Abstract
Objectives
The College of American Pathologists (CAP) Category 1 quality measures, tumor stage, Gleason score, and surgical margin status, are used by physicians and cancer registrars to categorize patients into groups for clinical trials and treatment planning. This study was conducted to evaluate the effectiveness of an application designed to automatically extract these quality measures from the postoperative pathology reports of patients having undergone prostatectomies for treatment of prostate cancer.
Design
An application was developed with the Clinical Outcomes Assessment Toolkit that uses an information pipeline of regular expressions and support vector machines to extract CAP Category 1 quality measures. System performance was evaluated against a gold standard of 676 pathology reports from the University of California at Los Angeles Medical Center and Brigham and Women's Hospital. To evaluate the feasibility of clinical implementation, all pathology reports were gathered using administrative codes with no manual preprocessing of the data performed.
Measurements
The sensitivity, specificity, and overall accuracy of system performance were measured for all three quality measures. Performance at both hospitals was compared, and a detailed failure analysis was conducted to identify errors caused by poor data quality versus system shortcomings.
Results
Accuracies for Gleason score were 99.7%, tumor stage 99.1%, and margin status 97.2%, for an overall accuracy of 98.67%. System performance on data from both hospitals was comparable. Poor clinical data quality led to a decrease in overall accuracy of only 0.3% but accounted for 25.9% of the total errors.
Conclusion
Despite differences in document format and pathologists' reporting styles, strong system performance indicates the potential of using a combination of regular expressions and support vector machines to automatically extract CAP Category 1 quality measures from postoperative prostate cancer pathology reports.
Introduction
A growing recognition of the inconsistencies in the quality of health care delivered 1 has led to a greater emphasis on the analysis of key quality measures contained within the medical record. 2–4 Unfortunately, many of the quality measures required for such analyses remain accessible only through time- and resource-intensive manual record review because of the predominant use of unstructured free text in the clinical record. 5
One disease for which patterns of care indicate differences in the quality of treatment is localized prostate cancer. 6,7 Prostate cancer is the second leading cause of cancer death in American men, with an estimated 234,460 new cases of prostate cancer diagnoses in the United States in 2006. 8 Despite the significance of this disease's effect on society, efforts to test whether and how the quality of care varies have historically been hindered by the lack of accessible and reliable quality measures. 9
As part of an effort to explore the challenges and opportunities of using automated approaches to facilitate records-based research, the University of California, Los Angeles (UCLA) and the Center for Surgery and Public Health at Brigham and Women's Hospital (BWH) have created the Clinical Outcomes Assessment Toolkit (COAT). COAT provides a framework for developers to create custom information pipelines to import, extract, transform, and analyze clinical record data and an integrated user interface to manage and audit the process. 10
The three quality measures the College of American Pathologists (CAP) identifies as Category I prognostic factors, or measures whose prognostic value has been demonstrated empirically in the literature, are Gleason score, tumor stage, and surgical margin status. 11 Surgical margin status also represents an important intermediate outcome because tumor inadvertently left in the patient, referred to as a positive surgical margin, is correlated with decreased cancer-specific and overall survival and a two to four times greater chance of biochemical cancer recurrence. 12,13 All three measures are reported in postoperative pathology reports and are used by physicians and cancer registrars to categorize patients into groups for clinical trials and treatment planning. 14 An application was developed using COAT that uses a combination of regular expressions and support vector machines to extract the three CAP Category 1 quality measures.
In this study, this technique was evaluated using a combined sample of UCLA and BWH prostatectomy pathology reports. To be considered a viable alternative to manual record abstraction, the pathology reports used in this study were identified using International Classification of Disease (ICD-9) and Common Procedural Terminology (CPT) codes. In addition, no manual review or filtering of the documents was conducted. The results produced by the system were compared to a manually abstracted gold standard to measure the sensitivity and specificity of the system's performance. A detailed failure analysis was conducted to identify the effects of data quality versus any shortcomings of the methods used.
