Abstract
The focus in protein folding has been very much on the protein backbone and sidechains. However, hydration waters make comparable contributions to the structure and energy of proteins. The coupling between fast hydration dynamics and protein dynamics is considered to play an important role in protein folding. Fundamental questions of protein hydration include, how far out into the solvent does the influence of the biomolecule reach, how is the water affected, and how are the properties of the hydration water influenced by the separation between protein molecules in solution? We show here that Terahertz spectroscopy directly probes such solvation dynamics around proteins, and determines the width of the dynamical hydration layer. We also investigate the dependence of solvation dynamics on protein concentration. We observe an unexpected nonmonotonic trend in the measured terahertz absorbance of the five helix bundle protein λ6–85* as a function of the protein: water molar ratio. The trend can be explained by overlapping solvation layers around the proteins. Molecular dynamics simulations indicate water dynamics in the solvation layer around one protein to be distinct from bulk water out to ≈10 Å. At higher protein concentrations such that solvation layers overlap, the calculated absorption spectrum varies nonmonotonically, qualitatively consistent with the experimental observations. The experimental data suggest an influence on the correlated water network motion beyond 20 Å, greater than the pure structural correlation length usually observed.
Keywords: solvation dynamics, THz spectroscopy, lambda repressor, molecular modeling
Water molecules interact with proteins on many length and time scales. Although the dynamics of the hydration water occurs on the picosecond time scale, “slaving” to fast solvent modes profoundly affects the slower but larger-scale protein motions (1). In return the protein influences the structure and dynamics of surrounding water molecules (2). X-ray crystallography has revealed ordered water structure around polar and charged sidechains (3), as well as cooperative insertion of water into hydrophobic cavities (4). Dielectric spectroscopy extends the time scale from microseconds down to 0.1 ns (5). Experiments have been extended to the THz range in films and crystals, probing motions on the picosecond time scale (6, 7). Hydrated protein powders probed by inelastic neutron scattering (0.1–100 ps) or solid-state NMR (nanoseconds) reveal that slower protein time scales and faster solvent time scales indeed show correlated dynamics (8). On the fastest time scales, 2D infrared spectroscopy and fluorescence of surface residues provide local probes of the dynamics in the femtosecond to picosecond range (9, 10). Coupling of modeling with experiments has revealed complex solvation structure around small biomolecules (11, 12), bridging our microscopic structural and thermodynamic understanding of biosolvation.
Terahertz absorption spectroscopy of biomolecules fully solvated in water yields direct information on the global dynamical correlations among solvent water molecules. Yet, THz spectroscopy is experimentally challenging (13) because of the strong THz absorption of water. With the advent of powerful table-top sources capable of penetrating the bulk of aqueous solutions (14), a new window between microwaves and the infrared is opening up onto the interaction of water molecules with proteins. At high protein concentrations, terahertz absorption decreases linearly when large concentrations of protein are added to the solution (15). Such behavior indicates that the solute molecule replacing the water has fewer low frequency modes than the solvent it replaces, leaving a hole in the spectral density. However, our precise measurements of terahertz absorbance of lactose solvation showed that the absorption coefficient levels off at low concentrations (11). This deviation from linearity is caused by competing contributions from hydration and bulk water to the total THz absorption spectrum and allows direct extraction of the size of the hydration layer. Here we report a more dramatic turnover behavior at low to moderate concentrations (0 to 2.3 mM) of the engineered five helix bundle protein λ6–85*, which has been investigated in respect of its folding before (16).
Experimental Results
We have determined the absorption of solvated λ6–85* from 2.25 to 2.55 THz with our table-top THz spectrometer (14). This frequency range probes the intermolecular collective modes of the hydrogen bonding network and some collective modes of the protein, such as skeletal and breathing modes. The experimental setup is described in ref. 14. The transmitted intensity was measured as a function of the layer thickness d (which was varied in steps of 5 μm) and fit to an exponential function according to Beer's law: The data were fit to the following expression: I(d) = I0 exp(−α d) + C, with I0, α, d, and C corresponding to the intensity before the probe, the absorption coefficient of the probe, the layer thickness of the probe, and the detector offset, respectively.
