Abstract
The Brca1 A complex contains Brca1/Bard1, Abraxas, Rap80, and Brcc36; however, with the exception of the Brca1–Abraxas interaction, how the A complex is assembled is not known. The A complex is localized to sites of DNA damage through the UIM domains of RAP80, which bind K63-linked polyubiquitin chains. In this study, we identified an FHA domain RING finger E3 ubiquitin ligase, RNF8, and an E2-conjugating enzyme known to form K63–polyubiquitin chains, Ubc13, each of which is required to recruit the Brca1 A complex to sites of DNA damage. Rnf8 localizes to sites of DNA damage through an FHA-domain-containing region. We found that Rap80 contains an Abraxas interaction domain [AIR (Abraxas-interacting region)], required for association of Rap80 with Abraxas, Brca1, and Brcc36. Abraxas and Brcc36 associate through coiled-coil domains on each protein. These data suggest a model through which Ubc13 and Rnf8 are recruited to sites of DNA damage through DNA-damage-induced phosphorylation of a chromatin-associated protein and generate polyubiquitin chains that then recruit Rap80 and the entire Brca1 A complex to DNA-damage foci. This sequential E3 ubiquitin ligase recruitment constitutes a ubiquitin ligase cascade required for DNA repair and checkpoint signaling.
Keywords: breast cancer, ubiquitination, Mdc1, 53BP1, K63
The Brca1 tumor suppressor is associated with hereditary breast and ovarian cancer and plays critical roles in DNA repair, cell cycle checkpoint control, and maintenance of genomic stability (1, 2). Brca1 contains two C-terminal BRCT repeats and a N-terminal RING domain. Bard1 forms a heterodimeric complex with Brca1 that exhibits E3 ubiquitin ligase activity (3–6). The Brca1 BRCT repeats constitutes a phosphopeptide-recognition domain that binds peptides containing a pSxxF motif (7–9). We and others have found that Brca1 forms at least three distinct complexes (the Brca1 A, B, and C complexes) by binding different adaptor proteins through their pSPxF motifs (10–12). The adaptor proteins are Abraxas (Abra1) for the A complex, Bach1/Brip1 for the B complex, and CTIP for the C complex, and each complex forms in a mutually exclusive manner, which is consistent with the fact that they occupy the same physical location on Brca1. In addition to Abraxas, the Brca1 A complex contains Rap80. A deubiquitinating enzyme, Brcc36, has been shown to be present in the Rap80 and Brca1 complex (11, 13). Thus, Brcc36 is also likely to be part of the Brca1 A complex. How Rap80, Abraxas, and Brcc36 assemble into the Brca1 A complex has not been resolved.
Rap80 contains two UIM domains that have been shown to bind K63-linked ubiquitin chains (11). The UIM domains of Rap80 are required for the localization of Rap80 and Abraxas to IR-induced foci (IRIFs) (10, 11, 14, 15). The fact that the UIMs of Rap80 bind to K63-linked polyubiquitin chains implies the existence of an upstream E3 ubiquitin ligase that should play a critical role in the DNA-damage response. An E2 ubiquitin-conjugating enzyme, Ubc13, has been implicated previously in Brca1 foci formation (16). Ubc13 forms a complex with an E2-related protein, Mms2, and is capable of catalyzing the formation of K63-linked polyubiquitin chains (17) and is a good candidate for the E2 enzyme that collaborates with the E3 ubiquitin ligase involved in Rap80 and, hence, BRCA1 A complex regulation. In this study, we found that Ubc13 and a Ubc13-associated E3 ubiquitin ligase, Rnf8, are required for Rap80 and Abraxas incorporation into IRIFs. Rap80 acts as a bridge to K63-linked polyubiquitin chains and, through its interaction with Abraxas, localizes Brca1 and Brcc36 to IRIFs.
Results
Ubc13 Acts with Rnf8 to Control Recruitment of the Brca1 A Complex to DNA-Damage Foci.
Ubc13, which forms a complex with a related E2-like protein Mms2, is so far the only known E2-conjugating enzyme that is capable of Lys-63 (K63)-linked ubiquitin-chain formation. The UIM domains of Rap80 have been shown to bind Lys-63-linked polyubiquitin chains (11). Because Ubc13 has been implicated in DNA-damage-induced ubiquitin foci formation and Brca1 foci formation (16), we examined whether Ubc13 is required for the recruitment of the Rap80–Abraxas–Brca1 A complex to the sites of DNA damage. By using siRNAs against Ubc13 to deplete Ubc13 in U2OS cells, we found that Ubc13 is required for Rap80, Abraxas, and 53BP1 IRIF formation, as well as ubiquitin foci formation (Fig. 1A). Therefore, the Rap80–Abraxas complex is involved in mediating Ubc13-initiated ubiquitin signaling to recruit the Brca1–Bard1 complex to DNA-damage sites.
