Abstract
The single-pass transmembrane domains (TMDs) of the syndecan family of cell surface adhesion molecules have been implicated in functional protein–protein interactions. Although each paralog contains a conserved GxxxG dimerization motif, we show here that the syndecan-1 TMD dimerizes weakly, the syndecan-3 and syndecan-4 TMDs each dimerize strongly, and the syndecan-2 TMD dimerizes very strongly. These markedly different levels of self-association suggest that paralog TMDs play different roles in directing functional interactions of each full-length syndecan family member. We further show that each syndecan TMD forms detergent-resistant heteromeric complexes with other paralogs, and that these interactions exhibit selectivity. Although heteromeric interactions among full-length syndecan paralogs have not been reported, we argue that the distinct hierarchy of protein–protein interactions mediated by the syndecan TMDs may give rise to considerable complexity in syndecan function. The demonstration that TMD homodimerization and heterodimerization can be mediated by GxxxG motifs and modulated by sequence context has implications for the signaling mechanisms of other cell surface receptors, including the integrins and the erbB family.
Keywords: homodimerization, heterodimerization, selectivity
Syndecans are cell surface receptors that participate in cell–cell and cell–matrix interactions critical to animal development. In mammals, where four syndecan paralogs exist (1–5), syndecan expression is tissue-specific and developmentally regulated, with most cells expressing one or more syndecans (6, 7). Syndecan-1 plays roles in early development (8–10) and wound healing (11), and syndecan-2 participates in neuronal dendritic spine morphogenesis (12), angiogenic sprouting (13), and Xenopus left/right axis formation (14). Syndecan-3 is involved in skeletogenesis (15), and syndecan-4 participates in the formation of integrin-containing focal adhesions (16). Although our knowledge of the mechanisms underlying syndecan biology is still sparse, it is already clear that understanding syndecan function has implications for treatment of cancer, wound healing, and viral and bacterial pathogenesis (reviewed in ref. 17).
Each syndecan contains an ectodomain, a transmembrane domain (TMD), and a short, C-terminal cytoplasmic domain. Syndecan family members interact with a wide array of partners: the divergent heparan sulfate-modified ectodomains bind matrix proteins and growth factors, and the cytosolic domains bind cytoskeletal proteins and PDZ-domain proteins through the C1 and C2 regions (reviewed in refs. 18 and 19). Syndecan TMDs are also thought to make functional interactions: some functions of syndecan-2 and syndecan-4 depend on self-association through their TMDs (20), and the TMD of syndecan-1 makes interactions that lead to the initial spreading response of Raji cells (21). The first evidence of TMD involvement in syndecan protein–protein interactions was the attribution of detergent-resistant noncovalent oligomerization of the syndecan-3 protein to the TMD (plus a four-residue juxtamembranous sequence), which led to the hypothesis that syndecan-3 exists as a dimer in the cell membrane (22). We seek to understand how the sequences of the syndecan TMDs give rise to interactions that underlie their biological functions.
Although TMD homodimerization has been implicated in syndecan function, the interaction strengths of the syndecan paralog TMDs have not been determined relative to one another, or by comparison to other TMDs known to self-associate (23). Here, we measure the association tendencies of the syndecan TMDs in both membranes and detergents, independent of the ectodomain and endodomains found in the native proteins. Results from a bacterial cell membrane assay, TOXCAT (24), agree closely with the oligomerization behavior of staphylococcal nuclease (SNase) fusion proteins (25) in detergents, providing a rank order of stability of human syndecan TMD self-association. Surprisingly, the TMDs of the four paralogs self-associate to very different degrees, and syndecan-2 associates more strongly than the well characterized TMD of glycophorin A (GpA) (24).
Based on the sequence similarity among the human syndecan paralog TMDs, including a conserved GxxxG dimerization motif, we hypothesized that syndecan paralogs might be able to make heteromeric interactions through their TMDs. Such interactions could have functional significance because multiple syndecans are often expressed within the same cell; for instance, all four paralogs are found in cells of the vasculature (26), and vascular phenotypes result from altered expression of three syndecans (reviewed in ref. 19). We show that each paralog TMD can form stable heteromeric complexes with other paralog TMDs, although not all pairwise interactions are strongly favored. The range of homodimerization interaction strengths and the selectivity of heteromeric complex formation support a role for paralog-specific protein–protein interactions by the TMDs.
Results
The Syndecan Paralog TMDs Self-Associate to Different Extents.
