Abstract
Posttranslational modification by small ubiquitin-like modifier (SUMO) controls diverse cellular functions of transcription factors and coregulators and participates in various cellular processes including signal transduction and transcriptional regulation. Here, we report that pontin, a component of chromatin-remodeling complexes, is SUMO-modified, and that SUMOylation of pontin is an active control mechanism for the transcriptional regulation of pontin on androgen-receptor target genes in prostate cancer cells. Biochemical purification of pontin-containing complexes revealed the presence of the Ubc9 SUMO-conjugating enzyme that underlies its function as an activator. Intriguingly, 5α-dihydroxytestosterone treatments significantly increased the SUMOylation of pontin, and SUMOylated pontin showed further activation of a subset of nuclear receptor-dependent transcription and led to an increase in proliferation and growth of prostate cancer cells. These data clearly define a functional model and provide a link between SUMO modification and prostate cancer progression.
Keywords: androgen receptor, SUMO, Ubc9, transcription, proliferation
Defining the signal integration pathways that involve diverse molecular mechanisms at the level of gene transcription remains an important goal in biology. The main downstream effects of a signaling pathway are the modulation of the function of transcription factors and coregulators in the nucleus (1). Thus, it is important to investigate the roles of transcription factors and coregulators in response to the upstream signaling pathway. The androgen receptor (AR) not only mediates prostate development but also serves as a key regulator of primary prostatic cancer growth (2). The effects of androgens are mediated through the AR, a member of the nuclear receptor family functioning as a ligand-inducible transcription factor that regulates the expression of target genes having an androgen response element. Analogous to other members of the nuclear receptor superfamily, the AR contains a DNA binding domain and a C-terminal ligand-binding domain functioning as ligand-dependent nuclear receptors with AF-1 and AF-2 transcription activation domains (3, 4). The ability of many nuclear receptors to bind a variety of ligands, including agonists or antagonists leading to either enhanced or diminished gene activation, raises intriguing issues about combinatorial transcriptional mechanisms mediated by coactivators and corepressors.
Gene expression is influenced by chromatin structure, and covalent modification of histones plays an important role in regulating transcription and chromatin dynamics (5). The transcription of most genes is regulated by the coordinate action of chromatin-remodeling complexes. Pontin and reptin have been reported to be components of the ATP-dependent chromatin-remodeling complexes and are closely related to the bacterial DNA helicase RuvB (6). Pontin has been demonstrated to bind and activate the β-catenin/TCF transcription complex, whereas reptin has been demonstrated to repress the Wnt/β-catenin signaling pathway (7). In mammals, they constitute parts of the Tip60 coactivator complex, which has intrinsic histone acetyltransferase activity (8). In zebrafish embryos, the reptin/pontin ratio serves to regulate heart growth during development via the β-catenin pathway (9).
Posttranslational modification of proteins plays an important role in the functional regulation of transcriptional coregulators. Numerous enzymatic activities have been demonstrated to be associated with coregulator complexes, including histone acetylation/deacetylation, phosphorylation/dephosphorylation, ubiquitination, and SUMOylation (10). Small ubiquitin-like modifier (SUMO) plays an important regulatory role in many cellular processes including signal transduction, transcriptional regulation, chromatin structure, and nuclear/cytoplasmic shuttling (11). SUMO is an ≈11-kDa protein that is structurally, but not functionally, homologous to ubiquitin. The enzymatic machinery that adds SUMO to, and removes it from, target proteins is distinct from the ubiquitination machinery. SUMOylation of target proteins is a multistep process involving the E1-activating molecules SAE1/SAE2, the E2-conjugating enzyme Ubc9, and E3 ligases in mammalian cells (12). Several SUMO-processing enzymes have been identified that hydrolyze the SUMO moiety from target substrates in mammals and yeast (13–15). Unlike ubiquitination, SUMO modification has not been found to be associated with protein degradation. Rather, it is similar to the nonproteolytic roles of ubiquitination including subcellular localization and the regulation of transcriptional activity. A growing number of SUMO-modified proteins have been identified including RanGAP1, p53, c-Jun, and reptin (16–18). Given that lysine is the site for a variety of modifications, it might be predicted that there exists elaborate modification code for both histone and nonhistone proteins.
