Abstract
7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxo-dG) is one of the most common DNA lesions induced by oxidative stress. This lesion can be bypassed by DNA polymerase eta (Pol η) using in vitro translesion synthesis (TLS) reactions. However, the role that Pol η plays in vivo contributing to 8-oxo-dG mutagenesis remains unclear. To clarify the role of Pol η in 8-oxo-dG mutagenesis, we have used an siRNA knockdown approach in combination with a supF shuttle vector (pSP189) which replicates in mammalian cells. The pSP189 plasmid was treated with methylene blue plus light (MBL), which produces predominantly 8-oxo-dG in DNA, and was then replicated in GM637 cells in presence of siRNA that knocks down the expression of Pol eta, or in XP-V cells, which lack functional Pol eta. The mutant frequencies were increased in the Pol η siRNA knockdown cells and in XP-V cells relative to control, meaning that Pol η plays an important role in preventing 8-oxo-dG mutagenesis. In the same system, knockdown of OGG1 also led to an increase in mutagenesis. Neither the type of mutations nor their distribution along the supF gene were significantly different between control and target specific siRNA-transfected cells (or XP-V cells) and were predominantly G to T transversions. These results show that Pol η has an important role in error-free 8-oxo-dG lesion bypass and avoidance of oxidative stress induced mutagenesis in vivo.
1. Introduction
The first line of defense against oxidative stress is afforded by antioxidants, such as glutathione peroxidase (GPX) [1]. But excess oxidative stress, which antioxidants cannot eliminate, can damage macromolecules, including DNA, proteins, or lipids. To date, over 20 specific oxidized modifications of DNA resulting from oxidative stress have been identified [2]. Of these, 8-oxod-G is one of the most common DNA modifications, and formation of 8-oxo-dG has been used as a biomarker of oxidative DNA damage.
DNA can be repaired by the direct reversal of damage or by damage excision. However, cells have evolved a variety of other strategies for coping with damaged DNA. Some of these do not meet the strict definition of DNA repair since they do not result in the removal of the original DNA lesion. Hence, these cellular responses to DNA damage are referred to as DNA damage tolerance mechanisms [3–6]. One important DNA damage tolerance mechanism in both prokaryotes and eukaryotes is translesion DNA synthesis, a process in which a DNA polymerase inserts nucleotides opposite noninstructional or misinstructional DNA lesions [4–8]. In certain cases, translesion synthesis across a particular lesion by a DNA polymerase can be relatively accurate. However, in other cases translesion synthesis can be highly mutagenic, and this is, in fact, the molecular basis of most DNA damage induced mutagenesis and may also contribute to a fraction of spontaneous mutations [8–10].
On the basis of phylogenetic relationships, DNA polymerases have been grouped into six families - A, B, C, D, X, and Y. The Y-family polymerases differ from others in having a low copying fidelity on undamaged DNA and in their ability to polymerize nucleotides across from DNA lesions. The Y-family polymerases share five conserved sequence motifs and a common structural architecture. However, they differ in many of their structural and functional features, and these differences may account for their lesion specificity [5,8].
Xeroderma pigmentosum is a genetic disorder characterized by extreme sensitivity of the skin to sunlight, together with profound sunlight-induced skin pigmentation changes and an increased incidence of skin cancer [11,12]. In most cases, these features result from a defect in nucleotide excision repair (NER), but in about 20% of the patients, NER is fully functional. These so-called XP variant (XP-V) patients are deficient in their ability to replicate DNA containing UV damage [13]. XP-V cells are, like their NER-defective counterparts, extremely hypermutable by UV light [14] and the patients are skin cancer prone. In XP-V, the gene coding for DNA polymerase eta (Pol η) is inactivated by mutation [15,16]. Pol η has the capacity to insert AA with relatively high fidelity when bypassing a thymine-thymine dimer [15,16]. In XP-V, its function is perhaps replaced by DNA polymerases, which introduce more frequently mutations opposite UV photoproducts leading to cancer. In other proposed models, the defect in replication of UV damaged DNA in XP-V cells allows more time for deamination of cytosine within pyrimidine dimers to occur resulting in an increased mutation frequency [17].
Interestingly, the related Y-family translesion polymerases Pol η and Pol ⌈ are both able (Pol η much more effectively than Pol ι) to insert a C opposite 8-oxo-dG and also proficiently extend from this base pair [18–20]. In in vitro systems, Pol η showed a much higher bypass efficiency for 8-oxo-dG lesions in presence of PCNA and RP-A proteins [21]. Absence of Pol η caused a synergistic increase in the rate of spontaneous mutations in yeast lacking Ogg1 [18], suggesting a potential role of this translesion polymerase in restoring the correct G:C base pairs. Recently, Avkin and Livneh [22], using Pol η-proficient cells as well as Pol η-deficient XP-V cells, obtained results consistent with a model where both Pol δ and η, when present, are involved in bypass of 8-oxo-dG. Yeast DNA Pol ζ was found to be unable to bypass 8-oxo-dG lesions, but efficiently extended from nucleotides inserted opposite 8-oxo-dG by Pol δ, suggesting a role for these polymerases in TLS over oxidized purines [23]. The exact mechanism underlying the choice of polymerase in lesion bypass, and whether different polymerases are used at replication forks and at other sites is not known. It should be noted, however, that the reported frequencies and types of misincorporation by the TLS polymerases across various types of DNA damage are primarily based on in vitro experiments using primer extension assays. Thus, their exact roles in living cells remain to be established.