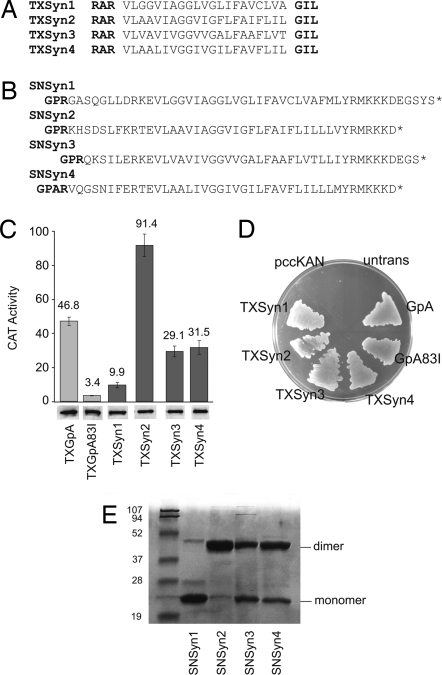
The four syndecan paralog TMDs each contain a GxxxG motif thought to mediate homodimerization (Fig. 1A). We tested this idea in bacterial membranes by using the TOXCAT assay (24), in which association between TMDs drives expression of the reporter gene chloramphenicol acetyltransferase (CAT) (reviewed in ref. 23). The TXSyn2 construct drives CAT levels to twice that of the positive control, TXGpA (24), which carries the strongly dimerizing GpA TMD (Fig. 1C). TXSyn3 and TXSyn4 also cause strong CAT expression, albeit slightly less than TXGpA. In contrast, the TXSyn1 construct gives low levels of CAT, about one-quarter that of the TXGpA control. Insertion of the fusion proteins into the membrane with the correct topology is confirmed by complementation of the malE phenotype of Escherichia coli NT326 cells (Fig. 1D) (24, 27). Western blots show that all ToxR(TMD)maltose binding protein (MBP) fusions are expressed at similar levels (Fig. 1C), so the different CAT levels can be interpreted to arise from differences in TMD self-association in the E. coli inner membrane. We conclude that the syndecan-2 TMD dimerizes more tightly than the TMDs of syndecan-3 or syndecan-4, and that the syndecan-1 TMD associates weakly. Note that the juxtamembranous residues initially implicated in syndecan-3 dimerization (22) are not required for dimerization in TOXCAT.
Fig. 1.
TOXCAT and SDS/PAGE analysis of syndecan TMD self-association. (A) Sequences of the ToxR(TMD)MBP fusions of human syndecan-1, syndecan-2, syndecan-3, and syndecan-4. (B) TMDs plus flanking regions of the human syndecans fused to the C-terminal end of SNase (bold) to yield the SNSyn1, SNSyn2, SNSyn3, and SNSyn4 fusion proteins. (C) (Upper) CAT activity from cells expressing the TXSyn1, TXSyn2, TXSyn3, and TXSyn4 constructs. Error bars show standard deviation of three independent cultures. GpA (TXGpA) and its disruptive mutant (TXGpA83I) serve as positive and negative controls. (Lower) ToxR(TMD)MBP expression levels measured by Western blot are similar for all cultures. (D) MalE complementation assay of ToxR fusions. Untransformed malE cells (untrans) or those carrying an empty vector (pccKAN) do not grow with maltose as the sole carbon source, but the control TXGpA or syndecan fusions permit robust growth, indicating that the MBP domains are directed to the periplasm. (E) SDS/PAGE of the purified SNSyn1, SNSyn2, SNSyn3, and SNSyn4 fusion proteins. Markers and migration positions consistent with the molecular masses of protein monomers and dimers are indicated.
Syndecans have been reported to form detergent-resistant oligomers, and the TMDs are implicated in this self-association (20, 22). Fusion proteins with SNase have been used to delineate the regions of GpA (25), phospholamban (28), and BNIP3 (29) that contribute to detergent-resistant oligomerization of these proteins. SNase/TMD fusion proteins can be readily expressed at high levels and purified in nonionic detergents, and the SNase domain shows little or no tendency to self-associate (25). Therefore, to test whether the TMD was sufficient for self-association in detergent micelles, we subcloned the TMDs and flanking regions of the four human syndecan paralogs as SNase fusions (Fig. 1B), purified the expressed proteins, and tested the resulting constructs for association in SDS micelles by electrophoresis. On SDS/PAGE (Fig. 1E), the SNSyn2 fusion migrates as >90% dimer with trace amounts of monomer, SNSyn3 migrates as ≈50% dimer, and SNSyn4 migrates as ≈60% dimer. [The SNGpA fusion forms ≈85% dimer under these conditions (30).] In contrast with the other SNase syndecan TMD fusions, SNSyn1 migrates as >90% monomer. We note that when the anomalous migration of the syndecan-1 core ectodomain (31) is considered, the syndecan-1 core protein migrates as a monomer on SDS/PAGE with no indication of TMD-driven detergent-resistant dimerization (3).
Together, these measurements establish a hierarchy of self-association strengths for the four human syndecan TMDs in biological membranes and detergents: syndecan-2 shows very strong dimerization, syndecan-3 and syndecan-4 display strong dimerization, and syndecan-1 shows only weak self-association tendencies. Characterizing all four paralog sequences under the same conditions demonstrates that despite the conserved GxxxG motif present in all four human syndecan TMDs, the intrinsic self-association tendencies of the syndecan TMDs vary tremendously from one paralog to another.