Background
Clinical Information Extraction
Several natural language processing (NLP) and information extraction (IE) techniques have been successfully applied to extract information from free text medical records. Robust natural language processing systems such as MedLEE 15 and the National Library of Medicine's MetaMap Transfer (MMTx) 16 use controlled vocabularies and grammatical rules to map free text to structured representations. The use of such systems is often supplemented with a degree of customization to achieve acceptable levels of performance in specific domains. 17,18 In contrast, many “lighter weight” clinical information extraction systems are designed to target specific clinical values by capitalizing on consistent patterns in text. Examples of techniques often used in such tasks include the use of numeric patterns and attribute labels to capture diabetes metrics, 19 regular expressions for identifying blood pressure and antihypertensive treatment intensification, 20 and statistical machine-learning techniques such as support vector machines and entropy-based approaches to identify key clinical findings. 21,22 The level of customization involved in such techniques can lead to high levels of performance, but can also result in systems that do not port well across medical subdomains. A third approach that has recently gained traction in the medical informatics community is the development of hybrid systems capable of applying several different extraction and natural language processing approaches, often in combination, to abstract targeted clinical information. 23,24 In consideration of the heterogeneity and complexity of clinical data 25 and the growing list of targeted quality measures, 26 a similar design was chosen for COAT. COAT combines a collection of reusable information extraction components and clinical data structures with integration to third-party open-source packages such as Weka and MetaMap, and a user interface to control and audit the results of custom information pipelines. 10
Prostate Cancer
CAP identifies three Category I prognostic factors: Gleason score, tumor stage, and surgical margin status. 11 All three variables are the results of microscopic analyses performed by a pathologist on specimens submitted after radical prostatectomy. Specimens typically include the prostate, lymph nodes, and some surrounding tissue. Examples of appearances of Gleason score, tumor stage, and surgical margin status can be found in ▶.
• Gleason score is assigned based on the appearance of cancer cells. A score of 1–5, with 5 being most aggressive, is assigned to the two most prevalent patterns of cancerous cells to create a combined Gleason score or sum.
• Tumor stage, otherwise referred to as TNM stage, is a composite score summarizing several key pathological findings.27,28 Tumor stage measures the volume and location of the tumor or tumors (T), possible spread to the lymph nodes (N), and whether disease has metastasized to other parts of the body (M).
• As part of the histological examination, the prostate is inked, sectioned, and inspected. If the inked margin of resection is found to contain tumor, it indicates that the surgeon inadvertently incised into the tumor, resulting in a positive surgical margin.29 Unlike Gleason scores and TNM stage, a widely adopted standardized format for reporting margin status does not exist. As a result, it is not uncommon for margin status to appear in sentence, phrase, or semistructured form.
Table 1.
Table 1 Typical Appearances of Gleason Score, TNM Stage, and Surgical Margin Status
| Gleason Score | Tumor Stage |
|---|---|
| Gleason grade 3 + 3 = 6 | T2a N0 MX |
| Gleason score (3 + 4) | T 3 a, No, Mx |
| (3 + 4 = 6) | T3 No Mx |
| Surgical margin status | |
| No tumor present at the soft tissue resection margin | |
| No carcinoma is present at the inked margin | |
| Surgical margins: negative | |
| Surgical margins involved at right apex | |
| Base margin positive focal left | |
| Tumor is present focally at the margin of resection | |
Data Collection and Sampling
Postoperative radical retropubic prostatectomy (RRP) pathology reports were gathered from two major hospitals, the UCLA Medical Center and BWH. Pathology reports were identified by the appearance of CPT code 55845, “Retropubic radical with bilateral lymph node dissection” 30 and an ICD-9 code of 185, “Malignant neoplasm of the prostate.” 31 The reports selected from BWH were drawn from RRP surgeries conducted at BWH between January 1, 1996, and June 1, 2006. The UCLA pathology reports were from surgeries performed between January 1, 1998, and June 1, 2006. The total number of patients identified as having RRPs during these time periods at both institutions is 3,483. From this data set, a random sample of 676 pathology reports was selected, 353 from BWH and 323 from UCLA. To conduct this study, institutional review board approvals were obtained from both institutions.
There are some noteworthy differences in the format of UCLA and BWH post-RRP pathology reports. Both BWH and UCLA reports feature a short summary of key findings in paragraph form. Unlike the BWH reports, most UCLA reports also contain a semistructured Microscopic Examination section that lists these and other pathological measures in bulleted form. As a result, UCLA reports feature considerable duplication of measures and tend to be longer than BWH reports.
Gold Standard
For this study, no manual preprocessing or filtering of documents was performed. This approach was taken to validate system performance with consideration of the current limitations of the clinical data environment. As shown in ▶, abstraction of the targeted quality measures does not require any degree of clinical interpretation. As a result, three nonphysician reviewers (medical informatics graduate students) were trained to identify and abstract Gleason score, TNM stage, and margin status to create a gold standard. Partial values were logged exactly as they appeared in the pathology reports. For example, a tumor stage value reported without a numbered stage associated with the T value was entered as “T N0Mx” as opposed to “T2N0Mx.” Margin status was recorded as positive, negative, or left blank if no margin status diagnosis was included in the report. Keeping with the majority position in the urology literature, tumor reported as being close but not at the margin (e.g., “less than a few millimeters”) was considered negative. 12 To measure interrater reliability, each of the three abstractors was asked to extract the three targeted values from a randomly selected subsample of 38 reports. From a total of 114 targeted values, there was only one inconsistency: a tumor stage transcribed by one of the abstractors as 2TbNxMx rather than T2bNxMx. The resulting Kappa coefficient for this analysis was 0.99. To create the gold standard, the sample was divided equally among the three abstractors.
