Abstract
Bacterial operons for F1Fo-ATP synthase typically include an uncI gene that encodes a function-unknown small hydrophobic protein. When we expressed a hybrid F1Fo (F1 from thermophilic Bacillus PS3 and Na+-translocating Fo from Propionigenium modestum) in Escherchia coli cells, we found that uncI derived from P. modestum was indispensable to produce active enzyme; without uncI, c-subunits in F1Fo existed as monomers but not as functional c11-ring. When uncI was expressed from another plasmid at the same time, active F1Fo with c11-ring was produced. A plasmid containing only uncI and c-subunit gene produced c11-ring, but a plasmid containing only c-subunit gene did not. Direct interaction of UncI protein with c-subunits was suggested from copurification of His-tagged UncI protein and c-subunits, both in the state of c11-ring and c-monomers. Na+ induced dissociation of His-tagged UncI protein from c11-ring but not from c-monomers. These results show that UncI is a chaperone-like protein that assists c11-ring assembly from c-monomers in the membrane.
Keywords: protein folding, membrane protein, Na+ transport
F1Fo-ATP synthase (F1Fo) is located in membranes of mitochondria and chloroplast thylakoid, and cytoplasmic membranes of bacteria, and it couples ATP synthesis/hydrolysis with transmembrane H+ or Na+ translocation (1, 2). F1Fo is composed of two domains, water-soluble F1 that has nucleotide-binding sites for ATP synthesis/hydrolysis, and membrane-integral Fo that mediates H+ translocation across the membrane. In the case of the simplest bacterial F1Fo, subunit compositions of F1 and Fo are α3β3γδε and a1b2c10–15, respectively. In Fo, the c-subunit takes a simple hairpin-like structure composed of two transmembrane helices (3), and multimeric c-subunits assemble into a ring architecture (c-ring), where the number of c-subunits differs in a range of 10–15 depending on the sources (4–7). F1Fo is a rotary motor enzyme in which a rotor composed of γεc10–15 rotates relative to a stator moiety of α3β3δab2 (8, 9). Downhill H+ flow through Fo drives rotation of the c-ring and hence a whole rotor, which induces sequential conformational changes of catalytic sites of F1 domain that result in ATP synthesis (10). In the reverse reaction, ATP hydrolysis at the F1 domain drives rotation of the γ-subunit and hence a whole rotor, which drives pumping H+ across membrane at the Fo domain. Structural studies revealed gearwheel-like architectures of chloroplast c14-ring (7) and Ilyobactor tartaricus c11-ring (11). In I. tartaricus and Propionigenium modestum F1Fos, the c11-ring is highly stable (even in SDS solution), and a strong acid-denaturant, trichloroacetic acid, is necessary to break the ring (12, 13). Although the c11-ring structure is highly stable, it cannot be spontaneously formed from c-subunit monomers; when P. modestum uncE (encoding the c-subunit) was solely expressed in Escherichia coli cells, c11-ring was not formed (14).
In 1981, Gay and Walker (15, 16) determined the structure of E. coli unc operon for F1Fo that contained nine open reading frames (uncIBEFHAGDC) in which uncB, E, F, H, A, G, D, and C encode a-, c-, b-, δ-, α-, γ-, β-, and ε-subunits, respectively. The uncI encodes a 14-kDa hydrophobic protein that is not a component of F1Fo, possibly corresponding to the unidentified protein synthesized by in vitro transcription–translation from a plasmid containing whole unc operon (17). The authors speculated that it must be a “pilot protein” necessary for F1Fo assembly. Later analysis confirmed that the unc promoter is located in front of uncI (18, 19), and therefore, uncI is indeed a member of the unc operon. However, uncI gene product protein (UncI) was not found in E. coli cells under the normal growth condition, and disruption of the uncI gene by insertions (20) or deletion (21) caused no significant effect on the functions of F1Fo, although growth yield was lowered. The UncI protein was detected later in E. coli minicells by using strong expression vectors (19, 22). Immunoblot analysis using anti-UncI antibody revealed the presence of UncI in preparations of Fo and F1Fo, although the amounts were far less than stoichiometric level (23, 24). Consistent with its high hydrophobicity predicted from the nucleotide sequence, UncI was purified by chloroform/methanol extraction (22). Despite of the accumulated information on UncI, its role remains unclear.
Although F1Fo from most organisms is a highly specific H+ pump, in some bacteria such as P. modestum, it operates on Na+ (25). In an attempt to make a hybrid F1Fo (F1 from Bacillus PS3 and Fo from P. modestum) in E. coli cells, we found the essential role of uncI to produce active hybrid F1Fo. The results show that UncI is a molecular chaperone-like protein that assists the c-ring assembly.
Results
Hybrid F1Fo Was Expressed in E. coli.