With SNSyn3, we observe the formation of a small fraction of trimers and tetramers in detergents. The significance of such species is not clear: the TMD of integrin αIIb forms dimers on SDS/PAGE, and also forms significant trimers at high concentrations (32), whereas SNase/TMD fusions of GpA (25, 29) and BNIP3 (25, 29) show strong dimer bands on SDS/PAGE and essentially no higher-order species. The possibility that the syndecan-3 TMD supports biologically relevant higher-order interactions is intriguing, especially because the other syndecan TMDs do not show higher-order complexes. Our findings suggest that the higher-order species previously noted for the syndecan-3 full-length protein under conditions of SDS/PAGE (22) are driven by the TMDs, but the involvement of nonphysiological antiparallel contacts cannot be ruled out. Because TOXCAT cannot distinguish between dimers and higher-order species, we cannot confirm the presence of higher-order syndecan-3 TMD oligomers in membranes. Further experiments are therefore needed to establish the significance of syndecan-3 TMD trimers and tetramers.
Mutations in the Conserved GxxxG Motifs Abolish Syndecan TMD Oligomerization.
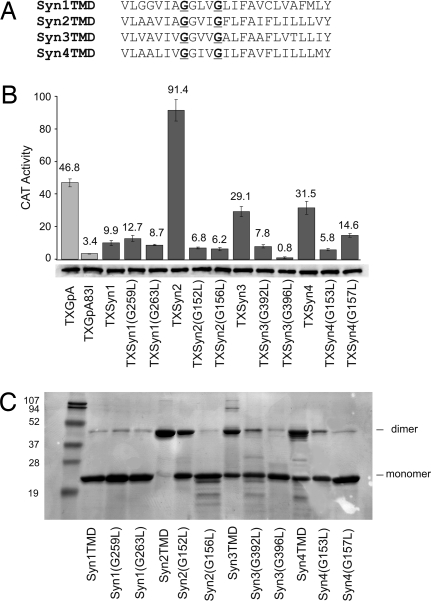
To determine the importance of the GxxxG dimerization motif (33, 34) in driving self-association of the syndecan TMDs, we mutated each motif glycine (Fig. 2A) and determined the effects on oligomerization in bacterial membranes (Fig. 2B; TOXCAT) and detergents (Fig. 2C; SDS/PAGE). All ToxR(TMD)MBP fusions are expressed at similar levels (Fig. 2B) and complement malE (data not shown). The disruptive effects of single-leucine substitutions in the conserved GxxxG motifs of the TMDs of syndecan-2, syndecan-3, and syndecan-4 implicate these motifs in syndecan TMD self-association, in agreement with previous reports (20, 22).
Fig. 2.
Role of GxxxG motifs in syndecan TMD self-association. (A) Syndecan TMD sequences showing the GxxxG motif residues targeted for mutagenesis (underlined). (B) (Upper) CAT activity from cells expressing wild-type and mutant syndecan ToxR constructs. Error bars show standard deviation of three independent cultures. GpA (TXGpA) and its disruptive mutant (TXGpA83I) serve as positive and negative controls. (Lower) ToxR(TMD)MBP expression levels measured by Western blot analysis are similar for all cultures. (C) SDS/PAGE of the wild-type SNase syndecan fusions and their glycine-to-leucine mutants. Bands consistent with the positions of protein dimers and monomers are indicated.
Our data for self-association in bacterial membranes (TOXCAT assay) suggest that the relative importance of the two motif glycines is different for each paralog. For syndecan-2, substitution of either motif glycine with leucine gives the same low TOXCAT dimerization signal. For syndecan-3, replacing the second glycine has a stronger effect than replacing the first, whereas the reverse is true for syndecan-4. By contrast, for the fusion proteins in detergent micelles as detected by SDS/PAGE, all substitutions at the second glycine have a more disruptive effect on dimerization than those at the first glycine.
Certain Syndecan TMDs Form Strong Heterooligomers in Detergent.
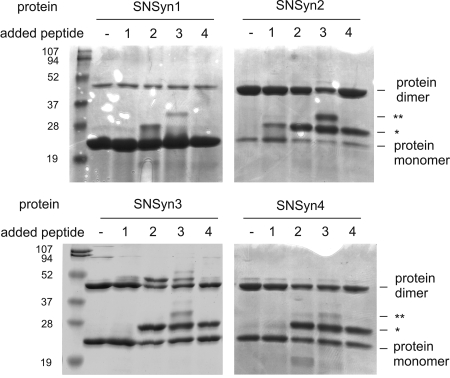
The role of the GxxxG motif in homodimerization of the TMDs of syndecans 2, 3, and 4 led us to wonder whether the syndecan TMDs might be able to make heterotypic and homotypic interactions. Such interactions could increase the diversity of syndecan complexes in cells where more than one syndecan is expressed. We tested this idea in detergent micelles by mixing the purified SN/TMD fusion proteins with each TMD peptide and resolving the mixtures by using SDS/PAGE (Fig. 3). We infer the formation of protein/peptide complexes from species that migrate at molecular masses distinct from those of the proteins alone (25, 29). Each SNase fusion construct forms heteromeric complexes with TMD peptides, but not all potential partners interact to the same degree. The SNSyn1 fusion construct, which self-associates weakly and does not interact with syndecan-1 TMD peptide (syn1pep), forms moderate levels of heterodimer with the syndecan-2 TMD peptide (syn2pep). SNSyn1 forms heterotrimers that contain two copies of the syndecan-3 TMD peptide (syn3pep), but no interaction is detected with the syndecan-4 TMD peptide (syn4pep).
Fig. 3.
