blot analysis using a cDNA fragment of AtCDC5 as a probe. As shown in Fig. 6, clear hybridization signals were detected in each lane in which EcoRI digested genomic DNA from each type of organism (e.g., human, mouse, frog, insect, and nematode) was loaded. This result suggests that all eukaryotes, yeast and plant through human have cdc5+-like genes. If this is the case, it would be interesting to determine if the structure of the Myb-like repeats of Cdc5-like proteins are conserved. We performed PCR using human cDNA mixture as templates with primers for Cdc5-like protein (see Materials and Methods). Sequencing of an obtained PCR fragment revealed that this fragment is derived from the cDNA for the human cdc5+-like gene. More than 80% of the amino acids in Myb-like repeats are identical among S. pombe Cdc5, Arabidopsis AtCDC5 and the human putative Cdc5 (Fig. 2C). There is also a cdc5+-like gene in the genomic sequence of budding yeast in the GenBank data base. These results strongly suggest that all eukaryotes have a cdc5+-like gene and that the functions of Cdc5-like proteins in the cell cycle are conserved among eukaryotes.
Figure 6.
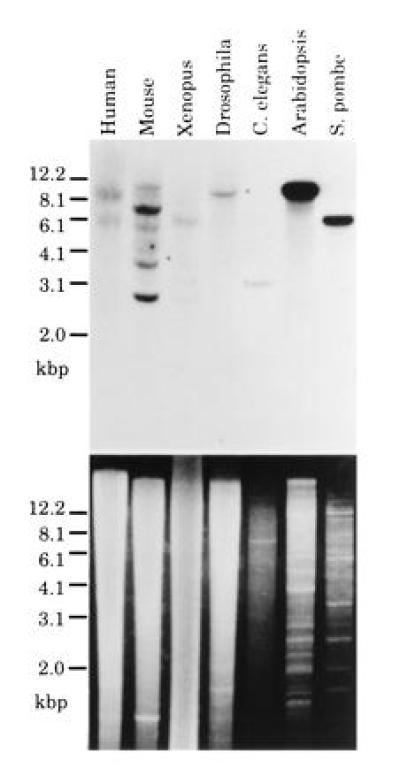
Zoo blot analysis. Genomic DNAs from various organisms such as human, mouse, Xenopus, Drosophila, Caenorhabditis elegans, Arabidopsis, and S. pombe were subjected to Southern blotting analysis. (Upper) The autoradiogram of the Southern hybridization using a cDNA fragment of AtCDC5 as a probe; (Lower) the photograph of the electrophoresed agarose gel stained with ethidium bromide.
DISCUSSION
In this study, we demonstrate that Arabidopsis has a cell-division-cycle-controlling gene, cdc5+ homolog, named the AtCDC5 gene. The cdc5+ gene of S. pombe encodes a Myb-like DNA binding protein, the function of which is required in cell-cycle-control especially in G2 phase (36). In the putative DNA binding domain, AtCDC5 protein and S. pombe Cdc5 share significant amino acid sequence identity (≈85%, Fig. 2C). Moreover, AtCDC5 cDNA could complement the cell growth defect of a S. pombe cdc5-120 under restrictive conditions (Fig. 3). The result of the in situ hybridization indicates that AtCDC5 may function in cell growth (Fig. 5A). These observations strongly suggest that AtCDC5 is a plant counterpart of cdc5+ of S. pombe. The result of the zoo blot analysis using AtCDC5 cDNA as a probe indicates that cdc5+-like genes are distributed in all eukaryotes (Fig. 6). Furthermore, we cloned the PCR fragment that corresponds to the Myb-like domain of the human Cdc5-like protein. The deduced amino acid of this PCR fragment exhibits high similarity to those of AtCDC5 and S. pombe Cdc5 (Fig. 2C). Taken together, these data suggest that cdc5+ homologs exist in all eukaryotes and that the cell-cycle-controlling mechanism that involves Cdc5-related protein may be highly conserved among eukaryotes.