In the gold standard there were a total of 12 reports (1.78% of the sample) that did not pertain to patients with prostate cancer who had undergone radical retropubic prostatectomies, despite being labeled with CPT code 55485 and ICD-9 code 185. If these 12 reports with incorrect administrative codes were excluded, the rates of quality measure inclusion would have been 100% for Gleason score, 98.2% for TNM stage, and 99.4% for margin status. As they were not excluded, the total number of missing quality measures was 12 for Gleason score for an inclusion rate of 98.2%, 24 for TNM stage (96.5%), and 16 for surgical margin status (97.6%). Considering partial or missing lymph node and metastasis information (the NM of TNM stage) in an all-or-nothing approach for TNM stage, the rate of inclusion for this measure decreased to 94.5%.
Methods
A pipeline was created using COAT to: (1) import data from UCLA and BWH; (2) select a random sample, and (3) extract CAP Category I prognostics. The COAT user interface was used to manage the flow of data through this pipeline and to evaluate the results of each step. Reports from both institutions were processed through the same pipeline. The heuristics used to extract the three quality measures were designed based on a qualitative analysis of the appearances of the values in a random subsample of 60 reports.
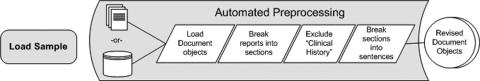
Automated Preprocessing
Automated preprocessing consisted of breaking reports into section and sentence-like structures and filtering out the Clinical History section. Sections were defined in this implementation as text appearing between double line breaks. Because much of the pathology report is formatted as attribute-value pairs (e.g., “Margin Status: Negative”), this method of partitioning sections also resulted in isolating sentence-like units. The Clinical History section of pathology reports often features a preliminary estimate of Gleason score. As a result, this section had the ability to produce inaccurate values for the pathological measurement of Gleason score. This section was therefore automatically identified and excluded from further processing by applying a regular expression to the first line of each section (“(?i)\\s*clinical.*[:.\n]\\s*\n”). Once the Clinical History section was removed, an attempt was made to break the remaining sections into smaller sentence-like units using Java's standard sentence boundary detector (Break Iterator). Each resulting string was then sent through the pipeline of three extraction components created for this study. The preprocessing components of the pipeline are shown in ▶.
Figure 1.
Automated preprocessing of clinical data.
Gleason Score, Tumor Stage, and Margin Status Extraction
Regular expressions were used to identify Gleason score and tumor stage. For identifying Gleason score (e.g., 3 + 4 = 7), each string was searched for the appearance of the numbers 1–5 followed by a “+” before another appearance of 1-5. In the case that the sum of the score was offered, it was also captured by searching for the optional appearance of “=” followed by a number between 1 and 10. The regular expression was designed to account for spaces and the use of parentheses. The pattern used to identify Gleason score is listed in ▶.
Table 2.
Table 2 Regular Expressions for Identifying Quality Measures in Pathology Reports
| Gleason score | “\\s\\(*[1-5]\\s*\\ + \\s*[1-5]\\s*\\)*(= \\s*[1-9]*0*)*” |
| TNM stage | “(?i)T[0-4]\\s*[abc]*(\\s*N[xO01]\\s*MX*[O01]*[abc]*)*” |
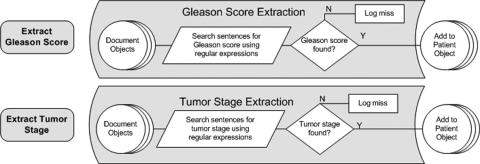
The tumor “T” portion of TNM stage (e.g., T2aN0Mx) was identified by conducting a case-insensitive search for the appearance of “t” followed optionally by the number 1 through 4, followed optionally by the letters a, b, or c. For both the N and M values, the letter “o” was accepted as a substitute for the number 0. The regular expression written in Java used for TNM stage is also provided in ▶. ▶ shows the similar processes for extracting Gleason score and tumor stage.
Figure 2.