In this study, we describe how 8-oxo-dG can be bypassed by the TLS system in vivo. To this end, we introduced oxidative damage into a supF shuttle vector. Predominantly 8-oxo-dG damage was introduced using methylene blue and visible light, a reaction that occurs by photosensitization. Plasmids containing 8-oxo-dG lesions were replicated in normal and Pol eta deficient cells and the mutation frequencies and spectra were determined.
2. Materials and Methods
2.1. siRNA
The 21-nucleotide siRNA sequences used for targeted silencing of Pol η and OGG1 were designed and synthesized by Ambion (Austin, TX). Two siRNA duplexes directed against each target were selected and combined for optimal knockdown efficiency. Sequences of synthetic siRNAs were as follows: Pol η A, 5′-UAAACCUUGUGCAGUUGUATT-3′ (sense), 5′-UACAACUGCACAAGGUUUATT-3′ (anti-sense); B, 5′-CCAUUGGCUGUA-GUAAGAATT-3′ (sense), 5′-UUCUUACUACAGCCAAUGGTT-3′ (anti-sense); OGG1 A, 5′-GCUUG-AUGAUGUCACCUACTT-3′ (sense), 5′-GUAGGUGACAUCAUCAAGCTT-3′ (anti-sense); B, 5′-GAGGUGGCUCAGAAAUUCCTT-3′ (sense), 5′-GGAAUUUCUGA-GCCACCUCTT-3′ (anti-sense). As a control, siRNA against LacZ was used (5′-CGUACGCGGAAUACUUCGATT-3′) (sense), (5′-UCGAAGUAUUCCGGCGUACGTT-3′) (anti-sense). Searches of the human genome database (BLAST) were carried out to ensure that the sequences would not target other gene transcripts.
2.2. In vitro plasmid treatment
The pSP189 plasmid (10 μg) was treated in 10 mM sodium phosphate, pH 7.5, with 10 μM methylene blue (RICCA Chemical Company; Arlington, Texas). The methylene blue treated plasmid was exposed for various time periods to a 150 W light bulb (Claudfar) at a distance of 10 cm on ice. The plasmid was washed three times by precipitation with ethanol at −80°C. The DNA was subjected to either Fpg or OGG1 digestion to confirm the formation of 8-oxo-dG. One μg of treated plasmid was incubated with either 100 ng of Fpg protein [24] in 1× Fpg buffer containing 70 mM Hepes, pH 7.2, 0.5 mM EDTA, 1 mM 2-mercaptoethanol, 70 mM NaCl, and 5% glycerol or OGG1 (1 unit, Sigma; O2135) in 1× OGG1 buffer, provided by the manufacturer for 20 min at 37°C. After phenol-chloroform extraction and ethanol precipitation, the plasmid was resolved in 1% alkaline agarose gels. Before electrophoresis, the OGG1-treated plasmid was incubated in 1.2 N NaOH for 20 min at 70°C to convert abasic sites produced by OGG1 into strand breaks.
2.3. HPLC-MS/MS for quantification of 8-oxo-dG
Quantification of 8-oxo-dG was performed using the previously published method of Singh et al. [25] with modifications [26]. Briefly, analytical grade 8-oxo-dG was purchased from Cayman Chemical (Ann Arbor, MI) and mass-labeled 8-oxo-dG was kindly provided by Dr. Miral Dizdaroglu of the National Institute of Standards and Technology. Instrumentation consisted of an Agilent 1100 Capillary LC system (Agilent Technologies; Palo Alto, CA) in line with a Micromass Quattro Ultima Triple Quadrupole Mass spectrometer. (Micromass Inc., Beverly, MA). The detector settings were as follows: capillary voltage = 2.20 kV; cone voltage = 16 V; collision cell voltage = 13 V; source temperature = 125 °C; desolvation temperature = 260 °C; cone gas flow = 130 L/h; desolvation gas flow = 500 L/h. The mass transitions monitored for 8-oxo-dG and internal standard were 284→168 and 286→170, respectively.
2.4. Cell lines and siRNA transfection
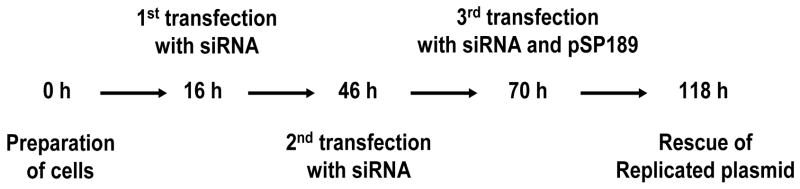
Two lines of human skin fibroblasts were used. GM637 (normal, repair-proficient) cells were obtained from the American Type Culture Collection (Manassas, VA). The XP-V cell line CTag was a generous gift from Dr. William Kaufmann (University of North Carolina, Chapel Hill) [27]. The cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and regularly passaged to maintain exponential growth. Synthetic siRNA was transfected using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) as described by the manufacturer. Briefly, 16 h before transfection, the cells were plated in 6-well plates at 60% confluence. Lipofectamine 2000 diluted in Opti-MEM (Gibco) was applied to the double-stranded RNA mixture. Per well, 125 pmol of siRNA mixed with 5 μl of Lipofectamine 2000 were applied. The final volume was 2.5 ml per well. To increase the siRNA knockdown efficiency, the cells were sequentially transfected three times. Cells were incubated for 30 h after the first transfection and then transfected with the same amount of siRNA (2nd transfection). Twenty-four hours after the second transfection, the cells were again transfected with siRNA (3rd transfection) and with methylene blue treated pSP189 plasmid. Forty-eight hours after the 3rd transfection, the expression of each target protein was examined.