Recently, we reported the dynamic role of a chromatin-remodeling complex in the regulation of the metastasis suppressor gene KAI1 (18, 19). Reptin chromatin-remodeling complexes contain SENP1 deSUMOylating enzyme as a component, and SUMO modification of reptin plays an important role for the transcriptional regulation of KAI1 (18). In this article, we provide evidence that pontin is modified by SUMO, but that this modification promotes the differential functional regulation of pontin having different downstream target genes compared with reptin. Biochemical purification of pontin-containing complexes revealed a Ubc9 SUMO-conjugating enzyme as a binding partner. Mutation that affects the attachment of SUMO to pontin decreased the transcriptional activation function of pontin in the regulation of AR target genes, such as PSA, in prostate cancer cells. SUMOylation of pontin is related to an active control mechanism for the transcriptional activation function of pontin through its nuclear retention and the enhancement of coactivator binding, whereas SUMOylation of reptin is involved in the potentiation of transcriptional repression. Further, SUMOylation of pontin increased proliferation and growth of prostate cancer cells. Taken together, we address a molecular mechanism in which SUMO modification of pontin mediates and elaborates the transcriptional regulation, thereby strongly activating a subset of AR target genes involved in tumor progression.
Results
Pontin Chromatin-Remodeling Complexes Contain a Ubc9 SUMO-Conjugating Enzyme.
We have previously reported the role of reptin chromatin-remodeling complex in regulating the expression of KAI1 (18). Although reptin and pontin exhibit structural similarity, they may play distinct roles in certain aspects of target-gene regulation. To explore the differential function of pontin, we used N-terminal Flag epitope-tag strategy to purify pontin-containing complexes in 293T cells. We used liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) to identify proteins in the pontin complexes purified from the Flag M2 affinity column (Fig. 1 A and B). The presence of reptin, β-catenin, and p400, well known binding partners of pontin, delineated the functional link between these molecules (7, 20).
Fig. 1.
Purification of pontin-containing complex. (A) The pontin-containing complex was immunoprecipitated with anti-Flag IgG-conjugated agarose beads from 293T cell extracts, and the bound proteins were eluted with Flag peptide and resolved by SDS/PAGE. (B) Peptide sequences of pontin-associated polypeptides from LC-MS/MS analysis. (C) Western blot analysis was performed by using the indicated antibodies, and reptin, β-catenin, p400, and Ubc9 were detected in the eluates. (D) Coimmunoprecipitation of endogenous pontin with Ubc9. Cell lysates were subjected to immunoprecipitation with either anti-Ubc9 IgG or control IgG, and the resultant precipitates were subjected to immunoblotting with anti-pontin IgG.
Of the remaining proteins identified by LC-MS/MS analysis, the presence of the SUMO-conjugating enzyme Ubc9 was striking (Fig. 1 A and B). The association of these polypeptides with pontin was confirmed by immunoblotting analysis of the eluates (Fig. 1C). To further validate the association of Ubc9 with pontin, an endogenous coimmunoprecipitation assay was performed; this revealed that pontin bound specifically to Ubc9 (Fig. 1D). Identification of the SUMO-conjugating enzyme in the pontin-containing complexes prompted us to examine whether SUMO modification is responsible for the functional regulation of pontin.
Lys 225 of Pontin Is Critical for SUMO Modification.
We applied an in vitro SUMO modification system to determine the possibility that pontin may be a substrate for the SUMO-conjugating enzyme Ubc9 (21). 35S-labeled, in vitro translated pontin protein was incubated in a SUMOylation mixture containing purified E1 (SAE1/SAE2) and E2 (Ubc9) in the presence or absence of purified SUMO. Pontin conjugates were formed after the addition of SUMO to the SUMOylation mixture (Fig. 2A). These data revealed that pontin undergoes SUMO modification along with the identity of the SUMO protein from LC-MS/MS analysis [supporting information (SI) Fig. 5]. To find the functional lysine residue(s) of pontin that serve as the SUMO acceptor sites, we searched the consensus SUMOylation sequence ψKxE (ψ represents a large hydrophobic amino acid) (Fig. 2B) (22). Each of the six lysines of pontin was changed independently to arginine, and the resultant mutants were tested for the ability of in vitro or in vivo SUMO modification (Fig. 2 C and D). Mutation of K2R, K108R, K171R, K231R, and K268R revealed little or no alteration in SUMO modification of pontin, whereas the K225R mutation abrogated pontin from modification by SUMO both in vitro and in vivo (Fig. 2 C and D). Therefore, only a single lysine residue K225 in pontin appears to function as a SUMO acceptor site, thereby conferring the modification function.