The processes for extracting Gleason score and tumor stage.
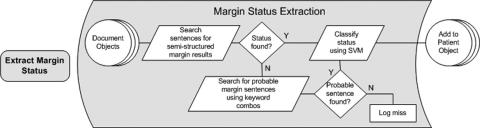
Surgical margin status was extracted in two steps. First, sentence-like strings were searched for indications that they contained a possible reference to margin status. Next, any strings identified as having a potential reference were classified using a trained support vector machine to arrive at a final diagnosis of margin status.
To capitalize on the consistency of any available semistructured margin status reporting, the algorithm used first sought out potential semistructured references to margin status. This initial pass searched for the appearance of the word “margin” or “margins” followed by a “:” and one or more words prior to a double line break using the regular expression as shown in ▶.
Table 3.
Table 3 Example of Semistructured Margin Status and the Regular Expression Used
| Example of semistructured surgical margin status reference: “Margin status: within 1 mm of ink“ |
| Java implementation of regular expression used: (?i)\\w + \\s*\\W*((margin)s*(:)\\s*\\W*\\w +) |
If no match was found, the algorithm continued searching strings for a series of two-word combinations discovered to appear regularly in text describing margin status. ▶ shows the heuristics used for extracting sentences describing margin status.
Table 4.
Table 4 Heuristics Used for Identifying Potential Margin Sentences
| resection and (margin or margins) |
| surgical and (margin or margins) |
| apical and (margin or margins) |
| tumor and (margin or margins) |
| carcinoma and (margin or margins) |
Each of the identified potential margin sentences was then automatically classified into one of three possible classes: (1) positive surgical margin, (2) negative surgical margin, and (3) not applicable or no explicit diagnosis provided. This third category was used to classify sentences extracted with no relevance to margin status (i.e., false positives). Examples of this third category of sentences are provided in ▶.
Table 5.
Table 5 Examples of Category 3 (False Positive) Sentences
| “the apical and basal margins are amputated and fixed separately” |
| “note benign prostate glands are focally present at an inked resection margin” |
A support vector machine (SVM) classifier was used to classify each potential margin sentence. SVMs have been used in the clinical domain for several NLP-related tasks including document classification 32 and complex concept identification in radiology reports. 22 To train the classifier, automatically extracted margin sentences were taken from a separate random sample of 782 pathology reports from the combined BWH/ UCLA collection. A total of 851 extracted sentences in the training set were manually categorized by the author (LWD) into one of the three categories: positive, negative, and not applicable. Extracted sentences were stripped of punctuation, converted to lower case, and their tokens and assigned class were passed to an implementation of an SVM classifier that used a polynomial kernel function and sequential minimal optimization (SMO) for training. 33 The resulting model was used to classify the margin status of sentences extracted from the 676 reports used in this study. For reports containing more than one classified margin sentence, an assignment of positive margin status was given if one of the sentences was classified as positive. In the case that no positive margin sentences were identified, the report was classified as negative if one of its sentences was classified as negative. Reports for which no potential margin sentence was found were classified as having no margin status and logged by COAT for review. ▶ illustrates the process used to extract and classify surgical margin status.
Figure 3.
Extraction and classification of surgical margin status.
Evaluations of system performance were conducted for the combined results as well as for each institution. A detailed analysis of failures was performed to identify areas of improvement as well as to measure the effects of poor data caused by incorrectly assigned administrative codes, missing values, and typographical errors. The COAT pipeline was run on a personal computer running Windows XP with a Pentium 4 3.06-GHz processor and 512 MB of RAM.
Results
The total number of quality measures targeted in this study was 2,028 (676 reports times three targeted measures per pathology report). Of these targets, 2,001 were accurately mapped to the gold standard for an overall accuracy of 98.7%. As shown in ▶, the accuracies for the extraction of Gleason score, TNM stage, and surgical margin status from the pathology reports of patients identified as having undergone prostatectomies as treatment for prostate cancer were 99.7%, 99.1%, and 97.2%, respectively.
Table 6.
Table 6 Overall System Accuracy
| Gleason score | 99.7% | 674/676 |
| TNM stage | 99.1% | 670/676 |
| Margin status | 97.2% | 657/676 |
| Overall accuracy | 98.7% | 2,001/2,028 |
System errors, or errors in which a quality measure existed in a report but was not accurately captured, were responsible for 74% (20 of 27) of all errors. The remaining 26% of errors were caused by poor data quality, including inaccurate administrative code assignment, typographical errors, and partial values. The distribution of system versus data quality failures is shown in ▶. There were 12 (1.8%) documents in the sample that should not have been assigned either CPT 55845 or ICD-9 code 185. However, only one of the false-positive extractions was caused by an inappropriately labeled report. Details of the system's performance along with analyses of system failures are provided in the following sections.