2.5. Western blotting
Cells were washed with PBS and collected by scraping. They were lysed in ice-cold RIPA buffer (1 × PBS, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS) and homogenized. After incubation on ice for 1 hour, the mixture was centrifuged at 15,000 × g for 20 min. The supernatant was used for protein determination by the Bradford procedure (Bio-Rad) and for Western blotting. The proteins (40 μg of the whole cell lysate) were resolved on 10% SDS-polyacrylamide gels and transferred onto PVDF membranes (Bio-Rad). The membranes were incubated with antibodies against Pol η (Santa Cruz Biotechnology; sc-17770) or OGG1 (Novus Biologicals; NB 100–106) and then with horseradish peroxidase-labeled secondary antibodies (Amersham Biosciences) at a 1 to 5,000 dilution. The signals were developed with the enhanced chemiluminescence technique (Amersham Biosciences). To confirm equal gel loading, blots were stripped and reprobed with an anti-β-tubulin antibody (Santa Cruz Biotechnology; sc-5274).
2.6. supF mutation assay
Twenty-four hours after the 2nd siRNA transfection, the methylene blue plus light treated-pSP189 plasmid (3 μg) was co-transfected with 125 pmol of siRNA using Lipofectamine 2000 (Invitrogen). After a 48 h incubation period, the plasmids were rescued from the cells by an alkaline lysis method. Cells were trypsinized, washed, and resuspended in suspension buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 100 μg/ml Rnase A), mixed with 1 volume of lysis buffer (0.2 M NaOH, 1% (w/v) SDS), and incubated on ice for 5 min, followed by the addition of neutralization buffer (3 M potassium acetate, pH 5.5). After 15 min incubation at room temperature, the mixture was centrifuged for 10 min at 16,000 × g, and the supernatant was extracted once with phenol/chloroform followed by ethanol precipitation. The retrieved plasmid DNA was digested with DpnI to remove unreplicated plasmid. The plasmids were then electroporated into MB7070 bacteria, which carry a lacZ gene with an amber mutation. The transformed bacteria were diluted in 1 ml of SOC medium and plated on agar plates containing 50 μg/ml ampicillin, 1 μM isopropyl-1-thio-β-d-galactopyranoside, and 100 μg/ml X-Gal. After an overnight incubation at 37 °C, wild-type (blue) and mutant (white) colonies were counted to determine the mutant frequency. The numbers were derived from three independent transfections. It should be noted that this assay does not score mutations derived in E. coli and MutY catalyzed excision of A misincorporated opposite 8-oxo-dG will not play a role. This is because replication of the pSP189 plasmid is of the runaway type and there will be an enormous dilution of adducted plasmids. 10’s of thousands of progeny molecules are recovered from a single cell. The plasmid replication cycle is about 15–20 minutes. So even if there are a few single cycle progeny they will be diluted by a huge excess of non-adducted progeny. In addition, direct transformation of MBM7070 with damaged plasmids does not lead to the recovery of mutations. Colonies containing a plasmid with a mutated supF gene were identified by the color screen. The plasmids were purified using plasmid purification kits (Qiagen, Valencia, CA) and sequenced using an automated DNA sequencer.
3. Results
3.1. Inhibition of target protein expression by siRNA
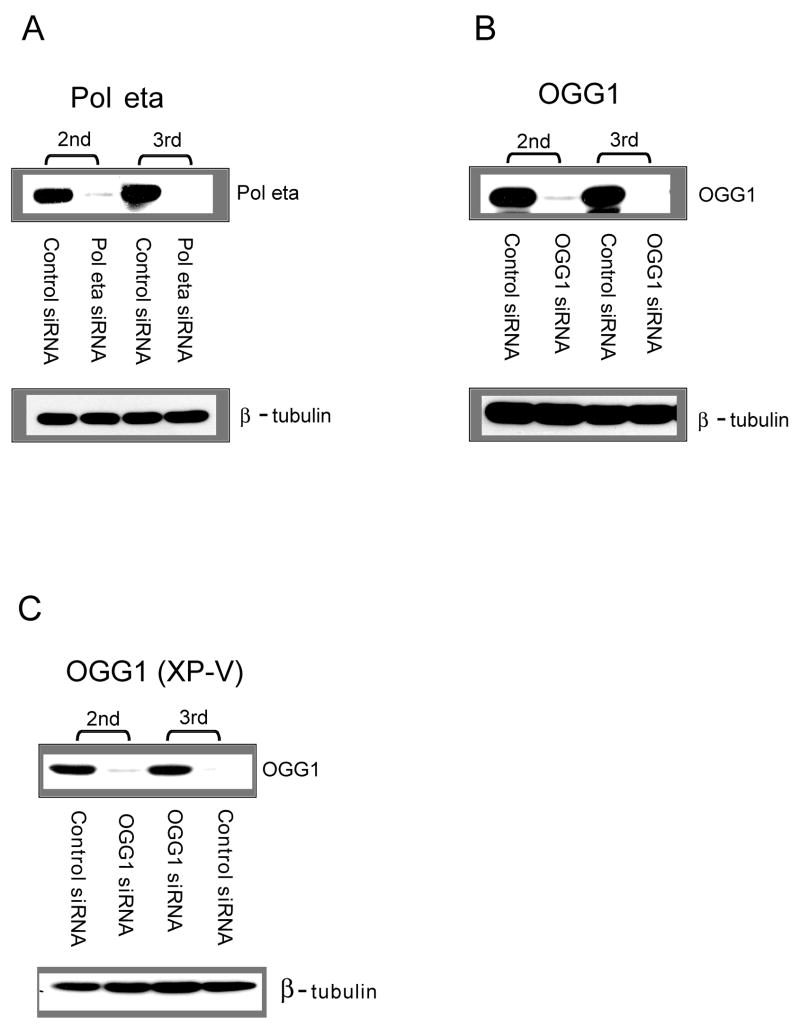
We attempted to inhibit Pol η or OGG1 expression in cells using siRNA and carried out a plasmid-based mutagenesis assay in the siRNA-treated cells (see Figure 1 for a general outline of this approach). To test how efficiently these siRNAs work for inhibition of targets, we performed Western blotting, as shown in Figure 2. For both OGG1 and Pol eta, we combined two siRNA duplexes for maximal knockdown efficiency. Western blotting analysis shows effective elimination of almost all target protein when the cells were transiently transfected with the synthetic siRNAs with or without pSP189 plasmid. Since we observed that the first transfection was not sufficient for complete inhibition (less than 50% reduction, data not shown), the cells were transfected three times with siRNA. Compared with the transfection with LacZ siRNA, the inhibition of target protein expression by siRNA was estimated to be more than 90% following the third transfection. For the third transfection, we co-transfected siRNA with the damaged or undamaged pSP189 plasmid. The lacZ siRNA, as a control, had no effect on the expression levels of target proteins, suggesting that the observed siRNA effect is specific.