Fig. 2.
Lysine 225 of pontin is crucial for SUMO modification. (A) In vitro modification of pontin by SUMO. 35S-labeled in vitro-translated pontin was incubated in a SUMOylation mix containing purified E1, E2, and ATP in the absence or presence of SUMO. (B) Search for consensus site (ψ KxE) for SUMOylation in pontin, where ψ is an aliphatic amino acid, and K is the lysine conjugated to SUMO. (C) Lysine 225 of pontin is a major SUMO conjugation site. In vitro SUMOylation assay was conducted with 35S-labeled in vitro-translated Gal4-fused wild type or the K225R mutant of pontin as in A. (D) 293T cells were cotransfected with plasmids expressing either Gal4-fused wild type or the K225R mutant of pontin in the presence of SUMO and Ubc9. Western blotting was performed with anti-Gal4 antibody. (E) Immunoblot analysis indicates expression of Flag-tagged pontin, SUMO-fused pontin, pontin K225R, or SUMO-fused pontin K225R and their schematic representations. (F) Subcellular localization of Flag-tagged pontin, SUMO-fused pontin, pontin K225R, or SUMO-fused pontin K225R in HeLa cells (green). Nuclei were visualized by DAPI staining (blue).
We covalently attached SUMO to the N terminus of pontin by gene fusion to generate a constitutively SUMOylated form of pontin to eliminate the complication of the indirect effects associated with overexpressing SUMO or Ubc9 (23, 24) and confirmed the expression by immunoblot analysis (Fig. 2E). Because SUMOylation has been demonstrated to regulate nuclear/cytoplasmic shuttling of many transcription factors and coregulators (25), we examined whether SUMO conjugation on pontin plays a role in the translocation between nucleus and cytoplasm. Pontin was both nuclear and cytoplasmic in its localization, whereas pontin K225R mutant displayed a more cytoplasmic localization than the wild-type pontin (Fig. 2F). On the other hand, the attachment of SUMO to wild-type pontin and pontin K225R mutant resulted in exclusive nuclear localization (Fig. 2F and SI Fig. 6). These data suggest that covalent modification by SUMO may explain the strong transcriptional regulatory function of pontin on a subset of target genes in the nucleus through its increased nuclear retention.
SUMO Modification of Pontin Is Required for Transcriptional Activation of Androgen-Receptor Target Genes.
It has been demonstrated, by using a TOPFLASH reporter assay, that pontin activates β-catenin-mediated transcriptional activity, whereas reptin is required for the repression of β-catenin-mediated transcriptional activation (7). Pontin is recruited on the KAI1 promoter along with the Tip60 coactivator but is not required for the transcriptional activation of KAI1, whereas reptin is crucial for conferring repression to KAI1 along with β-catenin (19). Transcriptional regulation by AR involves interaction with a variety of transcriptional coactivators in the presence of agonist, and these act in both a sequential and combinatorial manner (26). Because β-catenin and Ubc9 have been reported to function as AR coactivators (27) and their association with pontin was confirmed from identification of the pontin-containing complex as shown in Fig. 1C, we examined whether pontin contributes to 5α-dihydroxytestosterone (DHT)-dependent AR target-gene activation. Using reverse transcriptase (RT)-PCR analysis, we evaluated the effects of the shRNA-mediated knockdown of pontin on the DHT-induced transcriptional activation of AR target genes such as PSA, KLK2, and NKX3.1 (Fig. 3A). Knockdown of reptin did not affect expression of AR target genes (data not shown). Indeed, pontin was required for DHT-dependent AR target-gene activation.
Fig. 3.