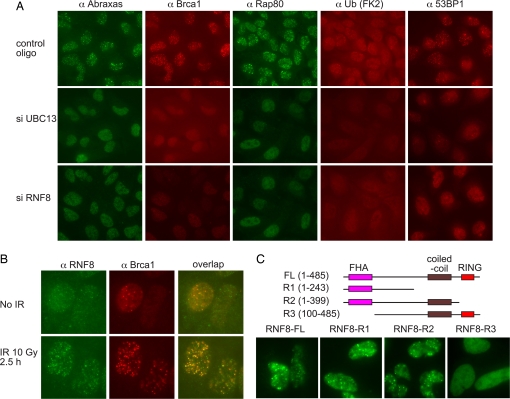
Fig. 1.
UBC13 and RNF8 are responsible for DNA-damage-induced foci formation of the Brca1 A complex. (A) U2OS cells were transfected with control or siRNA oligos against UBC13 or RNF8 for 48 h, irradiated with 10 Gy, and incubated at 37°C for 2 h before being fixed and immunostained with antibodies against Rap80, Abraxas, ubiquitin (FK2), 53BP1, and Brca1. Three different siRNA oligos against each gene were used, and similar results were obtained. (B) U2OS cells were untreated or treated with 10 Gy IR. Two hours later, cells were fixed and immunostained with antibodies against Rnf8 and Brca1. (C) Mutants of RNF8 were generated as a GFP-SV40 NLS N-terminal Rnf8 fusion protein. Plasmids expressing these deletion mutants were transiently expressed in U2OS cells for 48 h. Cells were then irradiated at 10 Gy and incubated for 2 h before being fixed and stained with antibodies against GFP.
A role for Ubc13- and K63-linked ubiquitin chains in Rap80 and Brca1 A complex IRIF formation implies the existence of an E3 ubiquitin ligase involved in this process. Presumably, that ligase might work together with Ubc13. Thus, to search for the putative E3 ligase involved in K63-linked ubiquitin signaling to Brca1, we examined several E3 ligases that have been shown previously to interact with Ubc13 (18) for roles in Rap80 IRIF formation by using siRNAs. Of these, we found that only Rnf8-depleted cells were compromised for Rap80, Abraxas, Brca1, 53BP1, and ubiquitin foci formation (Fig. 1A). The other Ubc13-interacting E3 ligases, KIAA0675 (Dzip3), Chfr, and Znrf2, did not affect Rap80–Abraxas–Brca1 foci formation [supporting information (SI) Fig. 6].
If Rnf8 catalyzes ubiquitination of a protein on chromatin at sites of DNA damage to directly recruit Rap80, then Rnf8 itself might be expected to localize to sites of DNA damage. Therefore we examined Rnf8 localization after DNA damage. We found that Rnf8 forms foci in response to IR (Fig. 1B), suggesting that its ubiquitination target is also localized to sites of DNA damage and is therefore likely to be involved directly in Rap80 recruitment. Furthermore, Rfn8 foci largely colocalize with Brca1 (Fig. 1B).
The protein structure of Rnf8 reveals an FHA domain at the N terminus and a RING domain at the C terminus. FHA domains are phospho-amino acid-interacting motifs. To determine which of the conserved portions of Rnf8 are required for IRIF formation, we generated a deletion series that eliminate the functional domains of Rnf8. We found that a region of Rnf8 containing the FHA domain is itself able to form IRIF, indicating that the FHA domain of Rap80 could localize Rnf8 to IRIF possibly through binding to phosphorylated proteins in response to IR (Fig. 1C). Deletion of the FHA domain blocks Rnf8 foci formation (Fig. 1C).
Optimal Recruitment of Rap80 to Foci Requires both the UIM Domains and Abraxas Interaction Domain of Rap80.