Figure 1.
Experimental approach to study 8-oxo-dG mutagenesis in cells depleted of lesion bypass polymerases.
Figure 2. Western blot analysis of cells transfected with target-specific siRNA duplexes.
The expression of Pol eta in GM637 cells (A), of OGG1 in GM637 cells (B), and of OGG1 in XP-V cells (C) was effectively reduced by the siRNA duplexes relative to control siRNA (LacZ siRNA). The blot was stripped and re-probed with a β-tubulin antibody to control for equal gel loading (bottom). 2nd, 24 h after the second transfection with siRNA. 3rd, 48 h after the third transfection with siRNA and pSP189.
3.2. Plasmid treatment with methylene blue plus light (MBL)
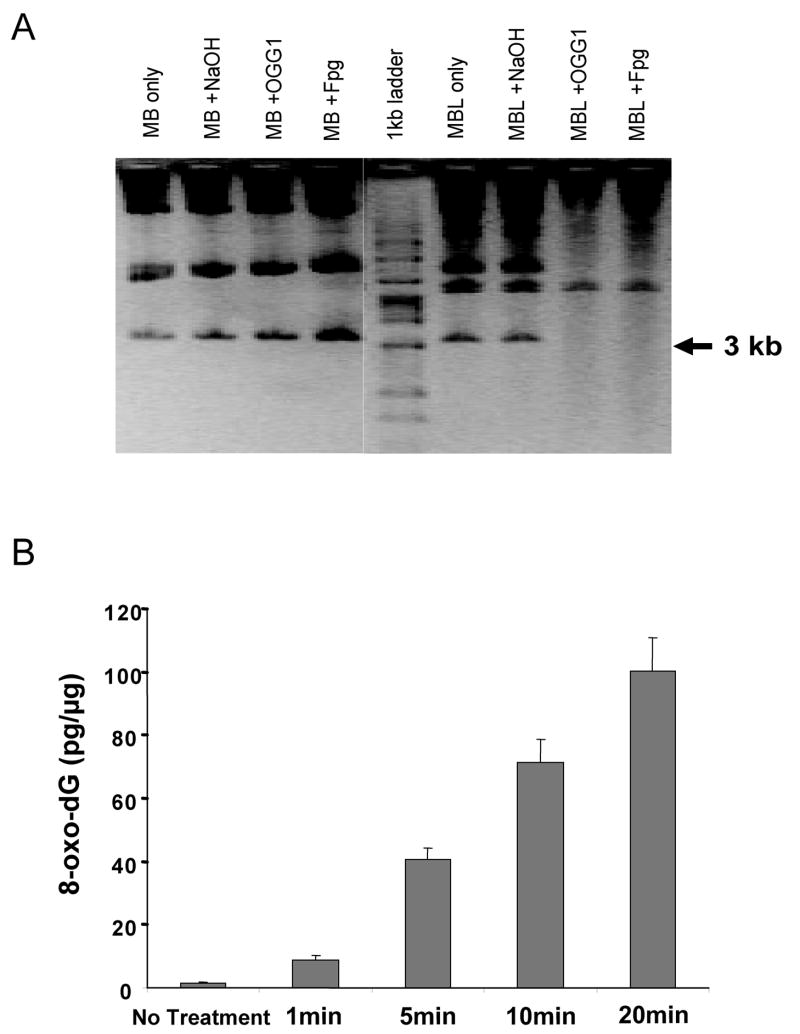
To determine the appropriate treatment conditions, the pSP189 plasmid was treated with methylene blue (10 μM) and was exposed to visible light for different time periods ranging from 0 to 30 min. Photoactivated methylene blue produces almost exclusively 8-oxo-dG in DNA without significant damage to the other DNA bases [28–30]. The treated plasmid was cleaved with either Fpg or OGG1 enzyme to evaluate 8-oxo-dG production. Methylene blue only did not damage the plasmid, but when exposed to light, DNA damage was observed. As shown Figure 3A, after 5 min of light exposure, it is clear that either Fpg or OGG1 cleaved the plasmid at random positions and the cleavage was almost equal, which means that most of the damage produced and cleaved by Fpg protein was 8-oxo-dG. OGG1 is a specific enzyme for removing 8-oxo-dG although both enzymes can also excise FapyG, which is however a minor product in MBL-treated DNA [28]. The cells were transfected with the plasmid, which was exposed to light for 5 min. The 8-oxo-dG production induced by the MBL treatment was also confirmed by HPLC-MS/MS analysis (Figure 3B). The extent of 8-oxo-dG generation increased with time of exposure to visible light. At 5 min exposure, the level of 8-oxo-dG was induced 20-fold over background, which increased to ~50-fold over background at 20 min exposure. We estimate that the 20 min MBL treatment produced approximately one 8-oxo-dG lesion per 10 kilobases of DNA.