Table 7.
Table 7 Reasons for Failures
| Types of Errors | Distribution of Error Types | Percentage of Total Error |
|---|---|---|
| System errors | 20/27 | 74% |
| Typos or inconsistencies | 6/27 | 22% |
| Incorrect billing code | 1/27 | 3.7% |
Gleason Score Extraction
In comparing the automatically extracted Gleason score results to the gold standard, the created pipeline accurately mapped 99.7% (674 of 676) of the values. The two inaccurately mapped measures were both the result of poor data quality. The first was due to a single text document from BWH containing two different pathology reports. The second inaccurate mapping was caused by the inclusion of two different Gleason scores in one UCLA pathology report. The 12 mislabeled reports in the gold standard did not adversely affect the automated extraction of Gleason score. The total time for extraction of Gleason scores from the 676 reports was 37 seconds.
Tumor Stage Extraction
Tumor stage was correctly mapped in 99.1% (670 of 676) reports in the sample. Of the six inaccurately mapped stage results, four were failures of the system to capture measures that existed in a report. Of these system errors, one was caused by an addendum added to the report that listed two different tumor stage results. The other three were caused by the inclusion of commas in the reported tumor stage, a pattern not accounted for in the regular expression used (e.g., “T2, No, Mx”). All four system errors occurred on UCLA reports. The two remaining errors were data quality errors. One was caused by an incorrectly formatted tumor stage in a BWH report (2Tc, rather than T2c). The other was a false positive caused by an incorrect administrative classification of the document in the UCLA collection. Although the document did feature a similarly formatted tumor stage result that was captured by the system, the pathology report was for a patient treated for bladder cancer, not prostate cancer. The total time for extraction of tumor stage was 44 seconds.
Surgical Margin Status Extraction
Surgical margin status was accurately mapped to the gold standard in 97% of the cases. Positive margin cases in the gold standard were accurately mapped for 93% (114 of 122) of cases. Negative margin classification was 99% (530 of 535) accurate. In reports that made no reference to margin status, the system accurately classified 81% (13 of 16) of the cases as having no margin status. A fourth category of margin status not anticipated in the design of the system that was discovered during the analysis of the results was cases in which the pathologist described being unable to make a definitive diagnosis of margin status. This occurred in 3 of the 676 reports. In these reports the pathologists cited poor specimen quality for their inability to make a diagnosis of margin status. Overall sensitivities and specificities for the combined sample are shown in ▶. Details of the sensitivities and specificities of the three categories of margin status for each institution are provided in ▶.
Table 8.
Table 8 Sensitivity and Specificity Results for Margin Status Identification
| Sensitivity | Specificity | |
|---|---|---|
| Positive margin | 93% | 99% |
| Negative margin | 99% | 91% |
| Missing margin status | 81% | 99% |
Table 9.
Table 9 Institution-specific Sensitivity and Specificity Results for Margin Status Identification
| Brigham and Women's Hospital |
UCLA Medical Center |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |
| Positive margin | 97% | 99% | 92% | 99.6% |
| Negative margin | 99% | 96% | 99% | 89.6% |
| Missing margin status | 89% | 99% | 72% | 99.4% |
Of the 19 errors in identifying surgical margin status, 16 (84%) were system errors and three (16%) were attributed to poor data quality. The three data quality errors were false positives mapped to reports that were in the sample because of incorrect administrative code assignment. Eleven (69%) of the sixteen system errors were from UCLA reports, and five were from BWH. Four system errors were caused by failure to capture sentences describing margin status. In all four cases the sentences describing margin status featured the words “margin or margins” and “ink,” a combination not accounted for in the original design of the algorithm used to identify potential margin sentences. The other 12 errors were caused by misclassification of extracted sentences. Of the eight missed positive margin diagnoses, seven were from UCLA reports. A confusion matrix with the distribution of errors per category of margin status is provided in ▶. The total time to extract potential margin sentences was 24 seconds. The trained model took 4 seconds to classify the extracted sentences.
Table 10.