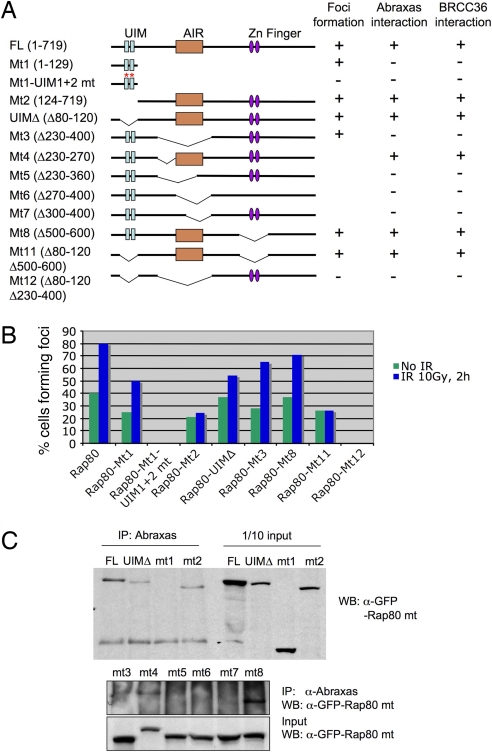
Previously we have shown that UIM domains alone can be recruited to DNA-damage sites, forming IRIF. However, Rap80 mutants that lack the UIM domains also form foci in the absence of exogenous damage (10). To examine the structural requirements for this, we generated a series of retroviral constructs for expression of Rap80 mutant proteins fused to GFP and investigated foci formation and Abraxas interaction for these mutants. We mapped a region of Rap80 (amino acids 270–400) we call “AIR” (Abraxas-interacting region) that is required for association with Abraxas (Fig. 2 A and B). This region is also responsible for the Rap80–Brca1 interaction (ref. 11 and data not shown), confirming that Abraxas mediates the interaction of Rap80 and Brca1. In addition, we found that both the UIM domains and AIR region were required for optimal recruitment of Rap80 to nuclear foci. Various Rap80 mutants that lacked either the UIM domains or the AIR region could still form foci, whereas only the mutant lacking both domains completely failed to form foci (Fig. 2 A and C).
Fig. 2.
Rap80 interacts with Abraxas through the AIR domain, and both UIM and AIR domains of Rap80 are required for optimal Rap80 foci formation. (A) The indicated Rap80 mutants were generated in a GFP–SV40 NLS N-terminal Rap80 fusion construct and used to examine foci formation as well as Abraxas and Brcc36 binding. (B) U2OS cells stably expressing GFP-tagged Rap80 and mutant proteins were either untreated or treated with IR, incubated at 37°C for 2 h, fixed, and stained with antibodies against GFP. At least 400 cells were counted for quantification. Cells that contained >10 foci were counted as positive. (C) 293T cells were transiently transfected with plasmids expressing the GFP-tagged Rap80 fusion proteins in A. After 40 h, cell lysates were prepared and immunoprecipitated (IP) with antibodies against Abraxas. WB indicates Western blot.
Abraxas and Rap80 Depend on Each Other for IRIF Formation.
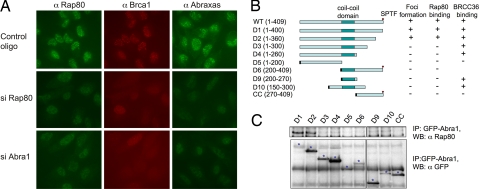
When Rap80 protein levels were depleted in U2OS cells by siRNAs, IR-induced foci formation of Abraxas and Brca1 was significantly decreased (Fig. 3A). We then investigated whether Abraxas affects Rap80 foci formation. We found that in Abraxas siRNA-treated cells, both Rap80 foci formation and Brca1 foci formation were compromised, indicating that Abraxas is also required for Rap80 and Brca1 foci formation (Fig. 3A). This is consistent with the mutational analysis of Rap80 that showed that both the UIM domains and AIR region are required for optimal foci formation of Rap80. We also mapped the region on Abraxas that is required for interaction with Rap80. Most of Abraxas, except for a short C-terminal region involved in Brca1 binding, is required for efficient binding to Rap80, as well as foci formation of Abraxas (Fig. 3 B and C).
Fig. 3.