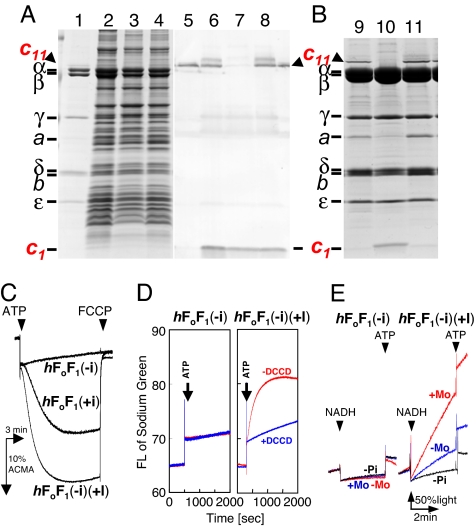
Both P. modestum and thermophilic Bacillus PS3 have a typical bacterial unc operon with nine genes in the order of uncIBEFHAGDC (26, 27). The hybrid F1Fo, composed of Bacillus PS3 F1 and P. modestum Fo, was generated by connecting P. modestum uncIBEF′ and Bacillus PS3 uncF′HAGDC. In bacterial F1Fos, the b-subunit has four domains: transmembrane, tether, dimerization, and δ-binding (28). Genetic connection to generate hybrid F1Fo was made at the region coding for the dimerization domain (position 73–74; P. modestum numbering) because this region has been assumed to have no interaction with other subunits. An F1Fo-deficient E. coli strain JM103Δunc was transformed with a pTR-hF1Fo(+i), an expression vector for the hybrid F1Fo. SDS/PAGE analysis showed expression of F1Fo in the membrane fraction of the host E. coli cells (Fig. 1A, lane 2). The expressed F1Fo [termed hF1Fo(+i) hereafter] has a histidine tag at the N terminus of β-subunit and therefore, could be purified to homogeneity by Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography after solubilization with Triton X-100 (Fig. 1 A, lane 1, and B, lane 9). N-terminal sequencing of the bands in the gel confirmed the expression of all F1Fo subunits except the a-subunit. The a-subunit band was identified by immunoblot analysis using anti-a antibody (data not shown) because of the difficulty of N-terminal sequencing of the a-subunit. As reported for P. modestum Fo§ (12), a high-molecular-mass band (≈60 kDa) was observed above the α-subunit band (indicated by c11 and an arrowhead in the figure). N-terminal sequencing gave a c-subunit sequence, and the band disappeared by treating the sample with trichloroacetic acid before electrophoresis (data not shown), confirming that this band corresponded to the c11-ring as in the case of P. modestum F1Fo.
Fig. 1.
Assembly of c11-ring in hybrid F1Fo assisted by uncI expressed in another plasmid and activities of the hybrid F1Fo thus made. Membrane fractions prepared from E. coli cells expressing the indicated plasmids (A) and purified hybrid F1Fos (B) were subjected to SDS/PAGE and stained with CBB (A, lanes 1–4, and B) or immnoblot staining with anti-c-subunit antibody (A, lanes 5–8). Lanes 1, 5, and 9, purified hF1Fo(+i) as a reference. Lanes 2 and 6, pTR-hF1Fo(+i). Lanes 3 and 7, pTR-hF1Fo(−i). Lanes 4 and 8, pTR-hF1Fo(−i) ± pST-I. Lane 10, hF1Fo(−i). Lane 11, hF1Fo(−i)(+I). (A) Membrane proteins (7 μg) were applied to each lane of the gels. (C) H+-pumping activities of PLs containing hybrid F1Fos, hF1Fo(+i), hF1Fo(−i), or hF1Fo(−i)(+I). (D) Na+-pumping activity of PLs containing hF1Fo(−i) and hF1Fo(−i)(+I). (E) ATP synthesis activity of membrane vesicles containing hF1Fo(−i) and hF1Fo(−i)(+I). (C–E) Experimental conditions were the same as in Fig. 2. Fifty percent light in E corresponds to half of the initial luminescence intensity in the cuvette before the addition of NADH.
Hybrid F1Fo Can Use Na+ as a Coupling Ion.
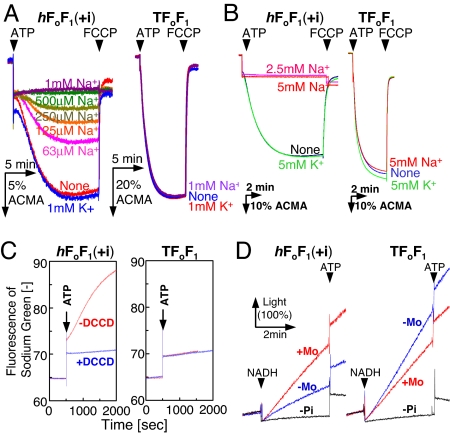
We examined the activities of hF1Fo(+i) in the membrane vesicles prepared from expressing E. coli cells (Fig. 2 A and D) and in the proteoliposomes (PLs) (Fig. 2 B and C). Activities of Bacillus PS3 F1Fo (TF1Fo) were shown as references (Fig. 2 A–D, Right). The hF1Fo(+i) exhibited H+-pumping activity upon addition of ATP in the absence of Na+, but not in the presence of 1 mM NaCl (Fig. 2A, Left). KCl at 1 mM had no effect. Titration of Na+ concentration indicated that approximately half of the H+-pumping activity was blocked by supplementing 0.2 mM NaCl. The inhibition of H+ pumping by Na+ was also observed for the purified hF1Fo(+i) in PLs (Fig. 2B, Left). The apparent inhibition of H+ pumping could be explained by increasing Na+ pumping as Na+ became available. The H+-pumping activity of TF1Fo, on the contrary, was unaffected by the presence of NaCl and KCl (Fig. 2 A and B, Right). Na+-pumping activity was measured directly with a Na+-indicator fluorescent dye, sodium green (Fig. 2C). In the presence of 2.6 mM NaCl, hF1Fo(+i) pumped Na+ upon addition of ATP, and this activity was totally lost by prior incubation with dicyclohexylcarbodiimide (DCCD) (Fig. 2C, Left). Such Na+-pumping activity was not observed for PLs of TF1Fo (Fig. 2C, Right). When the electrochemical potential of H+ was generated by respiratory proton pumps fueled by NADH, hF1Fo(+i) in the membrane vesicles synthesized ATP (Fig. 2D, Left). The vesicles showed the ATP synthesis activity of 3.4 milliunits/mg membrane protein in the presence of 2.5 mM NaCl (only outside the vesicles). The activity was increased 2.1-fold by the addition of a Na+/H+ antiporter, monensin (5 μM), which exchanged H+ inside the vesicle and Na+ outside the vesicles, transforming a significant magnitude of ΔpH into ΔpNa (Fig. 2D, Left). The vesicles containing TF1Fo synthesized ATP at a rate 17.6 milliunits/mg membrane proteins, and this activity decreased to ≈60% by monensin, likely because of the decrease in ΔpH (Fig. 2D, Right). ATP hydrolysis activity of hF1Fo(+i) in PLs was 1.2 units/mg of F1Fo in the absence of Na+. This activity was enhanced 4-fold by the addition of 5 mM NaCl (apparent Km value for Na+ = 0.7 mM at 42°C) as reported (12, 29). These results suggested that hF1Fo(+i) is very similar to the authentic P. modestum F1Fo in its Na+ dependence (29, 30).