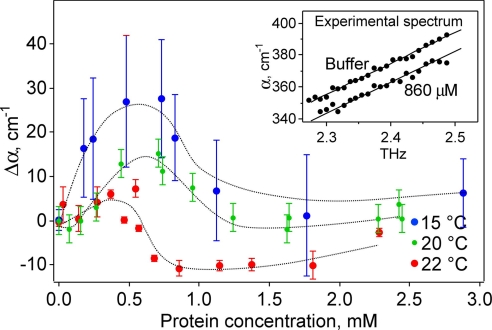
Within our measurement uncertainty, the absorption of the solvated protein increased linearly with frequency in this rather narrow frequency range (see Fig. 1Inset). Therefore, we used a linear fit in the measured frequency range between THz absorption and frequency to obtain accurate absorption coefficients at a given frequency. This procedure, together with averaging over multiple measurements, minimizes noise and allows a reliable comparison between the different THz absorption spectra for different protein concentrations. Fig. 1 displays the absorption coefficient relative to bulk water as a function of the concentration of λ6–85*. The absorption coefficient increases before dropping. At higher concentrations, it will decrease quasilinearly as discussed below (see Fig. 2) and in ref. 15.
Fig. 1.
Difference in the THz absorption coefficient at 2.25 THz relative to bulk water plotted against concentration to 3 mM at 15°C, 20°C, and 22°C (more extensive averaging was done at 22°C because of the slightly smaller effect). The absorbance depends nonlinearly on concentration in this region. Note that the THz absorption for bulk water (zero point) increases with increasing temperature. (Inset) The frequency dependence of the absorption coefficient is linear between 2.25 and 2.55 THz (22°C: comparison of buffer and at a protein concentration of 860 μM).
Fig. 2.
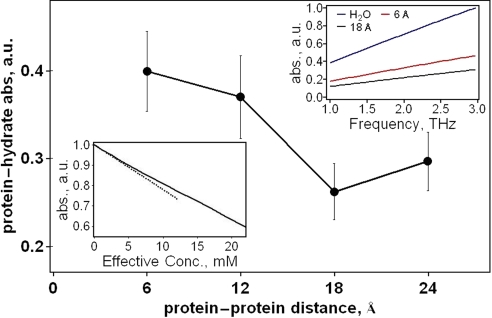
THz absorbance of protein and first hydration layer plotted against distance between the protein surfaces is nonmonotonic. (Upper Right Inset) Frequency dependence of the protein-hydration layer absorbance at low (6 Å) and high (18 Å) protein–protein separation, together with the absorbance computed for the same volume of bulk water. (Lower Left Inset) Total computed THz absorption against effective concentration of protein. The quasi-linear region at large protein concentration (>15 mM) reproduces the known behavior, and the nonlinearity at small protein concentration matches the experimental trend measured here (the dashed line is the linear fit to the low concentration trend).
We have measured the concentration dependence of the THz absorption at three different temperatures. Although the absolute differences relative to the bulk value differ for the three temperatures, the overall variation in the absorbance with concentration is the same at each temperature. When we compare the three curves one has to keep in mind that the zero point (the bulk water value at the given temperature) decreases with decreasing temperature (19). This partially explains the offset between the three curves. Whereas the absolute THz absorption coefficient of water (c = 0) is increased by approximately a factor of two for a temperature increase of 20°C at 2 THz (19), a less pronounced change of the THz absorption of the protein is expected. In this case Δα = α(c) − α(0), where α(c), the THz absorption coefficient for a given concentration, c, is expected to deviate at higher concentrations for different temperatures. The offset reflects the difference between the decrease for bulk water and protein + hydration water.