Table 10 Confusion Matrix of Surgical Margin Status Errors
| Extracted Results | |||||
|---|---|---|---|---|---|
| Could not be determined | Positive | Negative | Missing | Total False Negatives | |
| Could not be determined | 0 | 0 | 3 | 0 | 3 |
| Positive | 0 | 0 | 7 | 1 | 8 |
| Negative | 0 | 2 | 0 | 3 | 5 |
| Missing | 0 | 1 | 2 | 0 | 3 |
| Total False Positives | 0 | 3 | 12 | 4 | Total errors 19 |
Discussion
The overall accuracy of the system in mapping the three CAP Category 1 quality measures for prostate cancer (98.7%) shows promise for the use of automated extraction methods to facilitate prostate cancer surgical outcomes assessment. The use of regular expressions proved effective for extracting both TNM stage (99.1%) and Gleason score (99.7%). Performance in extracting these two quality measures also shows the significance of physicians' use of documentation standards to support automated quality measure extraction. Performance in the extraction and classification of margin sentences was also strong (97.2%), despite the lack of standards governing its format. The results of this study show that the described technique is capable of quickly and accurately determining important outcomes-related information including the number of prostate cancer surgery patients whose surgeries resulted in a positive surgical margin, the number and percentage of each surgeon's cases that resulted in a positive surgical margin, as well as the histological and staging characteristics of the cancers of treated patients. These results also indicate the potential to use these techniques to quickly identify the degree of adherence to prostate cancer–related pathology documentation requirements, recently cited as a measure of the quality of care delivered by the IOM's Committee on Assessing Improvements in Cancer Care 34 and an important component of accreditation by American College of Surgeons Commission on Cancer. 35 Equally important, the exclusion of any manual preprocessing in this study indicates that these measures can be gathered within minutes, given access to a collection of electronically formatted pathology reports, with minimal effects of poor data quality on the results produced.
There were some differences in system performance in extracting quality measures between UCLA and BWH reports. It was originally hypothesized that the performance on the UCLA collection would be stronger because of the inclusion of a semistructured Microscopic Examination section. However, classification of positive surgical margins from UCLA reports had a sensitivity of 91.95% versus 97.14% for BWH. On closer inspection of the seven missed UCLA positive margins responsible for this difference, only one was the fault of failure of the algorithm to extract the sentence containing the information; in the six other cases, the sentences were successfully extracted, but falsely classified as negative. This result was likely caused by insufficient training of the SVM classifier. Of the 851 sentences used to train the model, only 130 of the sentences were positive surgical margin sentences. Although the two institutions were equally represented in the training set, UCLA was discovered to have twice as many positive margin cases in the gold standard sample. Further, two of the seven misclassifications of margin status were discovered to have originated from a pathologist who dictated a total of five of the reports in the sample. All five of this pathologist's reports were created on a template that represented margin status in a format different than the other doctors at UCLA. Rather than feature one line with the margin status result coupled with the location (e.g., “surgical margins: positive at apex”), this pathologist featured one entry for each of the margin locations (e.g., “apical margin: positive”). This variation in the descriptions of positive surgical margins in UCLA reports was not significantly represented in the training set. Updating the classifier with more examples of positive surgical margins should increase its performance. Training the classifier to recognize cases in which the pathologist makes explicit reference to not being able to determine the margin status will be a greater challenge because only three of the 676 cases in the sample featured such a reference. Prevalence of this diagnosis should be analyzed in a larger sample to determine the clinical significance and technical feasibility of automatically extracting this result.
The effect of poor data quality on the final results was quite low in this case, causing only 0.3% of the 2,028 targeted values to be mapped incorrectly. However, poor data quality represented a large proportion of the failures, accounting for 25.9% of incorrect extractions. Poor data quality was also responsible for falsely inflating the number of patients believed to have had RRPs as a treatment of prostate cancer by nearly 2%. The inclusion of these reports did not, however, have a significant effect on the extraction of quality measures. Although the algorithm searched these twelve reports for a total of 36 potential measures, only one was found. In other words, there was only one pattern in the text of the twelve falsely classified reports similar enough to produce a false-positive quality measure (a tumor stage in an incorrectly coded bladder cancer operation). The issue of poor data quality and its effects on clinical outcomes assessment is a significant and largely unsolved issue. Although the effects of poor data quality in this study were relatively insignificant, such results are domain- and institution-dependent and should be accounted for in the evaluation of any automated system designed to learn from clinical data.
Several potential improvements to the technique were identified as a result of this study and are listed in ▶. These improvements should result in the capture of at least seven missed values and have the potential to improve overall system accuracy. To that effect, the ability to use COAT to log and review cases with missed values was useful in identifying potential improvements as well as in recognizing the cause of errors.
Table 11.