Fig. 2.
Activities of the hybrid F1Fo (Left) compared with TF1Fo (Right). (A) H+-pumping activity of inverted E. coli membrane vesicles containing the hybrid F1Fo [hF1Fo(+i)] and TF1Fo. NaCl or KCl was added into the assay mixture at the concentrations indicated. The reaction was initiated by adding 1 mM K+-ATP and terminated with FCCP. %ACMA shows the fluorescent intensity of ACMA, in which the initial intensity before the addition of ATP is calibrated as 100%. (B) H+-pumping activity of PLs containing hF1Fo(+i) and TF1Fo. The assay conditions were the same as A. (C) Na+-pumping activity of PLs containing hF1Fo(+i) and TF1Fo. As indicated, PLs were reacted with 50 μM DCCD for 1 h and used for the measurement. The reaction was initiated by the addition of 1.3 mM Na+2-ATP, and the increase in Na+ concentration inside the PLs was monitored with an increase in sodium green fluorescence at 540 nm. (D) ATP synthesis activity of membrane vesicles containing hF1Fo(+i) and TF1Fo. Electrochemical potential of H+ was generated by respiratory H+ pumps in the membrane vesicles driven by NADH oxidation. ATP synthesis was monitored in real time at 35°C by luciferase reaction at 560 nm. As indicated, a Na+/H+ antiporter, monensin (5 μM) was added to convert some amount of H+ gradient into Na+ gradient. Traces indicated by “-Pi” represent negative controls without addition of Pi. Light (100%) in D corresponds to the initial luminescence intensity in the cuvette before the addition of NADH.
uncI Is Necessary for c11-Ring Formation in Hybrid F1Fo.
To determine the function of uncI, we removed the uncI gene from the plasmid pTR-hF1Fo(+i) and generated an expression plasmid pTR-hF1Fo(−i). Also, we generated an expression plasmid pST-I that contained only uncI gene. Membrane fractions of E. coli cells transformed by pTR-hF1Fo(+i), by pTR-hF1Fo(−i), and by pTR-hF1Fo(−i) ± pST-I were analyzed with SDS/PAGE (Fig. 1A). They all produced F1Fos as demonstrated by appearance of the bands of α- and β-subunits in SDS/PAGE of membrane fractions (lanes 2–4). The immunoblot analysis by anti-c antibody revealed that c11-ring was present in membrane fractions from cells expressing pTR-hF1Fo(+i) (lane 6) and pTR-hF1Fo(−i) + pST-I (lane 8) but not in those from cells expressing pTR-hF1Fo(−i) (lane 7), whereas c-monomer existed in all cases. The hybrid F1Fos were purified to homogeneity and analyzed with SDS/PAGE. In SDS/PAGE analysis (Fig. 1B), the band of c-subunit appeared at the position of monomer in the case of F1Fo produced from pTR-hF1Fo(−i) [termed hF1Fo(−i)] (lane 10) but at the position of the c11-ring in hF1Fo(+i) (lane 9) and in F1Fo produced from pTR-hF1Fo(−i) ± pST-I [termed hF1Fo(−i)(+I)] (lane 11). Because F1Fos had a histidine tag at N termini of β-subunits and were purified by Ni-NTA affinity chromatography, c-subunits running as a monomer in SDS/PAGE should be components of the hF1Fo(−i). Although it is not known whether these c-subunits exist as monomers or as loosely associated oligomers in hF1Fo(−i), we call them “monomers” hereafter for simplicity. Comparison of immunoblot analysis (Fig. 1A, lanes 6 and 8) with protein staining of the purified F1Fos (Fig. 1B, lanes 9 and 11) suggested that c-monomers in membrane fractions of cells expressing pTR-hF1Fo(+i) and those expressing pTR-hF1Fo(−i) + pST-I would be free, which indicates that they were not incorporated into F1Fo complex and were removed during purification procedures. It was noticed that hF1Fo(−i) lost a significant amount of a-subunit (Fig. 1B, lane 10). These results clearly indicate that uncI, either in the same unc operon or in a different transcript, is necessary for assembly of the c11-ring. It should be noted that genomic DNA of the JM103Δunc strain lacks all of the structural genes for F1Fo but has an uncI gene of its own. However, it is clear from the results described above that the E. coli uncI gene cannot complement the P. modestum uncI gene.
Hybrid F1Fo Containing No c11-Ring Is Inactive.
The purified F1Fos were incorporated into liposomes, and activities were measured. hF1Fo(−i) was inactive in H+ pumping (Fig. 1C), Na+ pumping (Fig. 1D), and ATP synthesis (Fig. 1E). In contrast, hF1Fo(−i)(+I) showed good activities in all measurements. Actually, its H+-pumping activity was higher than that of hF1Fo(+i) (Fig. 1C). Na+-pumping activity of hF1Fo(−i)(+I) was sensitive to DCCD inactivation, and ATP synthesis was activated up to 5.1 milliunits/mg membrane proteins by monensin (Fig. 1 D and E).
UncI Can Assist c11-Ring Assembly Independent of Other Subunits of F1Fo.