The nonmonotonic behavior observed in Fig. 1 cannot be explained by an effective two-component model: λ6–85* displaces ≈7,400 Å3 of buffer, based on the solvation-free radius of gyration of 12.1 Å estimated from small angle x-ray scattering (17). If the protein itself were completely transparent at 2.4 THz, we would expect a linear decrease of the THz absorption coefficient α with increasing protein concentration cProtein in a solvent volume V according to
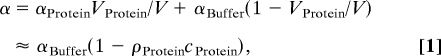 |
(ρProtein is the protein density, ≈ 1.4–1.5 g/cm3) (20). The 0.5–0.1 mM concentration at the turnaround in Fig. 1 corresponds to a water volume decrease of 1%. The measured THz absorption deviates strongly from a linear decrease as predicted according to Eq. 1. Thus, the hydration water around the protein must contribute in a nontrivial way to the total THz absorption. For solvated lactose, the total absorption could be decomposed into three components: the volume weighted average of the absorption of the solute, the solvation water, and the bulk water
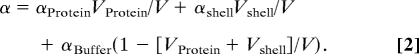 |
However, even this model is unable to describe adequately the experimentally observed concentration dependence in the THz absorption coefficient, unless the absorbance of the hydration water depends on the distance between protein molecules. To come to a microscopic understanding of the observed results, we carried out accompanying molecular modeling calculations, which reveal and quantify the protein distance dependence of the absorbance of the hydration layer.
Molecular Modeling Results
It has been known for some time that in effective media consisting of binary mixtures a linear variation of the measured changes of a specific property of the mixture with concentration does not necessarily hold (21). However, our goal here is to provide a direct microscopic picture in terms of solvation water structure and dynamics. To obtain such a picture, we studied the approximate dynamics of the protein and explicit solvent water by molecular dynamics (MD) simulations. We computed THz spectra (Fig. 2) from the dipole autocorrelation function from the simulation data for bulk water and for λ6–85* with a layer of hydration water. Because we are interested in how the absorbance of λ6–85* and hydration water changes as a function of the distance between proteins, we have carried out MD simulations for solvated λ6–85* over a range of protein–protein distances.
Fig. 2 shows the computed absorbance of the protein and first hydration layer at 2.5 THz as a function of the distance between protein surfaces. In accord with experiment, we find that the distance between the proteins significantly influences the absorbance of the protein and its first hydration layer (and layers beyond, see below). First, the absorbance decreases as the proteins are brought closer together from 24 to 18 Å. Then the absorbance turns over and flattens for the shortest distances, changing little with interprotein distance, mimicking the concentration-dependent turnover observed experimentally. This trend is especially strongly pronounced in the calculation because the bulk water, which contributes most at the measured protein concentration, is not included. The trend is less evident if we include the bulk water in the predicted total absorption because the simulation predicts water to have a higher absorbance than low concentrations of protein in water.
The nonmonotonic trend is seen over a broad range of frequencies in the THz region. Fig 2 Upper Right Inset displays the absorbance of the protein and first layer of water molecules for proteins separated by 6 Å and 18 Å. The increase in the absorbance with frequency resembles the corresponding increase observed in the experiments (Fig. 1 Inset). We note that we have also computed the absorbance in the harmonic approximation using normal modes, where we also find a significant dependence of the absorbance on protein–protein separation, and a trend similar to the trend we find here using the dipole autocorrelation function to compute the THz spectrum, which shows that the trend is robust.
We can computationally estimate the dependence of the absorption coefficient on protein concentration by using the surface-to-surface distances in Fig. 2. A given concentration c corresponds to a distribution of surface-to-surface distances, although in the simulated solution all protein molecules are oriented the same way due to the periodic boundary conditions. We have reached an approximation to the distribution of protein molecules for a given concentration by Monte Carlo sampling of hard-sphere 12.1 Å radius proteins, a good approximation to the radius of the globular λ6–85*. In this approximation, we assume that the proteins interact only as hard spheres. Sampling this distribution with the calculated absorbances, which were each used to represent a range of protein–protein separation distances ±3 Å of the distance given in the figure, yields the absorption of the protein and hydration shell. Combining this result for the concentration dependence of the absorbance of protein and hydration layer with the bulk water contribution yields an estimate for the total absorption as a function of effective concentration, i.e., the concentration is effective because the molecular dynamics periodic boundary conditions do not correspond to a bulk distance and orientation distribution of the protein.