Figure 3. Formation of 8-oxo-dG in methylene blue plus light (MBL) treated pSP189.
(A) Enzymatic cleavage and alkaline agarose gel analysis. After treatment of the plasmid with methylene blue only (MB) or methylene blue plus light (MBL) (5 min irradiation), the plasmid was subjected to treatment with NaOH, OGG1, or Fpg protein, which shows that the majority of DNA damage produced was 8-oxo-dG. The multiple bands represent different isoforms of the plasmid including concatemerized forms. Molecular size markers are indicated in kb. (B) 8-oxo-dG detection in MBL-treated pSP189 by HPLC-MS/MS. The amount of 8-oxo-dG was increased along with the increase in light exposure time. Results are expressed as averages of three independent experiments. Error bars = SD.
3.3. Mutant frequencies
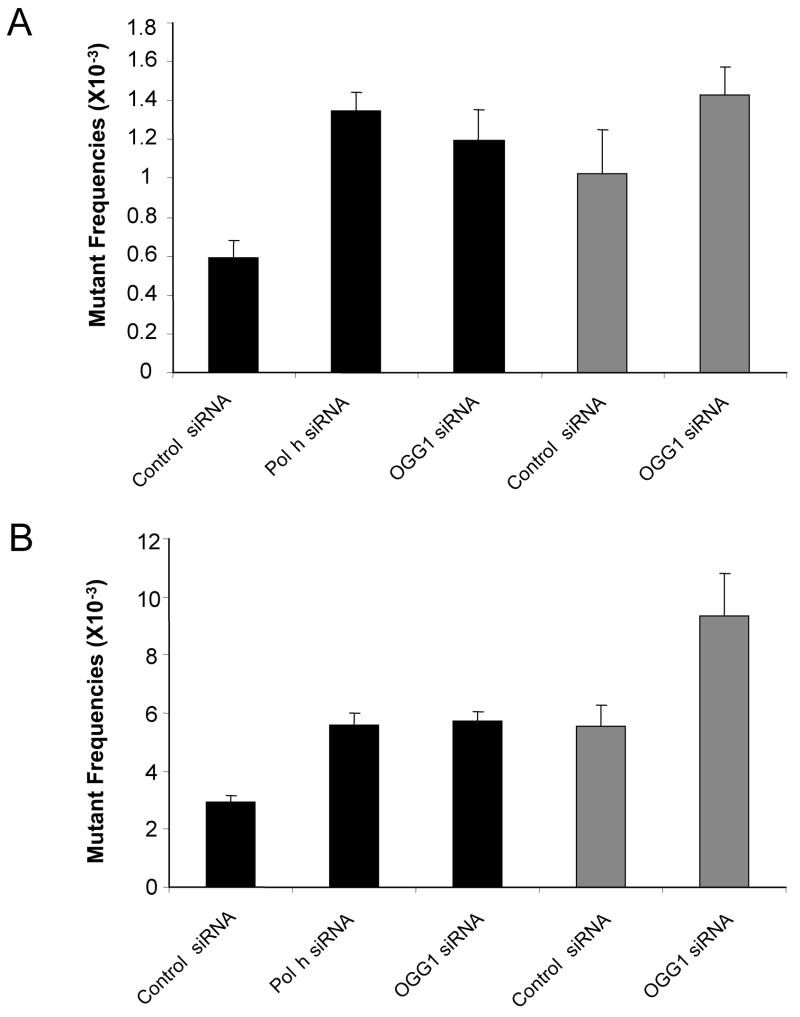
The SV40-based shuttle vector pSP189 was treated with MBL for 5 min on ice. The treated plasmids were co-transfected with siRNA duplexes into cells that had been pre-treated with the same siRNA duplexes twice. Cells were further incubated for 48 h to allow DNA replication and mutation fixation to take place (Figure 1). Then the plasmids were rescued, and the DNA was cleaved with DpnI to remove unreplicated plasmids. The rescued plasmids were then electroporated into MB7070 bacteria, which carry a lacZ gene with an amber mutation. Plasmids were isolated from white, mutant colonies, and the supF gene was sequenced. Siblings, i.e. plasmids originating from the same mutational event, are indicated by a repeated appearance of a signature sequence adjacent to the supF gene [31]. These siblings were about 5% of all sequenced plasmids and were excluded. As shown in Figure 4, in GM637 cells, the mutant frequency of untreated plasmid DNA with control siRNA was lower than all other samples (0.59 × 10−3 for control siRNA-treated cells, 1.35 × 10−3 for Pol η siRNA-treated cells, or 1.20 × 10−3 for OGG1 siRNA-treated cells). Compared to control siRNA-treated cells, the target siRNAs-treated cells showed an approximately 2-fold increase in background mutant frequencies. In XP-V cells transfected with control siRNA, the mutant frequency was similar as that in GM637 cells transfected with Pol η siRNA (~1 × 10−3).