Overexpression of AtCDC5 cDNA in S. pombe complemented the phenotype of the temperature-sensitive mutant allele (cdc5-120) but not that of the null mutant allele. The discrepancy between the results of two complementation analyses may be due to partial similarity of the function of AtCDC5 with S. pombe Cdc5. Cdc5 is thought to function as a homo-dimer complex (36). The products of the cdc5-120 allele has an amino acid substitution in its DNA binding domain and exhibits low DNA binding activity (R. Ohi and K. L. Gould, personal communication). We speculate that the AtCDC5 proteins can form a hetero-dimer complex with S. pombe Cdc5 and bind to the target sequence of Cdc5, however, other functions of S. pombe Cdc5, such as interactions with other yeast proteins, cannot be substituted correctly by AtCDC5 protein. This idea is supported by the structural similarity of AtCDC5 to S. pombe Cdc5 in its DNA binding domain and adjacent region but no significant similarity in the C-terminal two thirds. The N-terminal 232 amino acids of S. pombe Cdc5 is thought to contain the structure for the homo-dimer formation (36). The amino acid sequence of this region has high similarity with that of with AtCDC5. The region adjacent to the Myb-like domain may be required for the dimer formation.
It is noteworthy that there are many Thr-Pro di-amino acids in the middle regions of both AtCDC5 and S. pombe Cdc5 (Fig. 2B). Adjacent sequences of several T-P sequences have typical target sequences for Ser/Thr-kinases, such as p34cdc2 (Ser/Thr-Pro-Xaa-Arg/literys) or MAP kinases (Pro-Xaa(1–2)-Ser/Thr-Pro; refs. 48–50). These kinases are also found in higher plants and presumed to play pivotal roles in cell-cycle control. The phosphorylation of these threonine residues may modify the activity of AtCDC5 and S. pombe Cdc5.
S. pombe Cdc5, AtCDC5, and human Cdc5-like protein have a structure similar to Myb-like transcriptional factors (Fig. 2C). However, Cdc5-related proteins have several features distinct from Myb-related proteins. The amino acid residues that are involved in the recognition of the target DNA sequence and the interaction between the second and the third repeats of the DNA binding domain have been elucidated by the determination of the structure of DNA–protein complex of the human c-Myb DNA binding domain and the target oligonucleotide (51). Lys-128 in the second repeat, Lys-182 and Asn-183 in the third repeat of human c-Myb play a pivotal role in the recognition of target DNA sequence. Asp-178 and Asn-179 in the third repeat are involved in the interaction between the second and the third repeats. These residues are conserved among Myb-related proteins. In the case of Cdc5-related proteins, however, the corresponding residues are substituted for other amino acid residues (Fig. 2C). Thus, Cdc5-related proteins are thought to recognize different target sequences from those of Myb-related proteins. We determined the recognized sequence (CTCAGCG) of the GST-AtCDC5 protein by the random binding site selection, which is different from that of Myb-related proteins (YAACNG). Another difference is that Cdc5-related proteins may act as a homo-dimer (36) whereas there is no evidence for homo-dimer formation of Myb-related proteins. These characteristics lead us to speculate that the functions of Cdc5-related proteins have also diverged from those of Myb-related proteins. The identification of the target genes of AtCDC5 may allow us to understand the function of Cdc5-related proteins.