Heterologous interactions between syndecan TMDs revealed by peptide competition. SNase fusion proteins alone or mixed with equimolar amounts of each syndecan TMD peptide were resolved on SDS/PAGE and detected by staining with Coomassie blue. Bands consistent with protein dimers and monomers are labeled; protein–peptide heterodimers are indicated by *, and protein–peptide–peptide heterotrimers are indicated by **.
Mixing the SNase fusions pairwise with each of the four syndecan TMD peptides reveals a set of heterotypic interactions (Fig. 3) that are summarized in Table 1 by using the scale: −, no interaction; +, detectable interaction; ++, moderate interaction; +++, strong interaction; ++++, very strong interaction. SNSyn2, SNSyn3, and SNSyn4 interact with syn1pep to give complexes similar to those seen when SNSyn1 is mixed with syn2pep, syn3pep, and syn4pep. SNSyn2 and syn1pep form moderate levels of heterodimer, SNSyn3 and syn1pep form heterotrimers with two copies of SNSyn3 and one syn1pep, and SNSyn4 does not interact with syn1pep. SNSyn2, SNSyn3, and SNSyn4 each form strong heterodimers with any of syn2pep, syn3pep, or syn4pep. Higher-order species are also formed if SNSyn3 or syn3pep are involved, but as indicated previously, the biological significance of these higher-order complexes is not clear. In all instances, however, analogous heteromeric complexes form regardless of whether the partners are present as peptides or fusion proteins. The resulting symmetry of Table 1 about the diagonal demonstrates that the peptide competition assay is not strongly influenced by the presence of the SNase domain and indicates that the complexes resolved on SDS/PAGE arise from intrinsic oligomerization properties of the TMD sequences. The interactions of various syndecan SNase fusions with self-peptides (Fig. 3) parallel the earlier TOXCAT and SDS/PAGE homodimerization tendencies (Fig. 1), lending additional confidence to our interpretation of results from this peptide competition assay. Together, these peptide competition experiments show that the syndecan paralog TMDs support heteromeric interactions of different strengths and stoichiometries.
Table 1.
Relative strengths of syndecan TMD interactions
| Syndecan-1 | Syndecan-2 | Syndecan-3 | Syndecan-4 | |
|---|---|---|---|---|
| Syndecan-1 | + | ++ | −/++ | − |
| Syndecan-2 | ++ | ++++ | +++ | +++ |
| Syndecan-3 | −/++ | +++ | +++ | +++ |
| Syndecan-4 | − | +++ | +++ | +++ |
Homodimerization scores (bold) are the consensus from bacterial membranes, SDS/PAGE, and peptide competition. Heterodimerization scores are from peptide competition only; rows identify the SN/TMD protein, and columns correspond to the added peptide. The score −/++ indicates no heterodimer but moderate heterotrimer.
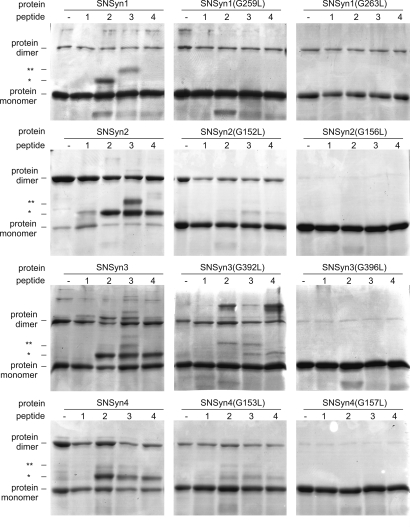
Heteromeric Interactions Between Syndecan TMDs Depend on the GxxxG Motifs.
To determine the importance of the conserved GxxxG motifs to the formation of heteromeric TMD complexes, we mixed wild-type TMD peptides for each syndecan paralog with SNase TMD fusions carrying glycine-to-leucine mutations. Fig. 4 shows the effects on heterooligomerization of replacing either glycine in the GxxxG motif with leucine. For the SNSyn1 fusion protein, either glycine-to-leucine substitution completely abolishes interaction with syn2pep or syn3pep. For the other syndecan fusions, replacing the second glycine of the motif with leucine completely abolishes interaction with any syndecan peptide, whereas replacing the first glycine abolishes interaction with syn1pep and significantly lowers interaction with peptides from syndecans 2, 3, and 4. Thus, the relative importance of the glycines to heteromeric association parallels the relative importance of these same glycines to homodimerization (Fig. 2).
Fig. 4.
Role of GxxxG motifs in syndecan TMD heteromeric interactions. Wild-type and mutant SNase fusion proteins alone or mixed with equimolar amounts of the indicated syndecan TMD peptide were resolved on SDS/PAGE and detected by staining with Coomassie blue. Bands consistent with protein dimers and monomers are labeled; protein–peptide heterodimers are indicated by *, and protein–peptide–peptide heterotrimers are indicated by **.