Fig. 2 Lower Left Inset shows the result, which qualitatively matches the trend in the experimental data at moderate protein concentrations: the absorbance drops (in the simulation up to 2.5 mM, in the experiment between 0.5 and 1.5 mM), then changes to a less steep slope. At high concentrations (>15 mM), the absolute value of the slope increases slightly again, and absorbance drops off linearly with increasing concentration. If one looks at a broad range of concentrations and neglects the change in curvature below ≈15 mM, the absorbance decreases approximately linearly with concentration, as observed in earlier measurements over a wide range of concentrations (15). Only by precise measurements of changes at low concentrations does the nonlinear variation, which is a direct probe of the dynamical hydration water, become apparent. As discussed below, the appearance of a change in the slope of the absorbance vs. concentration at low concentration implies a broad hydration water shell around each protein, despite the a priori assumption of a single hydration layer made in the preceding computational analysis.
Discussion
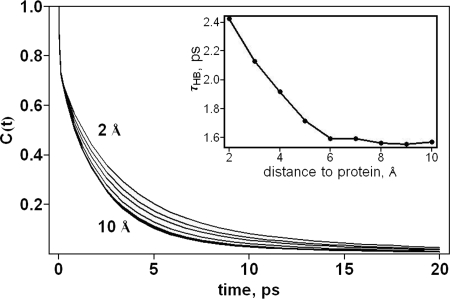
Whereas the existence of hydration layers of over 10 Å has not been reported experimentally, such large layers containing water dynamically distinct from water in the bulk have been found in earlier molecular dynamics simulations (22, 23). In addition to the dipole autocorrelation function and terahertz spectra discussed above, a hydration layer corresponding to water dynamics distinct from bulk water can be quantified by the hydrogen bond correlation function, C(t), which yields the probability that a hydrogen bond that exists between two water molecules at a given time, t = 0, is present at a later time, t, regardless whether the bond has been broken between 0 and t. MD simulations of solvated globular λ6–85* at 27°C reveals that the hydrogen bond correlation function for water molecules in 2-Å-thick layers of water up to 10 Å from globular λ6–85* is distinct from the hydrogen bond correlation function computed for bulk water (23), as we show in Fig. 3. Whereas hydrogen bonds between water molecules generally rearrange on the picosecond time scale, hydrogen bonds between water molecules out to ≈10 Å from a protein molecule survive longer than those between bulk water molecules. Thus, the THz dynamics of water molecules out to several layers around the protein are noticeably different from the THz dynamics of water in the bulk, consistent with the THz spectra, which reveal the influence of the protein on the water also out to several layers. Such a distinction between the hydration layer and bulk water seen upon examining THz dynamics of water molecules is not observed for structural parameters of the hydration water, e.g., orientational correlations or local density (24, 25), which is distinct from that of bulk water only out to ≈3 Å.
Fig. 3.
The hydrogen bond correlation function for the water molecules around λ6–85* shown, from top to bottom, within 2 Å of the protein, between 2 and 4 Å, etc., up to between 8 and 10 Å, which appears very close to the bulk water value. (Inset) The hydrogen bond lifetimes for water as a function of distance (Å) from the surface of the protein, which is simply defined as the time at which C(t) is 1/e.
The unexpected nonlinear absorbance vs. concentration is a collective dynamical property of the protein-hydration water system. Protein–protein distance-dependent changes in the collective dipole moment with time are evident upon examining the dipole autocorrelation function, which compares the dipole moment at time t = 0 with the dipole moment at later times. The absorbance obtained with the dipole autocorrelation function at 27°C decreases when the protein separation decreases from 24 to 18 Å by ≈15%, then increases by ≈40% when the distance between protein surfaces shrinks to 12 Å; there is still a modest increase in the absorbance when the protein–protein separation is reduced further still to 6 Å. The variation in the absorbance beyond a protein–protein separation of 18 Å indicates that the hydration shell around each protein extends to at least 9 Å, consistent with the differences observed between the hydrogen bond correlation function for water molecules out to 10 Å from the protein and the hydrogen bond correlation function computed for water in the bulk. The experimental THz spectra indicate a dynamical hydration layer that is even larger than the hydration layer of ≈10 Å that can be deduced from the MD simulations: the crossover to the plateau in the experiments occurs at 0.5–1 mM for measurements at 15, 20, and 22°C, respectively.