Three kinds of plasmids were made: pTR-c for expression of c-subunit, pST-IHis for expression of UncIHis protein with a His tag at the C terminus, and pTR-IHisc for expression of UncIHis protein and c-subunit (Fig. 3). Membrane fractions from cells expressing pTR-IHisc contained c11-ring (lane 2), whereas those from cells harboring pTR-c contained no c11-ring (lane 3). The c-monomer band was also faint in lane 3. It seems that in the absence of interactions with UncI and other subunits of F1Fo, newly synthesized c-subunit is unstable. As expected from the result of hF1Fo(−i)(+I), c11-ring was formed even when a gene for c-subunit and the uncI gene were on the different plasmids (pTR-c and pST-IHis), although the amount was relatively small (lane 4). These results suggest that the UncI protein can assist assembly of c-monomers into c11-ring without the involvement of other subunits of F1Fo.
Fig. 3.
c11-ring formation assisted by uncI without involvement of other subunits of F1Fo. SDS/PAGE of membrane fractions prepared from cells expressing pTR-IHisc (lane 2), pTR-c (lane 3), and pTR-c + pST-IHis (lane 4). Lane 1, purified hF1Fo(+i) as a reference. Membrane proteins (7 μg) were applied to each lane of the gels. Bands of c11-ring and c-monomer are indicated with arrowheads. It should be noted that expression of UncI from pST-IHis is weaker than that from pTR-IHisc because of a weaker transcription promoter.
UncI Protein Interacts Directly with c-Subunit.
The above results suggest a possibility of interaction of UncI protein with c subunit. E. coli cells expressing pTR-IHisc contained c11-ring (Fig. 4A, lane 2) as shown above, which was solubilized by Triton X-100 (Fig. 4A, lane 3). Solubilized proteins were applied to a Ni-NTA column, and, after washing the column, the proteins retained in the column were eluted by the high-imidazole buffer. The eluate contained several protein bands in SDS/PAGE (Fig. 4A, lane 4). From N-terminal amino acid sequencing, a 15-kDa protein band was identified as UncIHis and two bands below it as proteolysed products of UncIHis. Bands at ≈60 kDa and at 7 kDa gave the sequence of c-subunit, and treatment with 10% trichloroacetic acid converted the ≈60-kDa band to the 7-kDa band (Fig. 4A, lane 5). Therefore, they certainly corresponded to c11-ring and c-monomer. The fact that the c11-ring and c-monomer were copurified with UncIHis substantiates direct interaction between UncI and c-subunit, either in the state of c11-ring or c-monomer. UncI assists the ring assembly of c-subunits through a direct protein–protein interaction.
Fig. 4.
Association of UncI protein with c-subunit and release of c11-ring from UncI by Na+. (A) SDS/PAGE analysis of UncIHis-bound components. Lane 1, hF1Fo(+i). Membrane fractions of cells expressing pTR-IHisc (lane 2) were solubilized with Triton X-100. Solubilized proteins were isolated by ultracentrifugation (lane 3) and loaded on a Ni-NTA column. After washing the column, the proteins were eluted (lane 4), and the eluate was further treated with 10% trichloroacetic acid (lane 5). All of the buffers did not contain Na+. nc11, aggregated c11-ring; 2UncIHis, dimer UncIHis; digests, proteolytic products of UncIHis. Proteins on the gel were stained by Coomassie brilliant blue (CBB). (B) Effect of Na+ on the interaction between UncIHis and c-subunit. The eluate from the Ni-NTA column was directly subjected to SDS/PAGE without prior concentration procedures, and the proteins were stained with silver staining. UncIHis was only poorly stained by silver. Lane 6, the same as lane 4. Lane 7, the loaded Ni-NTA column was washed with the Na+-containing washing buffer (containing 100 mM NaCl) and eluted. (C) Detection of UncI in hF1Fo(−i)(+I). Membrane fractions of cells expressing pTR-hF1Fo(−i) ± pST-I were solubilized and loaded on a Ni-NTA column. The column was washed with the (Na+-free) washing buffer (lanes 8 and 10) or with the Na+-containing washing buffer (lanes 9 and 11). The hF1Fo(−i)(+I) was eluted, reacted with tetramethylrhodamine maleimide to label cysteine residues in UncI, and subjected to SDS/PAGE analysis. The proteins were visualized by CBB staining (lanes 8 and 9) or by fluorescence (excited at 302 nm, emission at >400 nm) (lanes 10 and 11). UncI proteins are indicated with arrowheads. Different from the gel in Fig. 1B, the band of UncI was visible even by CBB staining because the fluorescent dye labeling enhanced the sensitivity of UncI to the CBB staining.
Na+ Induces Dissociation of UncI from c11-Ring.
In this work, all experimental procedures, unless otherwise stated, were carried out under the conditions where Na+ was strictly omitted. To know the effect of Na+ on the UncI–c-subunit interaction, the Ni-NTA column, to which Triton X-100-solubilized membrane proteins from cells expressing pTR-IHisc were loaded, was washed with a Na+-containing washing buffer. Then, the proteins were eluted out with the high-imidazole buffer, and the eluate was analyzed directly by SDS/PAGE with silver staining. As seen, the eluate contained only c-monomer (Fig. 4B, lane 7), indicating that c11-ring was already washed out from the column during the wash with the Na+-containing washing buffer. In a control experiment where the column was washed with a (Na+-free) washing buffer, the eluate contained c11-ring and c-monomer (Fig. 4B, lane 6), as observed in the previous experiment (Fig. 4A, lane 4). The band of UncIHis was faint because it was stained only poorly by silver. These results suggest that Na+ induces dissociation of UncI from c11-ring but not from c-monomer.