Discussion
The ability of the TMD to drive detergent-resistant oligomerization of syndecan-3 (22) has led to the expectation that all syndecans self-associate strongly through their TMDs (7, 17–19, 35). Given the 48–72% pairwise sequence identity between paralog TMDs and the strict conservation of a GxxxG dimerization motif, it might seem that all syndecan TMDs would support the same interactions. In contrast with this prediction, our findings show that the syndecan paralog TMDs exhibit a graded tendency to homodimerize and that each paralog TMD is capable of a distinct set of heteromeric interactions (see Table 1). The syndecan-1 and syndecan-2 TMDs are at opposite ends of our scale for homodimerization, with syndecan-1 showing little tendency to self-associate, and syndecan-2 dimerizing more tightly than the well characterized protein GpA. Although syndecan-3 and syndecan-4 both show intermediate levels of homodimerization, they make different heteromeric interactions: syndecan-3 binds to syndecan-1 but syndecan-4 does not. Although the syndecan-1 TMD exhibits selectivity in binding syndecan-2 and syndecan-3 but not itself or syndecan-4, the three paralog TMDs that homodimerize strongly all form heterooligomers with one another. Measuring these effects in the absence of the extracellular or cytoplasmic domains demonstrates the wide range of stabilities and interaction selectivities that can be attributed to the TMDs alone.
The biological significance of these differences in paralog TMD oligomerization behavior is supported by the strong sequence conservation of syndecan TMDs across species: although human syndecan-2 TMD shares 48–72% sequence identity with other human syndecan TMDs, it is 100% identical to the TMDs of mouse, chicken, and zebrafish syndecan-2. Indeed, the high sequence conservation across species for the TMDs is comparable with the conservation seen across species for the syndecan cytoplasmic variable (V) regions (35), which are unique to each paralog and can support paralog-specific interactions (36). We propose that the conservation of TMD sequence across species is caused by evolutionary pressures that arise from distinct functional roles for TMD interactions of each syndecan paralog. The very strong association of the syndecan-2 TMD could drive constitutive dimerization of the full-length protein, whereas the weaker self-association tendency of the syndecan-4 TMD should allow dimerization to be modulated by phosphorylation of (37) and phosphatidylinositol biphosphate (PIP2) binding to (38) the cytosolic domain. The syndecan-3 TMD should support strong dimerization (and perhaps higher-order interactions) of the native protein, whereas the weak self-association of the syndecan-1 TMD in our assays suggests that homodimerization of the full-length protein would require a high effective concentration in the membrane (absent additional interaction partners). It is intriguing to consider that the GxxxG motif in syndecan-1, which does not confer strong homodimerization, may be evolutionarily conserved because of the functional significance of heterodimerization with syndecan-2 or syndecan-3.
More generally, syndecan TMD homoassociations and heteroassociations could help drive or block the formation of functional complexes depending on which paralogs are expressed, their relative abundance, and the availability of other effectors. Syndecan-3 and syndecan-4, the only paralogs detected in adult mouse skeletal muscle, are expressed in quiescent satellite cells (39). Wild-type satellite cells respond to basic fibroblast growth factor by phosphorylating extracellular signal-regulated kinases (ERK1/2), whereas explanted syndecan-4-null satellite cells do not activate ERK1/2 under these conditions (40), consistent with a role for syndecan-4 dimers in receptor tyrosine kinase signal transduction and/or activating protein kinase C (36, 37). Interestingly, explanted syndecan-3-null cells activate ERK1/2 more extensively than wild-type satellite cells. We suggest that this increased ERK1/2 phosphorylation could result from the relief of TMD-mediated syndecan-3 inhibition of syndecan-4 signaling: in wild-type cells, syndecan-3 and syndecan-4 could form heteromeric complexes that are less active than syndecan-4 dimers, whereas in syndecan-3-null cells nothing would interfere with syndecan-4 homodimerization. The divergence of syndecan-3 and syndecan-4 expression patterns during regeneration of wild-type muscle followed by strict coexpression in quiescent satellite cells (40) could similarly alter the availability of syndecan-4 dimers in proliferating and differentiating cells after injury. Interaction of the syndecan-3 cytosolic domain with cortactin/c-src (41) or of the syndecan-4 cytosolic domain with PIP2 (38), syndesmos (42), or α-actinin (43) could also modulate homooligomerization and heterooligomerization. Given that syndecan-4 is thought to be expressed more ubiquitously than other syndecans but at lower levels (7), this type of regulation could occur in many different cell types.
The fusion protein approaches reported here demonstrate that the syndecan TMDs drive paralog-specific homomeric and heteromeric interactions in the absence of any other syndecan domains. Further experiments must be done to address the extent to which TMDs can drive homodimerization or heterodimerization in the context of the full-length proteins in vivo. Just as some TMD interactions might cooperate with stabilizing contacts made by other domains of the native proteins, other TMD complexes might be disfavored because of steric hindrance. If steric interactions between the large syndecan ectodomains tend to prevent TMD association (perhaps especially the formation of higher-order complexes), then protease-mediated shedding of ectodomains (6) could actually enhance the ability of the liberated TMDs to interact with other full-length proteins.