Rap80 and Abraxas are mutually dependent for DNA-damage-induced foci formation. (A) U2OS cells were transfected with control or siRNA oligos against Rap80 or Abra1 for 48 h, irradiated with 10 Gy, and incubated at 37°C for 2 h before being fixed and immunostained with antibodies against Rap80, Abraxas, and Brca1. Three different siRNA oligos against each gene were used, and similar results were obtained. (B) Abra1 WT and mutants D1–4 were generated in a GFP–N-terminal Abra1 fusion construct, and mutants D5–10 and CC were generated in a GFP–SV40 NLS N-terminal Abra1 fusion construct and used to examine foci formation and Rap80 and Brcc36 binding. The SV40 NLS signal is indicated as a black rectangle. (C) 293T cells were transiently transfected with plasmids expressing cDNAs for GFP-tagged Abraxas and its mutants from B. After 40 h, cell lysates were prepared and immunoprecipitated with antibodies against Rap80. The asterisk indicates the correct size for the expressed fusion proteins.
The Brca1 A Complex Contains Brcc36, Which Is also Required for Rap80, Abraxas, and Brca1 Recruitment to DNA-Damage Sites.
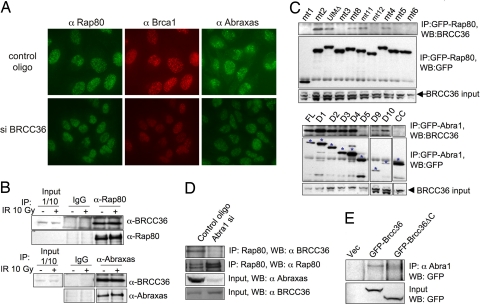
A JAMM domain containing deubiquitinase Brcc36 has been reported to be associated with Rap80 and Brca1 (11, 13). Brcc36 also plays a role in Brca1 foci formation in response to IR (19). We investigated whether Brcc36 is also involved in localizing Rap80–Abraxas to DNA-damage sites. In Brcc36-siRNA treated cells, foci formation of both the Rap80 and Abraxas proteins were significantly decreased (Fig. 4A). In addition, we found that Brcc36 associated with both Abraxas and Rap80 proteins (Fig. 4B). The Rap80 AIR domain was required for the interaction of Rap80 with Brcc36 (Fig. 4C, Upper), indicating that Abraxas is likely to mediate the interaction of Rap80 with Brcc36. Consistent with this interpretation, in Abraxas siRNA-treated cells, the Rap80–Brcc36 interaction was greatly decreased (Fig. 4D).
Fig. 4.
Abraxas mediates the interaction of Brcc36 with Rap80. (A) BRCC36 affects IRIF formation of Rap80, Abraxas, and BRCA1 proteins. U2OS cells were transfected with control oligos or siRNA oligos against BRCC36 for 48 h, irradiated with 10 Gy, and incubated at 37°C for 2 h before being fixed and immunostained with antibodies against Rap80 and Brca1. Three different siRNA oligos against BRCC36 were used, and similar results were obtained (data not shown). (B) Brcc36 associates with Rap80 and Abraxas. Cell lysates from 293T cells treated or not treated with 10 Gy IR were incubated for 2 h and then immunoprecipitated with antibodies against Rap80, Abraxas, or control rabbit IgG. These immunoprecipitates were then fractionated by SDS/PAGE and immunoblotted with anti-Brcc36, Rap80, or Abraxas antibodies. (C) The AIR domain of Rap80 and the coiled-coil domain of Abraxas are required for binding to Brcc36. 293T cells were transiently transfected with plasmid constructs expressing GFP-fused mutant proteins of Rap80 and Abraxas from Figs. 2 and 3. After 40 h, cell lysates were prepared and immunoprecipitated with antibodies against GFP. Immunoblots were carried out with antibodies against Brcc36 or GFP. The asterisk indicates the GFP fusion proteins. (D) The Brcc36 and Rap80 interaction is decreased in Abra1 siRNA-treated cells. 293T cells were transfected with control oligos or siRNA oligos against Abra1 for 48 h; cells lysates were then prepared and immunoprecipitated with antibodies against Rap80. The immunoprecipitates were then fractionated in SDS/PAGE gel and blotted with antibodies against Brcc36 or Rap80. The input protein level was also measured for Brcc36 and Abraxas by using WB. (E) The coiled-coil domain of Brcc36 is required for binding to Abraxas. Plasmid constructs expressing control empty vector or fusion proteins GFP–Brcc36 (1–316 aa) or GFP–Brcc36ΔC (1–249 aa) were transiently transfected into 293T cells for 36 h. Cell lysates were prepared and immunoprecipitated with antibodies against Abraxas and immunoblotted with antibodies against GFP.