Plants have a unique developmental process in which differentiation progresses during the cell division in apical and root meristems. In both meristems, the genes that are involved in the cell cycle progression, such as genes for p34cdc2 and cyclins, are highly expressed. In situ hybridization and Northern blot hybridization revealed that AtCDC5 is highly expressed in meristems and weakly expressed in the cells in other tissues, which suggests that AtCDC5 is involved in cell division (Fig. 5). The expression of the cyclin genes are closely correlated with cell division, suggesting that the expression of these genes might be one of the controlling factors for the activation of cell division (6, 11, 52). By contrast, although the expression of the CDC2a gene of Arabidopsis is highly correlated with the mitotic activity, its expression might determine the competence for the cell division rather than the mitotic activity of the cells (53, 54). The expression pattern of AtCDC5 is more similar to that of CDC2a than that of cyclin genes. These results suggest that the expression of AtCDC5 is necessary for cell division but it does not determine mitotic activity of the cells like CDC2a, and that the activity of AtCDC5 is regulated by posttranslational modifications. This idea is consistent with the fact that AtCDC5 has putative phosphorylation sites. S. pombe Cdc5 protein is thought to be a transcriptional regulator that functions in G2 phase progression. Thus, AtCDC5 might be involved in controlling the expression of a set of genes that are necessary for G2 phase progression in Arabidopsis. Lateral roots are generated from pericycle cells that are developmentally arrested in G2 phase (55). When lateral root formation was initiated, the concomitant induction of a cyclin gene, cyc1At, was observed in Arabidopsis (11), suggesting that the expression of a set of genes including cyc1At is necessary for lateral root initiation. If Cdc5-like proteins of multicellular organisms are involved in G2 phase progression as S. pombe Cdc5, AtCDC5 may have important role(s) in the regulation of the gene expression during the formation of lateral roots. We are investigating the possibility of the involvement of AtCDC5 in the G2 phase progression.
Acknowledgments
We are extremely grateful to Drs. K. L. Gould and R. Ohi for S. pombe strains, their valuable suggestions, and fruitful discussions. We also thank Dr. P. Nurse for providing S. pombe strains; Drs. T. Eki, T. Maekawa, J. Aruga, and Y. Andachi for materials for the zoo blot; Dr. F. Hanaoka for human cDNA mixture; Dr. T. Mizoguchi for RNA samples; and Drs. K. Ogata, T. Urao, and K. Mikami for their helpful advice. This work was supported by the Special Coordination Fund of the Science and Technology Agency of the Japanese Government and by a Grant-in-Aid from the Ministry of Education, Science and Culture of Japan to K.S. Further support was provided by a Grant for Biodesign Research Programs from RIKEN to T.H.
Footnotes
References
- 1.Hirt H, Pay A, Györgyey J, Bako L, Nemeth K, Bögre L, Schweyen R J, Heberle-Bors E, Dudits D. Proc Natl Acad Sci USA. 1991;88:1636–1640. doi: 10.1073/pnas.88.5.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira P C G, Hemerly A S, Villarroel R, Van Montagu M, Inzé D. Plant Cell. 1991;3:531–540. doi: 10.1105/tpc.3.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirayama T, Imajuku Y, Anai T, Matsui M, Oka A. Gene. 1991;105:159–165. doi: 10.1016/0378-1119(91)90146-3. [DOI] [PubMed] [Google Scholar]
- 4.Colasanti J, Tyers M, Sundaresan V. Proc Natl Acad Sci USA. 1991;88:3377–3381. doi: 10.1073/pnas.88.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miao G-H, Hong Z, Verma D P S. Proc Natl Acad Sci USA. 1993;90:943–947. doi: 10.1073/pnas.90.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fobert P R, Coen E S, Murphy G J P, Doonan J H. EMBO J. 1994;13:616–624. doi: 10.1002/j.1460-2075.1994.tb06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata S, Kouchi H, Suzuka I, Ishii T. EMBO J. 1991;10:2681–2688. doi: 10.1002/j.1460-2075.1991.tb07811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemerly A, Bergounioux C, Van Montagu M, Inzé D, Ferreira P. Proc Natl Acad Sci USA. 1992;89:3295–3299. doi: 10.1073/pnas.89.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirt H, Mink M, Pfosser M, Bögre L, Györgyey J, Jonak C, Gartner A, Dudits D, Heberle-Bors E. Plant Cell. 1992;4:1531–1538. doi: 10.1105/tpc.4.12.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day I S, Reddy A S N. Biochim Biophys Acta. 1994;1218:115–118. doi: 10.1016/0167-4781(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira P C G, Hemerly A S, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. Plant Cell. 1994;6:1763–1774. doi: 10.1105/tpc.6.