In the model, a noticeable change in slope occurs at an effective concentration between 2.5 and 8 mM, which has to be compared with the 5 times lower concentration where we have recorded a nonlinearity. Assuming “hard sphere” proteins, the experimental concentration range corresponds to an average separation of >20 Å (surface to surface distance) between proteins. Although lambda repressor shows no signs of irreversible aggregation at concentrations below 20 mM, a nonzero attractive interaction potential between proteins (transient aggregation) can shift the peak in absorbance toward smaller concentrations, because the actual distance is then smaller than expected for the assumed random distribution. However, any long-range interaction cannot explain the observed maximum in the THz absorption because it would only cause a “rescaling” of the concentration axis. The nonlinearity has to be attributed to the onset of overlaping dynamical hydration layers, which show an increased THz absorption compared with the buffer.
In summary, both experiment and simulations indicate a long-range dynamical hydration shell and reveal the dynamics of the hydration water to be sensitive to the distance between proteins.
Methods
Experiment.
The lambda repressor fragment 6–85 Tyr22Trp mutant gene, a gift from Terry Oas, was expressed in Escherichia coli BL-21 cells and purified, as described in ref. 16. The resulting λ6–85* protein was buffered in 50 mM magnesium acetate at concentrations up to 2.3 mM, where data could be taken without signs of precipitation in the 15–22°C range. Small-angle x-ray scattering data have shown that the protein does not cluster up to twice this concentration in aqueous ethylene glycol solvents, as described in Ref (17). A tabletop p-Ge laser system described in ref. 14 provided tunable light in the 2.2–2.6 THz region and transmission was detected with a liquid helium-cooled p-Ge detector (14). The THz absorption coefficient is obtained by scanning a variable-pathlength (d) cell and fitting the transmitted THz power I according to Beer's law for the absorption coefficient α, after subtracting a constant baseline
Molecular Modeling.
The absorption coefficient is computed from the dipole autocorrelation obtained from MD simulations as
where
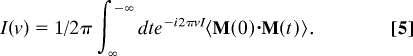 |
M is total dipole of system; n is index of refraction (taken as constant over the frequency range of the experiment); c is the speed of light, kB is Boltzmann's constant, and h is Planck's constant. MD simulations for the calculations of these quantities have been carried out using the GROMACS 3.2 package with the GROMOS96 force field where the protein is embedded in a box containing simple point charge (SPC) model water (18). Charges on both the protein atoms and the water molecules are fixed. MD simulations were run at 300 K using the NVT ensemble with periodic boundary conditions for 50 ns for proteins in cells that allowed for 3, 6, 9, and 12 Å water extending beyond the surface of the protein, thereby separating pairs of proteins by at least twice these distances. The absorption coefficients were calculated with the dipole correlation time averaged over 2,000, 25-ps segments of each MD trajectory, as longer times within the limits of the simulation did not change the results appreciably. Absorption coefficients reported for protein and a hydration layer correspond to the protein and the nearest 3 Å of water molecules. The absorption coefficient was also computed for the same volume of SPC water. The absorption coefficient computed for the water was not sensitive to box size. We computed hydrogen bond dynamics for water molecules around the protein by using a geometrical criterion for the hydrogen bonds, where bonds are taken to be formed when the distance between D and A is <3.5 Å and the angle between D, hydrogen, and A (D–H–A) is >120°. The results for the predicted distance-dependent autocorrelation function are shown in supporting information (SI) Fig. 4.
Supplementary Material
ACKNOWLEDGMENTS.
We thank E. Bründermann and G. Schwaab for the initial instrument setup, helpful discussions, and software development. B. Born helped with the purification of the protein. We thank Chr. Herrmann for access to his equipment and helpful advice. M.H., M.G. and D.M.L. gratefully acknowledge financial support from the Human Frontier Science Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709207104/DC1.