Through examination of the deletions series of Abraxas, we mapped the region that interacts with Brcc36 to the coiled-coil domain of Abraxas (Fig. 4C, Lower). A likely candidate region on Brcc36 that might interact with Abraxas is the C-terminal coiled-coil domain of Brcc36, because coiled-coil domains are known to interact. Therefore, we deleted the coiled-coil domain of Brcc36 and tested this mutant for interaction with Abraxas. The coiled-coil domain of Brcc36 was required for interaction with Abraxas (Fig. 4E). Therefore, Abraxas and Brcc36 are likely to heterodimerize through their coiled-coil domains.
Discussion
The elucidation of the function of Brca1 in the DNA-damage response is critical to understanding its role in cancer. We and others have shown that Brca1 exists in at least three complexes, the A, B, and C complexes (10–12). In this study, we explored the structure of the Brca1 A complex containing Rap80, Abraxas, Brca1/Bard, and Brcc36 and its ability to localize to sites of DNA damage. The two UIM motifs on Rap80 are required for foci formation of Brca1 and Abraxas (10, 11, 14). It has also been shown that the UIM domains of Rap80 bind to ubiquitin chains assembled through K63 linkages (11). Here we found that a K63 E2 ubiquitin-conjugating enzyme, Ubc13, was required for Rap80 and Abraxas to form foci in response to DNA damage. Ubc13 has been implicated previously in the control of homologous recombination and Brca1 function (16). Thus, Ubc13 is likely to act upstream of Rap80 in foci formation of the Brca1 A complex.
A key question concerns the nature of the E3 ubiquitin ligase that works with Ubc13 in the DNA-damage response. A number of RING E3 ligase proteins have been found to bind Ubc13 in two-hybrid experiments (18). Screening those candidates revealed that one, Rnf8, was required for Rap80, Abraxas, and Brca1 foci formation in response to DNA damage. Rnf8 was also required for the formation of ubiquitin foci on DNA in response to DNA damage. The role of Rnf8 in IRIF formation is likely to be direct because we found that Rnf8 also formed IRIF. Rnf8 contains a FHA phosphoprotein-binding domain, a coiled-coil domain, and a RING finger. The FHA domain is found in a number of proteins involved in the DNA-damage response, and the FHA domain of Rnf8 is necessary and sufficient to localize Rnf8 to IRIF.
We identified an Abraxas-interacting domain (AIR) on Rap80 between amino acids 270 and 400 and a Rap80-binding region spanning the N-terminal two thirds of Abraxas that overlaps with the ABR domain of Abraxas (10). Rap80 has two modes through which to form foci in cells. The first mode uses the UIM domains and localizes to IRIFs. The second mode uses the AIR domain. Whereas a Rap80 mutant lacking AIR still forms foci, a double mutant of the UIM and AIR domains completely abolished foci formation, implying a role for Abraxas in Rap80 foci formation. Furthermore, we found that siRNAs to Abraxas significantly reduced Rap80 foci in response to IR to levels below that of the Mt1 Rap80 mutant that contains only the UIM domains and does not bind to Abraxas (data not shown). This could be explained if Abraxas had a second role independent of Rap80 binding in Rap80 foci formation. Alternatively, the foci-forming ability of the Rap80 UIM domains might be inhibited in the context of the full-length Rap80 protein unless Abraxas binds and activates it. Nevertheless, contrary to two published reports (20, 21), we saw a role for Abraxas in Rap80 foci formation.
Brcc36 has been found to be associated with Rap80 and Brca1 (11, 13) and also has been suggested to be a positive regulator of Brca1/Bard1 E3 ligase activity (13). We found that it also associates with Abraxas and is therefore a component of the Brca1 A complex. Brcc36 forms a complex with Rap80 only when Rap80 is capable of binding Abraxas. Thus, it associates with Rap80 through Abraxas. Consistent with this, Brcc36 forms a complex with Abraxas only when the coiled-coil domains of Abraxas and Brcc36 are intact. Thus we conclude the topology of the Brca1 A complex is that Rap80 associates with Abraxas and that Abraxas, as an adaptor protein, binds directly to Brca1/Bard1 complex through its C-terminal pSPVF motif and associates with Brcc36 through its coil-coil domain.