Figure 4. supF mutant frequencies after replication of the MBL-treated plasmid in GM637 cells transfected with Pol eta-specific siRNA and in XP-V cells.
Mutant frequencies were determined from three independent experiments. (A) Mutant frequencies in cells co-transfected with siRNAs and untreated-pSP189. (B) Mutant frequencies in cells co-transfected with siRNAs and MBL-pSP189. The black bars indicate mutant frequencies in GM637 cells, while gray bars indicate mutant frequencies in XP-V cells.
After treatment of the plasmid with MBL, mutant frequencies were increased. In GM637 cells, the mutant frequencies were as follows; 2.92 × 10−3 for control siRNA-treated cells, 5.60 × 10−3 for Pol η siRNA-treated cells, and 5.74 × 10−3 for OGG1 siRNA-treated cells. In XP-V cells treated with control siRNA, the mutant frequency was 5.52 × 10−3, which was comparable to Pol η siRNA-treated GM637 cells. The mutant frequencies in target specific siRNA-treated cells were around two-fold higher than that in control siRNA-treated cells. By knocking down OGG1 in XP-V cells the mutant frequency was increased further, to 9.35 × 10−3 (Figure 4). These data show that the expression of Pol η or OGG1 in GM637 cells gives rise to a strong decrease in errors generated during replication of MBL-induced DNA lesions. The observed increase in MBL-induced mutagenesis also confirms that the knockdown of target proteins was efficient. Given the increases in mutant frequencies after knockdown of target proteins, we proceeded to determine the mutational spectra.
3.4. Mutation spectra
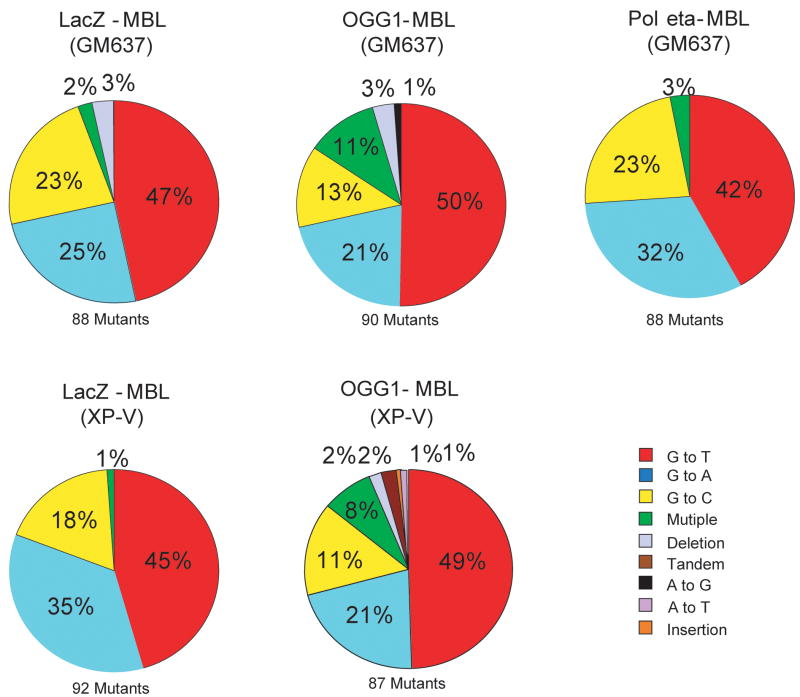
The sequence of the supF gene was determined in approximately 90 independently derived mutant plasmids from each transfection with MBL-treated DNA. Sequence analysis of mutants gave information on the types of mutations as shown in Figure 5, and the location of mutations as shown in Figure 6. Figure 5 compares the types of base-pair substitutions observed in the supF gene of MBL-treated plasmids replicated in target siRNA transfected cells with those in control siRNA transfected cells. Unexpectedly, the types of mutations induced by MBL were not substantially different between control siRNA-transfected cells and target siRNA-transfected cells. More than 90% of the mutants contained only a single base substitution. The majority of such mutations were G/C to T/A transversions (42–50%), G/C to A/T transitions (21–35%), and G/C to C/G transversions (11–23%) (Figures 5 and 6). Although G to T transversions are predominant mutations induced by 8-oxo-dG, bypass of this lesion in mammalian cells often also can lead to G to A and G to C mutations [32]. For the latter mutations, it is possible that they may be the result of further photooxidation of 8-oxo-dG leading hydantoin-type lesions [33,34].
Figure 5. Types of mutations in the supF gene after replication of the MBL-treated plasmid in control siRNA-treated cells, Pol η siRNA- or OGG1 siRNA- treated GM637 cells or XP-V cells.
The types of mutations introduced into the supF plasmid were determined by DNA sequencing. The total number of sequenced independent mutants was approximately 90 for each of the five mutation spectra.