The physical basis for the stability and specificity of the interactions described here is not entirely clear. Our data show that GxxxG motifs play a critical role in syndecan TMD association, and by analogy to the structure of the GpA TMD dimer (44) we expect that the motif glycines permit close packing of syndecan transmembrane helices and possibly intermonomer Cα-H ··· O hydrogen bond formation (45). The weak homodimerization of syndecan-1 in our assays nevertheless demonstrates that a GxxxG motif is not sufficient for robust self-association of syndecan TMDs: residues outside the motif must contribute to stability. The disruptive effects of a single glycine-to-leucine mutation in either partner of a heteromeric TMD complex, at either motif glycine, demonstrates the importance of GxxxG motifs (including that of syndecan-1) to heterodimerization. However, syndecan-1 does not interact with syndecan-4, even though the syndecan-4 TMD contains a GxxxG motif and is 78% identical to the tightly interacting syndecan-2 TMD. This selectivity of syndecan-1 heteromeric interactions shows that residues outside of the GxxxG must contribute to specificity. The role of sequence context in stabilizing helix–helix interactions mediated by a GxxxG motif has been studied quantitatively by Fleming and colleagues (46), who showed that single point mutations that leave the native GxxxG motif of GpA intact can modulate the free energy of dimerization in detergent micelles by −0.5 to +3.2 kcal·mol−1. We propose that the GxxxG motif permits close approach of syndecan transmembrane helices, but that the residues that flank the motif give rise to the distinct association properties of the four paralog TMDs. The strong sequence conservation across syndecan ortholog TMDs thus probably reflects the conservation of paralog-specific TMD interaction propensities. We expect that the close correspondence between measures of syndecan TMD dimerization from membranes and detergents, which has also been seen for GpA (47), will facilitate biochemical and structural investigations of syndecan TMD complex formation.
Functional roles for TMD interactions have also been identified in both integrin and ErbB signal transduction. The αIIb integrin TMD self-associates by using a GxxxG motif (48), sequence changes that modulate integrin TMD interactions influence activation of the αIIbβ3 integrin complex (49), and designed TMD peptides that interact specifically with αIIb or αv integrin TMDs selectively activate either αIIbβ3 or αvβ3 integrin signaling when added to platelets (50). The TMDs of the four ErbB receptors show moderate to strong levels of self-association in bacterial membranes that depend on motifs of small amino acids separated by three residues (51). No significant homodimerization of SN/erbB fusions is detected on SDS/PAGE or by sedimentation equilibrium in the detergent n-octyl pentaethylene glycol monoether (52), but a hierarchy of homomeric and heteromeric interactions has been identified for ErbB TMD peptides in low concentrations of the detergent lauryl dimethyl ammonium oxide (53), and these interactions may contribute to the stability of heteromeric ErbB signaling complexes (54). Signaling by receptor tyrosine kinases that lack GxxxG motifs has also been linked to TMD interactions: a mutation in the TMD of FGFR3 that gives rise to pathologies, including bladder cancer (55), enhances the weak homodimerization of the isolated wild-type TMD (56). For all of these systems, the ability of TMD interactions to influence the association states of the full-length proteins would be expected to depend on the intrinsic affinities of the TMDs for one another and on the effective concentrations of these species in cell membranes or membrane domains. We anticipate that the strong and specific interactions that we report here between syndecan TMDs will be found to contribute to interactions among the native proteins and to syndecan-dependent signal transduction.
Methods
Vectors and Constructs.
The parental TOXCAT construct, pccKAN, and derivatives carrying the GpA TMD (pccGpA) and a disruptive GpA mutant (pccGpA-G83I) have been described (24). DNA fragments coding for residues 252–272 of syndecan-1 [National Center for Biotechnology Information (NCBI) code gi:55749480], residues 145–166 of syndecan-2 [gi:39644970], residues 385–406 of syndecan-3 (NCBI code gi:57222247), and residues 146–167 of syndecan-4 (NCBI code gi:38201675), were generated by PCR amplification and ligated in-frame to the NheI and BamHI sites of pccKAN to generate the ToxR(TMD)MBP fusions. DNA fragments corresponding to residues 241–287 of human syndecan-1, residues 134–175 of syndecan-2, residues 376–418 of syndecan-3, and residues 135–176 of syndecan-4 were produced by PCR amplification and ligated in-frame to the ApaI/BamHI sites of the pT7SN/GpA vector (25) to generate plasmids encoding the SNase/TMD fusions.
Site-Directed Mutagenesis.
Mutagenesis was performed by using the QuikChange kit (Stratagene). All mutations were confirmed by automated sequencing.
TOXCAT Assay.
A colorimetric assay (57) was used to detect CAT activity in lysates prepared from aliquots of 3.0 OD420 of NT326 cells as described (29).
MalE Complementation.
E. coli NT326 cells expressing ToxR(TMD)MBP constructs were grown overnight in liquid M9 minimal medium containing 0.4% glucose, washed, and streaked onto M9 plates containing 0.4% maltose as the only carbon source. Plates were incubated for 2–3 days at 37°C and imaged with an Alpha Innotech FluoroChem 5500 digital camera system. Proteinase K digestion of NT326 spheroplasts (51) confirms that the fusion proteins expose their MBP domains to the periplasm (data not shown).
Expression and Purification of SNase Fusion Proteins.
Plasmids coding for SNase/syndecan-TMD fusion proteins (or mutants) were transformed into BL21(DE3) cells and plated onto LB plates (with 50 μg/ml ampicillin). Colonies were inoculated into LB medium (with 50 μg/ml carbenicillin), grown to A420 ≈0.2, and stored as glycerol stocks at −80°C. Expression and purification of the fusion proteins was performed as described (58).