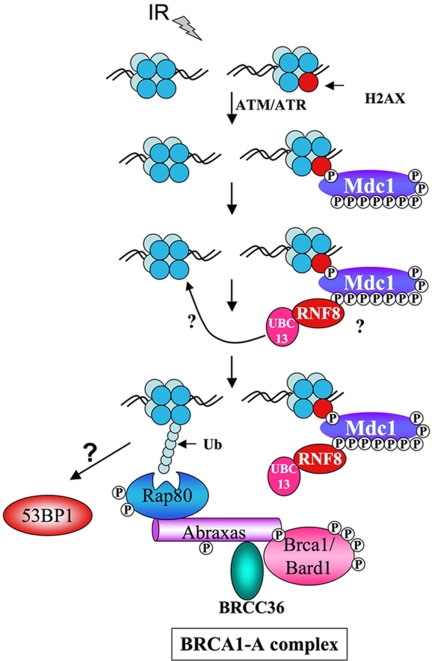
These data led us to the following model (Fig. 5) for IRIF formation of the Brca1 A complex. In response to DNA damage, ATM and ATR phosphorylates H2AX on Ser-139, which serves to recruit the Mdc1 protein to chromatin, where it is also phosphorylated (22–24). In an H2AX- and Mdc1-dependent manner (data not shown), Rnf8/Ubc13 complexes go to sites of DNA damage through their FHA domain and initiate the synthesis of K63 polyubiquitin chains on chromatin that recruit the Brca1 A complex through the UIM domains of Rap80.
Fig. 5.
A model describing the role of Ubc13 and Rnf8 in the generation of ubiquitin chains that recruit the Brca1 A complex to sites of DNA damage. See the text for details. Question marks indicate uncertainty in the target of the Rnf8 FHA domain and the K63 polyubiquitin chain receptor responsible for Rap80 binding.
The identity of the DNA-damage-induced phosphoprotein at the sites of DNA damage that is recognized by the FHA domain of Rnf8 for recruitment remains to be established. Two known phosphoprotein candidates exist that have been genetically linked to Brca1 foci formation, H2AX and Mdc1. Mdc1 has been shown additionally to be required for Rap80 foci (11, 14). Both proteins are phosphorylated by ATM, ATR, and possibly DNA PK in response to DNA damage, and both are required for Brca1 foci (22, 25, 26). We found that Mdc1 is phosphorylated on nine SQ/TQ sites in response to DNA damage and that these sites could serve as docking sites for Rnf8 (27).
The nature of the ubiquitin receptor is not yet known, but components of chromatin such as H2AX or other histones are good candidates. H2A is a likely candidate because it was recently found to become ubiquitinated in response to DNA damage in a Ubc13-dependent manner (28). A ubiquitinated target at the site of damage then localizes the Brca1 A complex to the sites of DNA damage, where Brca1 can then initiate further ubiquitination of proteins to facilitate repair, establishing a ubiquitin ligases cascade. Additional key questions remain, such as the role for the Dub Brcc36 in DNA damage foci formation of the Brca1 A complex, the nature of the ubiquitin acceptor for Rnf8 on chromatin, and the interplay between Rnf8 and Brca1 in formation of ubiquitin foci at sites of DNA damage. Nonetheless, the role of the Brca1 A complex assembly and regulation is becoming clearer.
Materials and Methods
Plasmids and siRNAs.
The cDNA fragments encoding various mutants of RAP80, ABRA1, RNF8, or BRCC36 were cloned into Gateway-compatible entry vectors and transferred into a MSCV vector carrying a GFP tag at the N terminus, followed or not followed by a fragment encoding a SV40 NLS sequence (a gift from J. Jin, Harvard University Medical School) by using a LR recombination kit (Invitrogen).