As shown in Fig. 1B (see lanes 9 and 11), UncI protein was not found in the purified hybrid F1Fo, and it is clear that UncI is not a stable component of the mature enzyme. However, weak, substoichiometric association of UncI to F1Fo has been reported for E. coli F1Fo (23, 24). To analyze such UncI–F1Fo interaction, we took advantage of the fact that hybrid F1Fo does not have cysteine residue, but UncI has three. hF1Fo(−i)(+I) was adsorbed to the Ni-NTA column through the His tag at the β-subunits, and the column was washed with either Na+-free or Na+-containing washing buffer. Then, the bound proteins were eluted by the high-imidazole buffer and analyzed by SDS/PAGE after treatment with a cysteine-reactive fluorescence dye, tetramethylrhodamine maleimide. In the protein-staining gel, the UncI band was seen faintly in Na+-unexposed hF1Fo(−i)(+I) but hardly seen in Na+-exposed hF1Fo(−i)(+I) (Fig. 4C, lanes 8 and 9). Consistently, fluorescence intensity of the UncI band was 1.4 (±0.1)-fold weaker in Na+-exposed hF1Fo(−i)(+I) than in Na+-unexposed hF1Fo(−i)(+I) (Fig. 4C, lanes 10 and 11). Although the amount of associated UncI decreased by Na+, a small amount of UncI still remained on F1Fo preparation.
Discussion
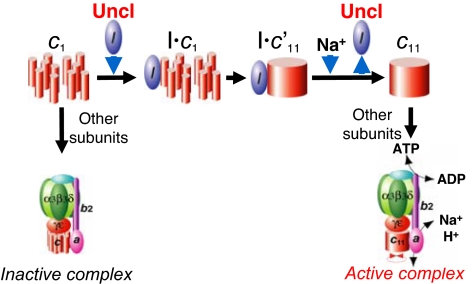
Ever since uncI gene was found 26 years ago (15, 16), its function has been elusive. It occupies the first position in the unc operon and is conserved in most bacteria, indicative of its importance. This work reveals an essential role of UncI protein for the ring assembly of c-subunits of P. modestum Fo (Fig. 5). In mitochondrial F1Fo, some proteins, such as yeast ATP10 (31, 32), ATP23 (33, 34), and OXA1 (35), have been shown to play a chaperone-like role to assist Fo assembly, although the detailed mechanism is not well understood. Although there is no sequence homology to eukaryotic proteins, UncI protein also plays a chaperone-like role; it associates with monomers of c-subunit, assists ring assembly, and dissociates from the assembled c-ring. Because molecular chaperone is not a final component of the mature protein (complex), it should leave its client protein at some stage either by using energy of ATP hydrolysis, by other factor(s), or just by reversible association/dissociation. UncI dissociates from c-ring but not from c-monomers in the presence of Na+ ion, thus “catalyzing” a forward reaction, assembling the ring. Because Na+ is a transporting ion for P. modestum Fo, binding of Na+ to the c-ring would be responsible for this reaction. This work also revealed that c-subunits that fail to form the functional ring can be incorporated into F1Fo complex, although the resultant F1Fo is inactive in coupling reactions. It awaits further study whether this inactive F1Fo is a dead-end complex or can be a precursor of an active F1Fo. Possibly related to this finding, a small amount of UncI protein remains bound in the purified hF1Fo(−i)(+I), especially in the Na+-unexposed F1Fo. This UncI could be associated with a small population of c-ring or of residual c-monomers in the complex. UncI protein has been found in mature E. coli F1Fo (23), and a possibility of interaction between UncI and Fo subunits other than c-subunit (24) is not eliminated by this work.
Fig. 5.
A model of UncI-assisted assembly of c11-ring. UncI binds to monomer (or some unassembled state of) c-subunit(s) in the membranes, assists their assembly into c11-ring, and leaves c11-ring as Na+ binds to c11-ring. Other subunits associate with c11-ring, and active F1Fo is formed. Without UncI, an inactive F1Fo complex is formed. In this complex, c-subunits are not assembled into c11-ring, and some fraction of a-subunit is lost. c′11 indicates an interim state of c11-ring formation.
A proposal of UncI function as an essential factor to assemble c-ring will raise immediate questions: why can an E. coli mutant that lacks uncI gene grow by oxidative phosphorylation, and why is F1Fo isolated from the mutant fully active (20, 21)? It is possible that chaperone-like function of UncI is restricted only in the case of P. modestum Fo. However, nonessential phenotype of E. coli uncI gene might be explained by another chaperone-like protein for membrane proteins in E. coli, YidC. Recent analysis of membrane-integration process of E. coli c-subunit showed that newly translated c-subunit is targeted by the signal recognition particles to the membranes and integrated into membranes by the assist of YidC (36). It was further demonstrated that YidC had an ability to assist the ring formation from c-monomers (37). Therefore, it is likely that YidC has an overlapping function with UncI and can complement the UncI deficiency. Such a role of YidC might be specific to E. coli c-ring and cannot assist P. modestum c11-ring assembly as implicated by this work.
Finally, aside from UncI issue, it is worth pointing out that the same F1 domain derived from a thermophilic Bacillus can dock with c11-ring in the hybrid as well as with c10-ring in the original TF1Fo. An analogous result has been also observed for another hybrid F1Fo, which is composed of E. coli F1 and P. modestum Fo (38). It appears that γ- and ε-subunits of thermophilic Bacillus F1 can manage to bind a c-ring with a slightly larger radius to make a body of rotor apparatus that is robust enough to transmit the large rotary torque between Fo and F1.
Materials and Methods
Expression Vectors for Hybrid F1Fo.
A DNA fragment, containing uncIBE and the 5′ half of uncF, was amplified from P. modestum genomic DNA by PCR. A DNA fragment, containing the 5′ half of uncF and uncHA′ was amplified by PCR from an expression plasmid for Bacillus PS3 F1Fo (TF1Fo), pTR19-ASDS (termed pTR-TF1Fo in this work) (10). The two fragments possess the same nucleotide sequence at the edge (b-subunit region), and therefore, the fragments were connected by PCR amplification without an additional primer. The resulting fragment composed of P. modestum uncIBEF′ and Bacillus PS3 unc′FA′ was digested with EcoRI and KpnI and replaced with the corresponding region of pTRN-ASDS¶ (EcoRI and KpnI sites) to obtain an expression plasmid, pTR-hF1Fo(+i). Also, pTR-hF1Fo(−i), in which uncI gene was removed from pTR-hF1Fo(+i), was made to test the role of uncI.