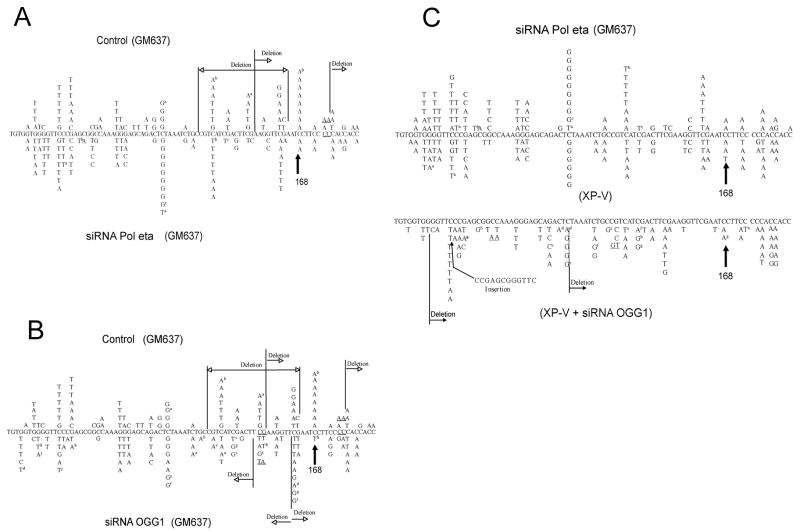
Figure 6. Mutation spectra induced by MBL-treatment in the supF plasmid.
Plasmid treated with MBL was transfected into GM637 or XP-V cells. The plasmid was introduced into cells after transfection with control siRNAs (A and B; upper) or target-specific siRNAs (A, Pol eta; B, OGG1; lower). A direct comparison of mutation spectra between GM637 cells treated with Pol eta siRNA and XP-V cells is shown in C, which also includes the mutation spectrum of XP-V cells treated with OGG1 siRNA (bottom). Tandem base substitutions are underlined. a to i indicate the occurrence of two mutations in the same plasmid.
There was no significant difference in the mutation spectra of XP-V cells versus that of Pol eta-siRNA transfected cells. The distribution of mutations along the gene was quite similar among all types of cells with similar hotspots (Figure 6). The only appreciable differences were the additional appearance of a C to A transversions hotspot at position 168, which was only observed in control-siRNA transfected cells, and the more frequent occurrence of multiple mutations when OGG1 was knocked down. The mutation distributions of XP-V cells treated with control siRNA are similar to that of GM637 cells treated with Pol η siRNA. These data indicate that loss of Pol η or OGG1, although leading to a substantial increase in mutant frequencies, had generally little effect on the types of mutations recovered in the MBL-treated target sequence.
4. Discussion
The 8-oxo-dG DNA lesion can be bypassed by Pol η in an error-free manner [18,20,21,35]. However, translesion synthesis studies of Y-family polymerases with oxidative DNA lesions have concentrated on in vitro assays, which prompted us to apply the in vivo methods reported here. Previously, our group provided proof of principle that siRNA technology can be used to dissect the in vivo roles of lesion bypass DNA polymerases in DNA damage-induced mutagenesis [17,36]. Our results provide in vivo evidence that Pol η prevents error-prone bypass during replication of MBL-damaged templates and affects the mutant frequencies. As shown in Figure 3, cleavage patterns of the MBL-treated plasmid were similar with Fpg protein, which recognizes a range of oxidized bases and with OGG1, which is quite specific for 8-oxo-dG. This indicates that the majority of lesions produced by MBL plus light were 8-oxo-dG bases, consistent with earlier reports [28–30], although we cannot exclude the possibility that small amounts of other base lesions not cleaved by Fpg protein are also formed. As shown in Figure 5, without MBL treatment, inhibition of Pol η caused an about two-fold increase in mutant frequencies. Thus, Pol η can protect against spontaneous mutations, perhaps caused by the presence of low levels of DNA damage present on the untreated plasmid DNA, as well as against MBL-induced mutations. The mutation spectrum of the MBL-treated supF plasmid was quite similar between all siRNA-transfected cells and XP-V cells. Moreover, the positions of mutational hotspots at such sites were similar in control siRNA-transfected cells and in target siRNA-transfected cells.
Since Pol η can bypass 8-oxo-dG DNA lesions in a mostly error-free manner in vitro, it was expected that the knockdown of this polymerase would result in an increase of G to T transversions, which are dominant mutations derived from 8-oxo-dG [35,37]. But unexpectedly, the percentages of G to T transversions were similar in all control and target siRNA-transfected cells. We recognize that more subtle differences in mutation spectra, which may exist, may be masked by the only 2-fold difference in mutant frequencies between the control and specific siRNA-treated cells. Avkin and Livneh [22] using a gapped plasmid with a single 8-oxo-dG lesion in the single-stranded region compared lesion bypass and mutational specificity in a human lung cancer cell line and in an XP-V cell line. They found that the lesion was bypassed with similar efficiency in both cell types and incorporation of A opposite the lesion was the predominant mutagenic event in the cancer cell line and in the XP-V cell line, which is consistent with our results.
The increase in mutant frequency may be due to increased error-prone lesion bypass in absence of Pol η but this error-prone lesion bypass is carried out by the same polymerase(s) regardless of the presence or absence of Pol η. In other words, in normal cells Pol η competes with more error-prone polymerases for bypass of 8-oxo-dG and keeps the mutant frequency low. Although correct nucleotide incorporation and bypass efficiency of DNA polymerase iota (Pol ι) on 8-oxo-dG templates is much lower than that of Pol η in vitro [19,20,38], Pol ι may still be a possible candidate as a substitute for Pol η in vivo. Pol lambda is another candidate polymerase for 8-oxo-dG bypass, although so far this activity has been demonstrated only by in vitro assays [21]. Alternatively, mutations at 8-oxo-dG may be caused most frequently by yet other lesion bypass or by replicative DNA polymerases. Data on the roles of Pol ι, Pol λ, Pol κ, Rev1, and Pol ζ in 8-oxo-dG mutagenesis within cells are not yet available. The successful use of the siRNA approach in this chapter suggests that a similar siRNA-based method would have value for examining the function of many other lesion bypass polymerases in vivo.