siRNAs used in the experiments were purchased from Invitrogen: Stealth RNAi Negative Control Med GC (12935-300), Stealth RNAi for Abra1 (HSS130669, HSS130670, HSS130671) (CGUUUAGAGAGAGGCUGCUUCACAA, CCAAGUAUAAUAACAGAAAGCUGCU, ACUGUAUCAGGUUCCUGUAUGUCCA), Rap80 (HSS122566, HSS122567, HSS122568) (UUUAAUUGAGCUUUCCUGGAAAUCC, UUGUGAAGCAGGUACAGAGUUUCCC, GCGUAGACUUGAGGAUUGCAUUCAUU), RNF8 (HSS113323, HSS113324, HSS113325) (GGGUUUGGAGAUAGCCCAAGGAGAA, GCAGCAAGAAGGACUUUGAAGCAAU, GGAGAAUGCG G A GUAUGAAUAUGAA), Ubc13 (HSS144400, HSS144401, HSS144402) (UUCUGGAAGGAAUAGUUCAAGUUUA, UUCCCAACUUGUCUACAUUAGGAUG, AUUGGGAGCACUUAACAAGGCCUGG), BRCC36 (HSS128338, HSS128339, HSS128340) (GCCGUCAGAAUUGUUCACAUUCAUU, GAGAGAAUCGAAAUCCCAAUCCAUA, GCGUAUAGGAGGAUCCACAGCCUUA), ZnRF2 (HSS137665, HSS137666, HSS137667) (GGAUUUGCAUCUUGUAAUGUGUUUA, GCAUAGAUGAAUGGUUUGAAGUAAA, AAGGCAGUCGUGCUAUAGUAUCUCC), DZIP3 (HSS114461, HSS114462, HSS114463), (GCAGAGCUUUAACAGCCGAGGUGUA, CCCUCCAUCAUGAAUUGGGAGAGAA, GGAAAUUGAAGGAUGCUUAUGGAAA), Chfr (HSS124930, HSS124931, HSS124932) (GCAGUGAAGAAGAUGUGCAAAGUAU, GGCAAGUGAUGAAGUCUCCAGCUUU, CCCAGACAGAAAGACUGCGUCCUUU). Cells were transfected with siRNAs (100 nM) by using Oligofectamine (Invitrogen) according to the protocol of the manufacturer.
Cell Lysis and Immunoprecipitation.
Cells were lysed in NETN buffer [50 mM Tris·HCl (pH 8.0), 0.15 M NaCl, 1 mM EDTA, 0.5% Nonidet P-40] with protease inhibitors (Roche) and protein phosphatase inhibitors, 10 mM NaF, and 50 mM β-glycerophosphate. Immunoprecipitations were carried out in the same buffer with appropriate antibodies for 3 h to overnight at 4°C. The antibodies used in the experiments were anti-Brca1 D9 (Santa Cruz Biotechnology), anti-Rap80 (Bethyl), anti-GFP (Invitrogen), anti-BRCC36 (Invitrogen), anti-RNF8 (Abcam), anti-Abraxas [as previously described elsewhere (10)], and anti-Ub (FK2) (BioMol International).
Cell Culture.
U2OS cells were grown in McCoy's 5A medium supplemented with 10% FBS. 293T and HeLaHeLa cells were grown in DMEM supplemented with 10% FBS.
Immunofluorescence.
All cells were cultivated at 37°C in a humidified incubator with 5% CO2. Cells were fixed with 3% paraformaldehyde/2% sucrose for 10 min, permeabilized with 0.5% Triton X-100 solution, and then immunostained with primary antibodies against various proteins and the appropriate Alexa 488-conjugated (green; Molecular Probes) and Cy3-conjugated (red; Amersham Biosciences) secondary antibodies. Images were taken with a Zeiss microscope.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Jianping Jin (Harvard University Medical School) for the MSCV–N-GFP gateway expression construct and Dr. Mark Vidal (Dana–Farber Cancer Institute, Boston) for ORfeome clones for RNF8 and BRCC36. This work was supported by Children's Memorial Research Center and National Institutes of Health grants (to S.J.E.). B.W. is a recipient of National Cancer Institute Howard Temin Award 1KO1, CA116275-01. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 20645.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710061104/DC1.
References
- 1.Venkitaraman AR. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 2.Narod SA, Foulkes WD. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 3.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Proc Natl Acad Sci USA. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer R, Ludwig T. Curr Opin Genet Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 5.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 6.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 7.Manke IA, Lowery DM, Nguyen A, Yaffe MB. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Chini CC, He M, Mer G, Chen J. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M, Yu X, Chen J, Songyang Z. J Biol Chem. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Chen J. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R. Mol Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Chen J, Yu X. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 15.Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann RM, Pickart CM. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 18.Plans V, Scheper J, Soler M, Loukili N, Okano Y, Thomson TM. J Cell Biochem. 2006;97:572–582. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK. Cancer Res. 2006;66:5039–5046. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- 20.Kim H, Huang J, Chen J. Nat Struct Mol Biol. 2007;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Wu J, Yu X. Nat Struct Mol Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 22.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 23.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 25.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, et al. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lou Z, Chini CC, Minter-Dykhouse K, Chen J. J Biol Chem. 2003;278:13599–13602. doi: 10.1074/jbc.C300060200. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 28.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.