Preparation of Membrane Vesicles and the Hybrid F1Fo.
The plasmid pTR-hF1Fo(+i) was used for transformation of an F1Fo-deficient E. coli strain, JM103Δ(uncB-uncD) (hereafter, JM103Δunc) (39). The transformants were aerobically grown in 2×YT medium supplemented with 100 μg/ml ampicillin for 21 h. In the case of transformants harboring pSTV28 derivative plasmid (mentioned later), 30 μg/ml chloramphenicol was also supplemented to maintain the plasmid in the cells. Cells (≈30 g) harvested from a 12-liter culture were washed twice with buffer PA3 [10 mM Hepes/KOH buffer (pH 7.5) containing 5 mM MgCl2 and 10% glycerol] and used for the preparation of inverted membrane vesicles as described in ref. 40 except cell disruption by sonication instead of the French press. Membrane vesicles were used for analyses of F1Fo. Purification of the hybrid F1Fo was carried out as follows. The membrane vesicles (≈15 mg of proteins per ml, 16 ml) was washed with 16 ml of 10 mM Hepes/KOH buffer (pH 7.5) and then solubilized in 10 mM Hepes/KOH buffer (pH 7.5) containing 1% Triton X-100 in the presence of a protease inhibitor (Complete EDTA-free; Roche) for 30 min on ice with occasional mixing. After a centrifugation (153,000 × g, for 20 min, at 4°C), the supernatant was diluted 4-fold with the buffer M [20 mM potassium phosphate (KPi)i buffer (pH 7.5) containing 100 mM KCl] supplemented with 20 mM imidazole. The diluted supernatant was mixed with 12 ml of Ni-NTA Superflow (Qiagen) and calmly stirred for 30 min on ice. The resin was poured onto an appropriate column and washed with 3 vol of the buffer M supplemented with 0.05% Triton X-100 and 20 mM imidazole. F1Fo was eluted with the buffer M supplemented with 0.05% Triton X-100 and 200 mM imidazole. F1Fo in the elution was mixed with 50 mM MgCl2 and 2.7% PEG 6000, incubated on ice for 15 min, and collected by ultracentrifugation (153,000 × g, for 20 min, at 4°C). The pellet obtained was dissolved in a small volume of 10 mM Hepes/KOH buffer (pH 7.5) containing 0.05% Triton X-100. After a brief centrifugation, the supernatant was frozen by liquid N2 and stored at −80°C until use. PLs that contained the purified hybrid F1Fo in the membrane were prepared by the freeze–thaw method as used for TF1Fo (10).
Expression Vectors for uncI and c-Subunit.
The uncI and uncE (c-subunit gene) were individually amplified by PCR from pTR-hF1Fo(+i). For Ni-NTA purification of uncI gene product, a nucleotide sequence coding for a His tag (6 residues) was introduced in front of the stop codon of uncI in the PCR step (termed uncIHis). The amplified fragments of uncI and uncIHis were digested with EcoRI and PstI and introduced into the plasmid pSTV28 (Takara) previously digested with both restriction enzymes. The resultant plasmids were named pST-I and pST-IHis. By the same manner, the PCR product of uncE was introduced into pTrc99A (Amersham Pharmacia) to obtain plasmid pTR-c. For simultaneous expression of uncI and uncE, uncI was amplified by PCR, digested with EcoRI and BamHI, and introduced upstream of uncE in pTR-c to obtain pTR-IHisc. Nucleotide sequences of the regions amplified by PCR were verified by sequencing. Procedures for cultivation of JM103Δunc transformed with the plasmids, membrane preparation, and membrane solubilization with Triton X-100 were the same as described above for hybrid F1Fo, but when the A600 reached 1, 2 mM isopropyl-β-thiogalactopyranoside was supplemented to the culture. After that, the cultivation was continued for 3 h. Ni-NTA column chromatography was performed as described for hybrid F1Fo except that (i) the Ni-NTA column was washed with 10 bed-vol of the washing buffer [20 mM KPi buffer (pH 7.5), 100 mM KCl, 30 mM imidazole, and 0.05% Triton X-100] and was eluted with the high-imidazole buffer [20 mM KPi buffer (pH 7.5), 100 mM KCl, 200 mM imidazole, and 0.05% Triton X-100]; (ii) the eluate from the Ni-NTA column was concentrated by a 100-kDa centrifugal concentrator (Ultra YM-100; Amicon) instead of PEG precipitation. In the experiments to test the effect of Na+ on the UncI–c-subunit interaction, the loaded Ni-NTA column was washed with 5 bed-vol of the Na+-containing washing buffer [20 mM KPi buffer (pH 7.5), 10 mM KCl, 100 mM NaCl, 30 mM imidazole, 0.05% Triton X-100] and subsequently with 5 bed-vol of the (Na+-free) washing buffer. Then, the proteins were eluted with the high-imidazole buffer.
Analytical Procedures.