Table 11 Identified Improvements to Increase System Performance
| Area of Improvement | Solution |
|---|---|
| Identifying potential margin sentences | “ink” and (“margin” or “margins”) |
| Classifier training | Addition of more examples of positive surgical margins |
| Tumor stage regular expression | ”(?i)T[0-4]\\s*,*[abc]*(\\s*,*N[xO01]\\s*,*MX*[O01]*[abc]*)*” |
There were several limitations of this study. First, both UCLA and BWH are American College of Surgeons Commission on Cancer (ACoS CoC)–approved hospitals, which indicates that both have achieved a baseline of quality in regard to their oncology services. 35 The format and inclusion of quality measures in nonapproved facilities may differ systematically. This study also did not consider data quality issues introduced by properly formatted but inaccurate measurements. Although this approach offers insights into the effects of poor specificity of claims data on outcomes assessment, it is unable to measure the effects of the poor sensitivity of claims data, an important obstacle to the foundational task of target population identification. 36
Conclusion
In this study, automated extraction proved capable of identifying the three CAP Category 1 quality measures for prostate cancer surgery from pathology reports with high levels of accuracy. Interinstitutional differences in data format were largely mitigated, as were the effects of poor data quality on the final results. Despite the small number of failures, data quality was responsible for 26% of failures and is a topic that deserves greater attention. The existence of documentation standards contributed significantly to strong system performance. However, in the absence of standards, the machine-learning approach still performed with an accuracy of >97%. Future directions in the urology domain include the application of COAT as part of an RRP outcomes assessment effort. We are also planning to test the extensibility of the COAT architecture to facilitate outcomes assessment in other medical subdomains.
Acknowledgments
The authors thank Dr. Jim Sayre for statistics guidance, Drs. David Miller and Jim Hu for clinical expertise, and Emily Watt, Lewellyn Andrada, and Vijay Bashyam for their reviews and insights.
Footnotes
This work was supported in part by the National Library of Medicine Medical Informatics Training Grant LM07356 and National Institutes of Health grant R01 EB00362.
References
- 1.Corrigan J, Kohn L, Donaldson M. To err is human: building a safer health systemWashington, DC: National Academy Press; 1999. [PubMed]
- 2.Chassin M, Galvin R. The urgent need to improve health care quality: Institute of Medicine National Roundtable on Health Care Quality JAMA 1998;280:1000-1005. [DOI] [PubMed] [Google Scholar]
- 3.Jencks S, Cuerdon T, Burwen D, et al. Quality of medical care delivered to Medicare beneficiaries: a profile at state and national levels JAMA 2000;284:1670-1676. [DOI] [PubMed] [Google Scholar]
- 4.Mardon R, Shih S, Mierzejewski R, Halim S, Gwet P, Bost JE, National Committee for Quality Assurance The state of health care qualityWashington, DC: NCQA, Research and Analysis; 2002.
- 5.Tange H, Schouten A, Kester D, Hasman A. The granularity of medical narratives and its effect on the speed and completeness of information retrieval J Am Med Inform Assoc 1998;5:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krupski T, Kwan L, Afifi A, et al. Geographic and socioeconomic variation in the treatment of prostate cancer J Clin Oncol 2005;23:7881-7888. [DOI] [PubMed] [Google Scholar]
- 7.Potosky A, Davis W, Hoffman R, et al. Five-year outcomes after prostatectomy or radiotherapy for prostate cancer: the prostate cancer outcomes study J Natl Cancer Inst 2004;96:1358-1367. [DOI] [PubMed] [Google Scholar]
- 8.American Cancer Society Overview: Prostate Cancer. 2006http://www.cancer.org/docroot/CRI/CRI_2_1x.asp?dt = 36 2004. Accessed May 2006.
- 9.Miller D, Spencer B, Ritchey J, et al. Treatment choice and quality of care for men with localized prostate cancer Med Care 2007;45:401-409. [DOI] [PubMed] [Google Scholar]
- 10.D'Avolio L, Bui A. The Clinical Outcomes Assessment Toolkit: A framework to support automated clinical records-based outcomes assessment and performance measurement research J Am Med Inform Assoc 2008;15:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srigley J, Amin M, Humphrey P. College of American Pathologists web site. Prostate Gland Checklist: Prepared for Members of the Cancer Committee College of American Pathologists; 2005. http://www.cap.org/apps/docs/cancer_protocols/2005/prostate05_pw.pdf 2008. Accessed March 2006.