Syndecan TMD Peptides.
Syndecan TMD peptides were produced by overnight trypsin digest (1:40 weight ratio) of the purified SNase/TMD fusions. Soluble tryptic fragments were removed by dialysis against buffer containing 20 mM Tris (pH 8.0), 2 mM EDTA, 0.1 M NH4OAc, and 0.2% Thesit. Peptides were purified by RP-HPLC, and the identities of syn1pep (4,976.4 Da; 47 residues), syn2pep (4,577.6 Da; 40 residues), syn3pep (4,689.9 Da; 42 residues), and syn4pep (4,714.1 Da; 42 residues) were confirmed by MALDI-TOF mass spectroscopy.
SDS/PAGE of SN/TMD Fusion Proteins.
Protein (0.3 mg/ml; 14 μM) in SDS/PAGE sample buffer (with 2% SDS) was heated to 90°C for 5 min, resolved on 15% polyacrylamide gels (Bio-Rad) with running buffer containing 0.2% SDS, and visualized by Coomassie blue staining. Band intensities were quantified by using an Alpha Innotech gel documentation system.
ACKNOWLEDGMENTS.
We thank members of K.R.M.'s laboratory and Yousif Shamoo's laboratory at Rice University for constructive comments and R. B. Hill for critical reading of the manuscript. I.C.D. was supported by National Institutes of Health Molecular Biophysics Training Grant T32 GM008280. K.R.M. received support from National Institutes of Health Grant R01 GM-067850.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Saunders S, Jalkanen M, O'Farrell S, Bernfield M. J Cell Biol. 1989;108:1547–1556. doi: 10.1083/jcb.108.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marynen P, Zhang J, Cassiman JJ, Van den Berghe H, David G. J Biol Chem. 1989;264:7017–7024. [PubMed] [Google Scholar]
- 3.Kojima T, Shworak NW, Rosenberg RD. J Biol Chem. 1992;267:4870–4877. [PubMed] [Google Scholar]
- 4.Carey DJ, Evans DM, Stahl RC, Asundi VK, Conner KJ, Garbes P, Cizmeci-Smith G. J Cell Biol. 1992;117:191–201. doi: 10.1083/jcb.117.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould SE, Upholt WB, Kosher RA. Proc Natl Acad Sci USA. 1992;89:3271–3275. doi: 10.1073/pnas.89.8.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim CW, Goldberger OA, Gallo RL, Bernfield M. Mol Biol Cell. 1994;5:797–805. doi: 10.1091/mbc.5.7.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couchman JR. Nat Rev Mol Cell Biol. 2003;4:926–937. doi: 10.1038/nrm1257. [DOI] [PubMed] [Google Scholar]
- 8.Solursh M, Reiter RS, Jensen KL, Kato M, Bernfield M. Dev Biol. 1990;140:83–92. doi: 10.1016/0012-1606(90)90055-n. [DOI] [PubMed] [Google Scholar]
- 9.Trautman MS, Kimelman J, Bernfield M. Development. 1991;111:213–220. doi: 10.1242/dev.111.1.213. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland AE, Sanderson RD, Mayes M, Seibert M, Calarco PG, Bernfield M, Damsky CH. Development. 1991;113:339–351. doi: 10.1242/dev.113.1.339. [DOI] [PubMed] [Google Scholar]
- 11.Elenius K, Vainio S, Laato M, Salmivirta M, Thesleff I, Jalkanen M. J Cell Biol. 1991;114:585–595. doi: 10.1083/jcb.114.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ethell IM, Yamaguchi Y. J Cell Biol. 1999;144:575–586. doi: 10.1083/jcb.144.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen E, Hermanson S, Ekker SC. Blood. 2004;103:1710–1719. doi: 10.1182/blood-2003-06-1783. [DOI] [PubMed] [Google Scholar]
- 14.Kramer KL, Yost HJ. Dev Cell. 2002;2:115–124. doi: 10.1016/s1534-5807(01)00107-1. [DOI] [PubMed] [Google Scholar]
- 15.Shimo T, Gentili C, Iwamoto M, Wu C, Koyama E, Pacifici M. Dev Dyn. 2004;229:607–617. doi: 10.1002/dvdy.20009. [DOI] [PubMed] [Google Scholar]
- 16.Woods A, Couchman JR. Mol Biol Cell. 1994;5:183–192. doi: 10.1091/mbc.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fears CY, Woods A. Matrix Biol. 2006;25:443–456. doi: 10.1016/j.matbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Tkachenko E, Rhodes JM, Simons M. Circ Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 19.Alexopoulou AN, Multhaupt HA, Couchman JR. Int J Biochem Cell Biol. 2007;39:505–528. doi: 10.1016/j.biocel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Choi S, Lee E, Kwon S, Park H, Yi JY, Kim S, Han IO, Yun Y, Oh ES. J Biol Chem. 2005;280:42573–42579. doi: 10.1074/jbc.M509238200. [DOI] [PubMed] [Google Scholar]
- 21.McQuade KJ, Rapraeger AC. J Biol Chem. 2003;278:46607–46615. doi: 10.1074/jbc.M304775200. [DOI] [PubMed] [Google Scholar]
- 22.Asundi VK, Carey DJ. J Biol Chem. 1995;270:26404–26410. doi: 10.1074/jbc.270.44.26404. [DOI] [PubMed] [Google Scholar]