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renaudin J-P, Colasanti J, Rime H, Yuan Z, Sundaresan V. Proc Natl Acad Sci USA. 1994;91:7375–7379. doi: 10.1073/pnas.91.15.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meskiene I, Bögre L, Dahl M, Pirck M, Ha D T C, Swoboda I, Heberle-Bors E, Ammerer G, Hirt H. Plant Cell. 1995;7:759–771. doi: 10.1105/tpc.7.6.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soni R, Carmichael J P, Shah Z H, Murray J A H. Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl M, Meskiene I, Bögre L, Ha D T C, Swoboda I, Hubmann R, Hirt H, Heberle-Bors E. Plant Cell. 1995;7:1847–1857. doi: 10.1105/tpc.7.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nurse P. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 17.Andrews B J, Herskowitz I. J Biol Chem. 1990;265:14057–14060. [PubMed] [Google Scholar]
- 18.Johnston L H, Lowndes N F. Nucleic Acids Res. 1992;20:2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews B J, Herskowitz I. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 20.Taba M R M, Muroff I, Lydall D, Tebb G, Nasmyth K. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- 21.Lowndes N F, Johnson A L, Johnston L H. Nature (London) 1991;350:247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- 22.Lowndes N F, Johnson A L, Breeden L, Johnston L H. Nature (London) 1992;357:505–508. doi: 10.1038/357505a0. [DOI] [PubMed] [Google Scholar]
- 23.Dirick L, Moll T, Auer H, Nasmyth K. Nature (London) 1992;357:508–513. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- 24.Aves S, Durkacz B, Carr T, Nurse P. Gene. 1985;114:59–66. [Google Scholar]
- 25.Lowndes N F, Mclnerny C J, Johnson A L, Fantes P A, Johnston L H. Nature (London) 1992;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 28.Müller R. Trends Genet. 1995;11:173–178. doi: 10.1016/S0168-9525(00)89039-3. [DOI] [PubMed] [Google Scholar]
- 29.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 30.Nurse P. Nature (London) 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 31.Forsburg S L, Nurse P. Annu Rev Cell Biol. 1991;7:227–256. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- 32.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 33.Ghiara J B, Richardson H E, Sugimoto K, Henze M, Lew D J, Wittenberg C, Reed S I. Cell. 1991;65:163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- 34.Nasmyth K, Seddon A, Ammerer G. Cell. 1987;49:549–558. doi: 10.1016/0092-8674(87)90457-0. [DOI] [PubMed] [Google Scholar]
- 35.Moreno S, Nurse P, Russell P. Nature (London) 1990;344:549–552. doi: 10.1038/344549a0. [DOI] [PubMed] [Google Scholar]
- 36.Ohi R, McCollum D, Hirani B, Haese G J D, Xhang X, Burke J D, Turner K, Gould K L. EMBO J. 1994;13:471–483. doi: 10.1002/j.1460-2075.1994.tb06282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maundrell K. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 38.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 39.de Almeida Engler J, Van Montagu M, Engler G. Plant Mol Biol Rep. 1994;12:321–333. [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. Proc Natl Acad Sci USA. 1995;92:3903–3907. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirayama T, Oka A. Plant Mol Biol. 1992;20:653–662. doi: 10.1007/BF00046450. [DOI] [PubMed] [Google Scholar]
- 44.Oppenheimer D G, Herman P L, Sivakumaran S, Esch J, Marks M D. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 45.Paz-Ares J, Ghosal D, Weinand U, Peterson P A, Saedler H. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majello B L, Kenyon L C, Dalla-Favera R. Proc Natl Acad Sci USA. 1986;83:9396–9640. doi: 10.1073/pnas.83.24.9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzen A L, Kornberg T B, Bishop J M. Cell. 1985;41:449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- 48.Moreno S, Nurse P. Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 49.Clark-Lewis I, Sanghera J S, Pelech S L. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 50.Gonzalez F A, Raden D L, Davis R J. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 51.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 52.Doerner P, Jørgensen J-E, You R, Steppuhn J, Lamb C. Nature (London) 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- 53.Martinez M C, Jørgensen J-E, Lawton M A, Lamb C J, Doerner P W. Proc Natl Acad Sci USA. 1992;89:7360–7364. doi: 10.1073/pnas.89.16.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemerly A S, Ferreia P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blakely L M, Evans T A. Plant Sci Lett. 1979;14:79–83. [Google Scholar]