- 12.Wieder J, Soloway M. Incidence, etiology, location, prevention, and treatment of positive surgical margins after radical prostatectomy for prostate cancer J Urol 1998;160:299-315. [PubMed] [Google Scholar]
- 13.Hull G, Rabbani F, Abbas F, Wheeler T, Kattan M, Scardino P. Cancer control with radical prostatectomy alone in 1,000 consecutive patients J Urol 2002;167:528. [DOI] [PubMed] [Google Scholar]
- 14.Miller D, Spencer B, Shah R, et al. The quality of surgical pathology care for men treated with radical prostatectomy in the United States Cancer 2007;109:2445-2453. [DOI] [PubMed] [Google Scholar]
- 15.Friedman C, Alderson P, Austin J, Cimino J, Johnson S. A general natural language text processor for clinical radiology J Am Med Inform Assoc 1994;1:161-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aronson A. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program AMIA Symp 2001:17-21. [PMC free article] [PubMed]
- 17.Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports MedInfo 2004:565-572. [PubMed]
- 18.Chapman WW, Fiszman M, Dowling JN, Chapman BE, Rindflesch TC. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap Medinfo 2004;11:491. [PubMed] [Google Scholar]
- 19.Voorham J, Denig P. Computerized extraction of information on the quality of diabetes care from free text in electronic patient records of general practitioners J Am Med Inform Assoc 2007;14:349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turchin A, Kolatkar N, Grant R, Makhni E, Pendergrass M, Einbinder J. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes J Am Med Inform Assoc 2006;13:691-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreyer K, Mannudeep K, Hurier A, et al. Application of a recently developed algorithm for automatic classification of unstructured radiology reports: validation study Radiology 2005;234:323-329. [DOI] [PubMed] [Google Scholar]
- 22.Bashyam V, Taira RK. Identifying Anatomical Phrases in Clinical Reports by Shallow Semantic Parsing Methods. IEEE Symposium on Computational Intelligence and Data Mining, Honolulu, Hawaii, 1 March–5 April 2007, pages 210–214. DOI: 10.1109/CIDM.2007.368874 Posted online: 2007-06-04.
- 23.Zeng Q, Goryachev S, Weiss S, Sordo M, Murphy S, Lazarus R. Extracting principle diagnosis, co-morbidity, and smoking status for asthma research: evaluation of a natural language processing system BMC Med Inform Decis Mak 2006;6:30DOI: 10.1186/1472-6947-6-30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Biomedical Informatics Grid About caTIES. 2007http://caties.cabig.upmc.edu/overview.html 2006. Accessed March 10, 2007.
- 25.Cios K, Moore G. Uniqueness of medical data mining Artif Intell Med 2002;26:1-24. [DOI] [PubMed] [Google Scholar]
- 26.Tang P, Ralston M, Fernandez Arrigotti M, Qureshi L, Graham J. Comparison of methodologies for calculating quality measures based on administrative data versus clinical data from an electronic health record system: implications for performance measures J Am Med Inform Assoc 2006;14:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.In: Sobin L, Wittekind C, editors. TNM classification of malignant tumors. 6th ed.. New York: Wiley; 2002.
- 28.In: Greene F, Page D, Fleming I, et al. editors. AJCC cancer staging manual. New York: Springer; 2002.
- 29.Richie J. Management of patients with positive surgical margins following radical prostatectomy Urol Clin North Am 1994;21:717. [PubMed] [Google Scholar]
- 30.American Medical Association Staff CPT 2006Standard ed.. Chicago, IL: American Medical Association; 2005.
- 31.American Medical Association Staff AMA ICD-9-CM: Physician, International Classification of Diseases: Clinical ModificationChicago, IL: American Medical Association; 2007.
- 32.Yetisgen-Yildiz M, Pratt W. The effect of feature representation on MEDLINE document classificationProceedings of the American Medical Informatics Association, 2005; Washington, DC AMIA Annu Symp Proc 2005:849-853. [PMC free article] [PubMed]
- 33.Platt J. Machines using sequential minimal optimizationIn: Schölkopf B, Burges CJC, Smola AJ, editors. Advances in Kernel Methods—Support Vector Learning. Cambridge, MA: MIT Press; 1998.
- 34.Institute of Medicine Assessing the quality of cancer care: an approach to measurement in GeorgiaWashington, DC: National Academies Press; 2005.
- 35.American College of Surgeons Commission on Cancer Categories of Approvalhttp://www.facs.org/cancer/coc/categories.html#3 2005. Accessed Feb 24, 2007.
- 36.Fowles J, Fowler E, Craft C. Validation of claims diagnoses and self-reported conditions compared with medical records for selected chronic diseases J Ambul Care Manage 1998;21:24-34. [DOI] [PubMed] [Google Scholar]