- 23.MacKenzie KR. Chem Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 24.Russ WP, Engelman DM. Proc Natl Acad Sci USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 26.Cizmeci-Smith G, Langan E, Youkey J, Showalter LJ, Carey DJ. Arterioscler Thromb Vasc Biol. 1997;17:172–180. doi: 10.1161/01.atv.17.1.172. [DOI] [PubMed] [Google Scholar]
- 27.Langosch D, Brosig B, Kolmar H, Fritz HJ. J Mol Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 28.Arkin IT, Adams PD, MacKenzie KR, Lemmon MA, Brunger AT, Engelman DM. EMBO J. 1994;13:4757–4764. doi: 10.1002/j.1460-2075.1994.tb06801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sulistijo ES, Jaszewski TM, MacKenzie KR. J Biol Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 30.Mingarro I, Whitley P, Lemmon MA, von Heijne G. Protein Sci. 1996;5:1339–1341. doi: 10.1002/pro.5560050712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weitzhandler M, Streeter HB, Henzel WJ, Bernfield M. J Biol Chem. 1988;263:6949–6952. [PubMed] [Google Scholar]
- 32.Li R, Babu CR, Lear JD, Wand AJ, Bennett JS, DeGrado WF. Proc Natl Acad Sci USA. 2001;98:12462–12467. doi: 10.1073/pnas.221463098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russ WP, Engelman DM. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 34.Senes A, Gerstein M, Engelman DM. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 35.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 36.Oh ES, Woods A, Couchman JR. J Biol Chem. 1997;272:8133–8136. doi: 10.1074/jbc.272.13.8133. [DOI] [PubMed] [Google Scholar]
- 37.Horowitz A, Tkachenko E, Simons M. J Cell Biol. 2002;157:715–725. doi: 10.1083/jcb.200112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh ES, Woods A, Couchman JR. J Biol Chem. 1997;272:11805–11811. doi: 10.1074/jbc.272.18.11805. [DOI] [PubMed] [Google Scholar]
- 39.Cornelison DD, Filla MS, Stanley HM, Rapraeger AC, Olwin BB. Dev Biol. 2001;239:79–94. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- 40.Cornelison DD, Wilcox-Adelman SA, Goetinck PF, Rauvala H, Rapraeger AC, Olwin BB. Genes Dev. 2004;18:2231–2236. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinnunen T, Kaksonen M, Saarinen J, Kalkkinen N, Peng HB, Rauvala H. J Biol Chem. 1998;273:10702–10708. doi: 10.1074/jbc.273.17.10702. [DOI] [PubMed] [Google Scholar]
- 42.Baciu PC, Saoncella S, Lee SH, Denhez F, Leuthardt D, Goetinck PF. J Cell Sci. 2000;113:315–324. doi: 10.1242/jcs.113.2.315. [DOI] [PubMed] [Google Scholar]
- 43.Greene DK, Tumova S, Couchman JR, Woods A. J Biol Chem. 2003;278:7617–7623. doi: 10.1074/jbc.M207123200. [DOI] [PubMed] [Google Scholar]
- 44.MacKenzie KR, Prestegard JH, Engelman DM. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 45.Senes A, Ubarretxena-Belandia I, Engelman DM. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doura AK, Kobus FJ, Dubrovsky L, Hibbard E, Fleming KG. J Mol Biol. 2004;341:991–998. doi: 10.1016/j.jmb.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 47.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li R, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS. J Biol Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 49.Li R, Mitra N, Gratkowski H, Vilaire G, Litvinov R, Nagasami C, Weisel JW, Lear JD, DeGrado WF, Bennett JS. Science. 2003;300:795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 50.Yin H, Slusky JS, Berger BW, Walters RS, Vilaire G, Litvinov RI, Lear JD, Caputo GA, Bennett JS, DeGrado WF. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 51.Mendrola JM, Berger MB, King MC, Lemmon MA. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 52.Stanley AM, Fleming KG. J Mol Biol. 2005;347:759–772. doi: 10.1016/j.jmb.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 53.Duneau JP, Vegh AP, Sturgis JN. Biochemistry. 2007;46:2010–2019. doi: 10.1021/bi061436f. [DOI] [PubMed] [Google Scholar]
- 54.Riese DJ, II, Stern DF. BioEssays. 1998;20:41–48. doi: 10.1002/(SICI)1521-1878(199801)20:1<41::AID-BIES7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 55.Meyers GA, Orlow SJ, Munro IR, Przylepa KA, Jabs EW. Nat Genet. 1995;11:462–464. doi: 10.1038/ng1295-462. [DOI] [PubMed] [Google Scholar]
- 56.Li E, You M, Hristova K. J Mol Biol. 2006;356:600–612. doi: 10.1016/j.jmb.2005.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw WV. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 58.Sulistijo ES, MacKenzie KR. J Mol Biol. 2006;364:974–990. doi: 10.1016/j.jmb.2006.09.065. [DOI] [PubMed] [Google Scholar]