References
- 1.Ansari A, Berendzen J, Bowne SF, Frauenfelder H, Iben IET, Sauke TB, Shyamsunder E, Young RD. Proc Natl Acad Sci USA. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Despa F, Fernández A, Berry RS. Phys Rev Lett. 2004;93:228104. doi: 10.1103/PhysRevLett.93.228104. [DOI] [PubMed] [Google Scholar]
- 3.Burling FT, Weis WI, Flaherty KM, Brunger AT. Science. 1996;271:72–77. doi: 10.1126/science.271.5245.72. [DOI] [PubMed] [Google Scholar]
- 4.Collins M, Hummer G, Quillin ML, Matthews BW, Gruner SM. Proc Natl Acad Sci USA. 2005;102:16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant EH, Sheppard RJ, South GP. Dielectric Behaviour of Biological Molecules in Solution. Oxford: Clarendon; 1978. [Google Scholar]
- 6.Chen J-Y, Knab JR, Cerne J, Markelz AG. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.040901. 040901. [DOI] [PubMed] [Google Scholar]
- 7.Siegrist K, Bucher CR, Mandelbaum I, Walker AR, Balu R, Gregurick SK, Plusquellic DF. J Am Chem Soc. 2006;128:5764–5775. doi: 10.1021/ja058176u. [DOI] [PubMed] [Google Scholar]
- 8.Zanotti JM, Bellissent-Funel MC, Parello J. Biophys J. 1999;76:2390–2411. doi: 10.1016/S0006-3495(99)77395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochstrasser RM. Nature. 2005;434:570–571. doi: 10.1038/434570a. [DOI] [PubMed] [Google Scholar]
- 10.Zhong DP, Pal SK, Zhang DQ, Chan SI, Zewail AH. Proc Natl Acad Sci USA. 2002;99:13–18. doi: 10.1073/pnas.012582399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heugen U, Schwaab G, Bründermann E, Heyden M, Yu X, Leitner DM, Havenith M. Proc Natl Acad Sci USA. 2006;103:12301–12306. doi: 10.1073/pnas.0604897103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rösgen J, Pettitt BM, Bolen DW. Biophys J. 2005;89:2988–2997. doi: 10.1529/biophysj.105.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beard MC, Turner GM, Schmuttenmaer CA. J Phys Chem B. 2002;106:7146–7159. [Google Scholar]
- 14.Bergner A, Heugen U, Bründermann E, Schwaab G, Havenith M, Chamberlin DR, Haller EE. Rev Sci Inst. 2005;76 063110(1)–063110(5) [Google Scholar]
- 15.Xu J, Plaxco KW, Allen SJ. Prot Sci. 2006;15:1175–1181. doi: 10.1110/ps.062073506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang WY, Gruebele M. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 17.Dumont C, Matsumura Y, Kim SJ, Li J, Kondrashkina E, Kihara H, Gruebele M. Protein Sci. 2006;5:2596–2604. doi: 10.1110/ps.062257406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Gunsteren WF, Daura X, Mark AE. Encyclopaedia of Computational Chemistry. Vol. 2. London: Wiley; 1998. pp. 1211–1216. [Google Scholar]
- 19.Rønne C, Thrane L, Åstrand P-O, Wallqvist A, Mikkelsen KV, Keiding SR. J Chem Phys. 1997;107:5319–5331. [Google Scholar]
- 20.Fischer H, Polikarpov I, Craievich AF. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weingärtner H. Ullmann′s Encyclopedia of Industrial and Technical Chemistry. Vol A28. Wiley-VCH: Weinheim; 1985–1996. pp. 4–19. [Google Scholar]
- 22.Makarov VA, Feig M, Andrews BK, Pettitt BM. Biophys J. 1998;75:150–158. doi: 10.1016/S0006-3495(98)77502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitner DM, Havenith M, Gruebele M. Int Rev Phys Chem. 2006;25:553–582. [Google Scholar]
- 24.Svergun D, Richard S, Koch MHJ, Sayers Z, Kuprin S, Zaccai G. Proc Natl Acad Sci USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merzel F, Smith JC. Proc Natl Acad Sci USA. 2002;99:5378–5383. doi: 10.1073/pnas.082335099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.