In summary, using an siRNA-based mutagenesis assay and XP-V cells with a genetic defect in DNA polymerase η, we present evidence that Pol η protects against mutations induced by 8-oxo-dG DNA lesions. We conclude that in the absence of Pol η, incorrect replication of 8-oxo-dG lesions by polymerases other than Pol η produces similar mutations as when Pol η is present but at a higher frequency.
Acknowledgments
This work was supported by NIH grant ES06070 to G.P.P.
Abbreviations
- 8-oxo-dG
7,8-dihydro-8-oxo-2′-deoxyguanosine
- Pol η
DNA polymerase eta
- Pol ι
DNA polymerase iota
- XP-V
XP variant
- TLS
translesion synthesis
- BER
base excision repair
- NER
nucleotide excision repair
- MBL
methylene blue plus light
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee DH, Esworthy RS, Chu C, Pfeifer GP, Chu FF. Mutation accumulation in the intestine and colon of mice deficient in two intracellular glutathione peroxidases. Cancer Res. 2006;66:9845–9851. doi: 10.1158/0008-5472.CAN-06-0732. [DOI] [PubMed] [Google Scholar]
- 2.Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat Res. 1992;275:331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- 3.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, Feaver WJ, Gerlach VL. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci U S A. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg EC, Fischhaber PL, Kisker C. Error-prone DNA polymerases: novel structures and the benefits of infidelity. Cell. 2001;107:9–12. doi: 10.1016/s0092-8674(01)00509-8. [DOI] [PubMed] [Google Scholar]
- 6.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 9.Abdulovic AL, Minesinger BK, Jinks-Robertson S. Identification of a strand-related bias in the PCNA-mediated bypass of spontaneous lesions by yeast Poleta. DNA Repair (Amst) 2007 doi: 10.1016/j.dnarep.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci U S A. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 16.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 17.Choi JH, Pfeifer GP. The role of DNA polymerase eta in UV mutational spectra. DNA Repair (Amst) 2005;4:211–220. doi: 10.1016/j.dnarep.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 19.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. Embo J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 22.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat Res. 2002;510:81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connor TR, Graves RJ, de Murcia G, Castaing B, Laval J. Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J Biol Chem. 1993;268:9063–9070. [PubMed] [Google Scholar]
- 25.Singh R, McEwan M, Lamb JH, Santella RM, Farmer PB. An improved liquid chromatography/tandem mass spectrometry method for the determination of 8-oxo-7,8-dihydro-2′-deoxyguanosine in DNA samples using immunoaffinity column purification. Rapid Commun Mass Spectrom. 2003;17:126–134. doi: 10.1002/rcm.883. [DOI] [PubMed] [Google Scholar]
- 26.Besaratinia A, Synold TW, Chen HH, Chang C, Xi B, Riggs AD, Pfeifer GP. DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 tumor suppressor gene. Proc Natl Acad Sci U S A. 2005;102:10058–10063. doi: 10.1073/pnas.0502311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bullock SK, Kaufmann WK, Cordeiro-Stone M. Enhanced S phase delay and inhibition of replication of an undamaged shuttle vector in UVC-irradiated xeroderma pigmentosum variant. Carcinogenesis. 2001;22:233–241. doi: 10.1093/carcin/22.2.233. [DOI] [PubMed] [Google Scholar]
- 28.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 29.Floyd RA, West MS, Eneff KL, Schneider JE. Methylene blue plus light mediates 8-hydroxyguanine formation in DNA. Arch Biochem Biophys. 1989;273:106–111. doi: 10.1016/0003-9861(89)90167-7. [DOI] [PubMed] [Google Scholar]
- 30.Ravanat JL, Cadet J. Reaction of singlet oxygen with 2′-deoxyguanosine and DNA. Isolation and characterization of the main oxidation products. Chem Res Toxicol. 1995;8:379–388. doi: 10.1021/tx00045a009. [DOI] [PubMed] [Google Scholar]
- 31.Canella KA, Seidman MM. Mutation spectra in supF: approaches to elucidating sequence context effects. Mutat Res. 2000;450:61–73. doi: 10.1016/s0027-5107(00)00016-6. [DOI] [PubMed] [Google Scholar]
- 32.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchko GW, Wagner JR, Cadet J, Raoul S, Weinfeld M. Methylene blue-mediated photooxidation of 7,8-dihydro-8-oxo-2′-deoxyguanosine. Biochim Biophys Acta. 1995;1263:17–24. doi: 10.1016/0167-4781(95)00078-u. [DOI] [PubMed] [Google Scholar]
- 34.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 35.Klungland A, Bjelland S. Oxidative damage to purines in DNA: role of mammalian Ogg1. DNA Repair (Amst) 2007;6:481–488. doi: 10.1016/j.dnarep.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Choi JH, Besaratinia A, Lee DH, Lee CS, Pfeifer GP. The role of DNA polymerase iota in UV mutational spectra. Mutat Res. 2006;599:58–65. doi: 10.1016/j.mrfmmm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 37.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RE, Haracska L, Prakash L, Prakash S. Role of hoogsteen edge hydrogen bonding at template purines in nucleotide incorporation by human DNA polymerase iota. Mol Cell Biol. 2006;26:6435–6441. doi: 10.1128/MCB.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]