ATPase-driven H+-pumping activity was assayed with quenching of fluorescence of 9-amino-6-chloro-2-methoxyacridine (ACMA) (excitation at 410 nm, emission at 480 nm) at 32°C as described in ref. 10. Inverted membrane vesicles or PLs were added to the assay mixture at a final concentration of 0.2 mg of membrane protein per ml or 20 μg of FoF1 per ml. To assess the sensitivity toward Na+, either NaCl or KCl was supplemented into the assay mixture at the concentration indicated. The reaction was initiated by adding 1 mM K+-ATP and terminated with 0.25 μg/ml carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP). ATPase-driven Na+-pumping activity was measured by the method of von Ballmoos and Dimroth (41) with some modifications as follows. Soybean l-α-phosphatidylcholine (300 mg, type II-S; Sigma) was suspended by a gentle mixing in 10 ml of buffer PA3 supplemented with 30 μg/ml sodium green (Molecular Probes) and sonicated with a large tip for 30 s. Purified hybrid F1Fo or TF1Fo (1 mg) was mixed with 500 μl of the sodium green liposomes, rotated for 5 min at 25°C, frozen with liquid N2, and thawed on a desk. After water bath sonication (45 s), the PLs were subjected to a cartridge gel filtration column NAP5 (Amersham Pharmacia) previously equilibrated with buffer PA3 to remove the outside sodium green. Thus, prepared PLs were used for the measurement. When indicated, PLs were reacted with 50 μM DCCD in buffer PA3 for 1 h at room temperature before use. Na+-pumping activity was measured at 32°C in 50 mM Hepes/KOH buffer (pH 7.5) containing 100 mM KCl, 5 mM MgCl2, 4 mM phosphoenolpyruvate, 0.1 mg/ml pyruvate kinase, and 0.22 ng/ml FCCP in the presence of PLs (final lipid concentration is 3.3 mg/ml). The reaction was initiated by the addition of 1.3 mM Na2-ATP. The increase in Na+ concentration inside the PLs was monitored by an increase in fluorescence of sodium green (excitation at 488 nm and emission at 540 nm).
ATP synthesis activity of the membrane vesicles was measured at 35°C in buffer PA3 supplemented with 2.5 mM KPi, 0.5 mM ADP, 2.5 mM NaCl, membrane vesicles (85 μg and 53 μg of membrane proteins/ml for vesicles containing hybrid F1Fo and TF1Fo, respectively), and 1/10 vol of the CLSII solution containing luciferin/luciferase (ATP bioluminescence kit; Roche). If stated, a Na+/H+ antiporter, monensin, was supplemented to the solution at a final concentration of 5 μM. The reaction was initiated by adding 1.7 mM NADH to generate electrochemical potential gradient of H+ across the membrane. The ATP production was monitored in real time by the light from luciferase reaction at 560 nm. The amounts of ATP synthesized were calibrated with a defined amount of ATP at the end of the measurement. The activity that synthesized 1 μmol of ATP per min was defined as 1 unit. ATPase activities were analyzed in 50 mM Hepes/KOH buffer (pH 7.5) containing 100 mM KCl, 5 mM MgCl2, 0.2 μg/ml FCCP, 0.8 mM K+-ATP, and the ATP-regenerating system (10), and, in the case of membrane vesicles, 2.5 mM KCN to prevent NADH oxidation by E. coli respiratory chain. Average hydrolysis rates in a time period from 3 to 6 min after initiation of the reactions at 42°C were measured. The activity that hydrolyzed 1 μmol of ATP per min was defined as 1 unit. Protein concentrations were determined by BCA protein assay kit from Pierce, with BSA as a standard. Fluorescence labeling of UncI in the F1Fo preparation was performed in 100 mM Mes/KOH (pH 6.5) containing 1% SDS, 1 μg/ml tetramethylrhodamine maleimide (Molecular Probes), and 0.05% Triton X-100 for 30 min at room temperature. All SDS/PAGE in this work used a gradient polyacrylamide gel (10–20%). All data shown in the present study were measured at least in triplicate.
ACKNOWLEDGMENTS.
We thank our colleagues Drs. T. Hisabori, N. Mitome, P. Kahar, and M. Fujikawa for helpful discussion; and Mrs. J. Suzuki, T. Kamita, and A. Tatsuguchi for excellent technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
In a report of P. modestum F1Fo (12), a band of c11-ring in SDS/PAGE was observed just below β-subunit, not above α-subunit as observed here. The difference is a result of the gel used. We used gradient gels (10–20% polyacrylamide) in this work. When gels with constant polyacrylamide concentration were used, the band was observed below β-subunit.
Plasmid pTRN-ASDS is a derivative of pTR19-ASDS (10). In the plasmid, its lac operator regulation system does not function by unknown mutation, and therefore, the gene introduced is strongly expressed without an inducer, isopropyl-β-thiogalactopyranoside.
References
- 1.Boyer PD. J Biol Chem. 2002;277:39045–39061. doi: 10.1074/jbc.X200001200. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida M, Muneyuki E, Hisabori T. Nat Rev Mol Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 3.Girvin ME, Fillingame RH. Biochemistry. 1993;32:12167–12177. doi: 10.1021/bi00096a029. [DOI] [PubMed] [Google Scholar]
- 4.Stock D, Leslie AG, Walker JE. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 5.Mitome N, Suzuki T, Hayashi S, Yoshida M. Proc Natl Acad Sci USA. 2004;101:12159–12164. doi: 10.1073/pnas.0403545101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vonck J, von Nidda TK, Meier T, Matthey U, Mills DJ, Kuhlbrandt W, Dimroth P. J Mol Biol. 2002;321:307–316. doi: 10.1016/s0022-2836(02)00597-1. [DOI] [PubMed] [Google Scholar]
- 7.Seelert H, Poetsch A, Dencher NA, Engel A, Stahlberg H, Muller DJ. Nature. 2000;405:418–419. doi: 10.1038/35013148. [DOI] [PubMed] [Google Scholar]
- 8.Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CA, Graber P. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 9.Ueno H, Suzuki T, Kinosita K, Jr, Yoshida M. Proc Natl Acad Sci USA. 2005;102:1333–1338. doi: 10.1073/pnas.0407857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T, Ueno H, Mitome N, Suzuki J, Yoshida M. J Biol Chem. 2002;277:13281–13285. doi: 10.1074/jbc.M111210200. [DOI] [PubMed] [Google Scholar]
- 11.Stahlberg H, Muller DJ, Suda K, Fotiadis D, Engel A, Meier T, Matthey U, Dimroth P. EMBO Rep. 2001;2:229–233. doi: 10.1093/embo-reports/kve047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laubinger W, Dimroth P. Biochemistry. 1988;27:7531–7537. doi: 10.1021/bi00419a053. [DOI] [PubMed] [Google Scholar]
- 13.Neumann S, Matthey U, Kaim G, Dimroth P. J Bacteriol. 1998;180:3312–3316. doi: 10.1128/jb.180.13.3312-3316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthey U, Kaim G, Dimroth P. Eur J Biochem. 1997;247:820–825. doi: 10.1111/j.1432-1033.1997.t01-1-00820.x. [DOI] [PubMed] [Google Scholar]
- 15.Gay NJ, Walker JE. Nucleic Acids Res. 1981;9:3919–3926. doi: 10.1093/nar/9.16.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gay NJ, Walker JE. Nucleic Acids Res. 1981;9:2187–2194. doi: 10.1093/nar/9.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downie JA, Langman L, Cox GB, Yanofsky C, Gibron F. J Bacteriol. 1980;143:8–17. doi: 10.1128/jb.143.1.8-17.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter AC, Brusilow WS, Simoni RD. J Bacteriol. 1983;155:1271–1278. doi: 10.1128/jb.155.3.1271-1278.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brusilow WS, Porter AC, Simoni RD. J Bacteriol. 1983;155:1265–1270. doi: 10.1128/jb.155.3.1265-1270.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Meyenburg K, Jorgensen BB, Nielsen J, Hansen FG. Mol Gen Genet. 1982;188:240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]
- 21.Gay NJ. J Bacteriol. 1984;158:820–825. doi: 10.1128/jb.158.3.820-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneppe B, Deckers-Hebestreit G, Altendorf K. J Biol Chem. 1990;265:389–395. [PubMed] [Google Scholar]
- 23.Solomon KA, Hsu DK, Brusilow WS. J Bacteriol. 1989;171:3039–3045. doi: 10.1128/jb.171.6.3039-3045.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneppe B, Deckers-Hebestreit G, Altendorf K. FEBS Lett. 1991;292:145–147. doi: 10.1016/0014-5793(91)80853-u. [DOI] [PubMed] [Google Scholar]
- 25.Gerike U, Dimroth P. Arch Microbiol. 1994;161:495–500. doi: 10.1007/BF00307770. [DOI] [PubMed] [Google Scholar]
- 26.Kaim G, Ludwig W, Dimroth P, Schleifer KH. Eur J Biochem. 1992;207:463–470. doi: 10.1111/j.1432-1033.1992.tb17072.x. [DOI] [PubMed] [Google Scholar]
- 27.Ohta S, Yohda M, Ishizuka M, Hirata H, Hamamoto T, Otawara-Hamamoto Y, Matsuda K, Kagawa Y. Biochim Biophys Acta. 1988;933:141–155. doi: 10.1016/0005-2728(88)90064-3. [DOI] [PubMed] [Google Scholar]
- 28.Revington M, McLachlin DT, Shaw GS, Dunn SD. J Biol Chem. 1999;274:31094–31101. doi: 10.1074/jbc.274.43.31094. [DOI] [PubMed] [Google Scholar]
- 29.Kaim G, Dimroth P. Eur J Biochem. 1993;218:937–944. doi: 10.1111/j.1432-1033.1993.tb18450.x. [DOI] [PubMed] [Google Scholar]
- 30.Laubinger W, Dimroth P. Biochemistry. 1989;28:7194–7198. doi: 10.1021/bi00444a010. [DOI] [PubMed] [Google Scholar]
- 31.Ackerman SH, Tzagoloff A. J Biol Chem. 1990;265:9952–9959. [PubMed] [Google Scholar]
- 32.Tzagoloff A, Barrientos A, Neupert W, Herrmann JM. J Biol Chem. 2004;279:19775–19780. doi: 10.1074/jbc.M401506200. [DOI] [PubMed] [Google Scholar]
- 33.Osman C, Wilmes C, Tatsuta T, Langer T. Mol Biol Cell. 2007;18:627–635. doi: 10.1091/mbc.E06-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng X, Neupert W, Tzagoloff A. Mol Biol Cell. 2007;18:617–626. doi: 10.1091/mbc.E06-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altamura N, Capitanio N, Bonnefoy N, Papa S, Dujardin G. FEBS Lett. 1996;382:111–115. doi: 10.1016/0014-5793(96)00165-2. [DOI] [PubMed] [Google Scholar]
- 36.van Bloois E, Jan Haan G, de Gier JW, Oudega B, Luirink J. FEBS Lett. 2004;576:97–100. doi: 10.1016/j.febslet.2004.08.069. [DOI] [PubMed] [Google Scholar]
- 37.van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. J Cell Biol. 2004;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laubinger W, Deckers-Hebestreit G, Altendorf K, Dimroth P. Biochemistry. 1990;29:5458–5463. doi: 10.1021/bi00475a008. [DOI] [PubMed] [Google Scholar]
- 39.Monticello RA, Angov E, Brusilow WS. J Bacteriol. 1992;174:3370–3376. doi: 10.1128/jb.174.10.3370-3376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki T, Murakami T, Iino R, Suzuki J, Ono S, Shirakihara Y, Yoshida M. J Biol Chem. 2003;278:46840–46846. doi: 10.1074/jbc.M307165200. [DOI] [PubMed] [Google Scholar]
- 41.von Ballmoos C, Dimroth P. Anal Biochem. 2004;335:334–337. doi: 10.1016/j.ab.2004.08.011. [DOI] [PubMed] [Google Scholar]