SUMOylation of pontin is required for transcriptional activation of androgen-receptor target genes. (A) Knockdown of pontin diminishes the PSA, KLK2, and NKX3.1 transcripts in the presence of DHT in LNCaP cells. (B) Fold change of AR target gene KLK2 transcripts after introduction of pontin K225R or SUMO-fused pontin K225R. (C–E) SUMOylation of pontin enhances the transcriptional activation function of pontin. Luciferase assay was conducted after cotransfection of ARE-luciferase reporter in the presence of DHT (C). Expression of pontin activated an RORα 2E-luciferase reporter (D) but not a RARE-luciferase reporter (E). (F) In vivo association experiments between pontin and nuclear receptors in 293T cells. (G) Coimmunoprecipitation assay to verify interaction of Flag-tagged pontin or pontin K225R with β-catenin, CBP, or Tip60. Coimmunoprecipitation assay was performed with anti-Flag IgG, and precipitated materials were detected with either β-catenin, CBP, or Tip60 antibody, respectively.
Pontin and reptin are closely related members of the ATPases associated with diverse cellular activities and are components of many different complexes, including the Tip60, INO80, and p400 complexes that are involved in transcriptional regulation (6, 8, 20). To gain an insight into the role of the SUMO modification of pontin, we assessed the effects of SUMO modification on the transcriptional properties of pontin. Introduction of pontin K225R decreased AR target-gene transcripts, whereas SUMO-fused pontin K225R, which mimics constitutive SUMOylation, activated AR target-gene transcripts in the presence of DHT (Fig. 3B). Consistent with the data shown in Fig. 3B, the expression of pontin K225R mutant reversed the activation function of pontin on an ARE-luciferase reporter in the presence of DHT, whereas SUMO-fused pontin K225R mutant further activated the androgen response element (ARE)-luciferase reporter (Fig. 3C). Further, expression of pontin activated an RORα 2E-luciferase reporter (Fig. 3D), but not retinoic acid response element (RARE) or estrogen receptor response element (ERE) luciferase reporters (Fig. 3E and data not shown). Coimmunoprecipitation data confirmed that pontin bound to AR and RORα 2 but not to RAR and ER (Fig. 3F and data not shown). These data suggest that pontin is specifically involved in a subset of nuclear receptor-regulated transcription function, and the SUMO conjugation of pontin appears to be related to an active control mechanism regulating the transcriptional activation function of pontin.
In most cases, SUMO modification of certain transcription factors and coregulators are responsible for the potentiation of transcriptional repression (18, 22). SUMOylated reptin binds to histone deacetylase 1 (HDAC1) stronger than non-SUMOylated reptin, and this explains, at least to some extent, why SUMO modification of reptin increases transcriptional repression function of reptin (18). To examine whether SUMO modification of pontin changes binding preference toward other coactivators for the enhanced transcriptional activation in addition to modulating transcriptional activity, we examined whether SUMO-fused pontin exhibits altered binding specificity toward AR coactivators (Fig. 3G). Both wild-type pontin and pontin K225R mutant bound to Tip60 coactivator, whereas pontin K225R mutant exhibited very weak binding to β-catenin and CBP coactivators compared with that of wild-type pontin (Fig. 3G). SUMOylation status of pontin did not affect binding affinity toward reptin (SI Fig. 7). These data suggest that SUMO modification of pontin is important for maintaining and exerting a subset of coactivator-mediated AR transcriptional activation function.
SUMOylation of Pontin Increases the Proliferation and Growth of Prostate Cancer Cells.
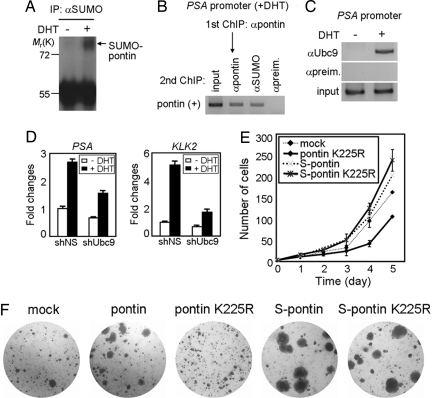
Androgen stimulates prostate cell growth, and LNCaP cell growth is repressed by androgen deficiency (28). To examine whether androgen affects the SUMOylation of pontin and to assess the potential effects of the SUMOylation of pontin on tumor cell growth and proliferation, we performed an in vivo SUMOylation assay of endogenous pontin in the absence and presence of DHT (Fig. 4A and SI Fig. 8). Intriguingly, DHT treatment significantly increased the SUMOylation of pontin. We next examined whether SUMOylated pontin is present on the PSA promoter under active conditions. Two-step ChIP assay was performed to monitor whether SUMOylated pontin is present on the PSA promoter in the presence of DHT (Fig. 4B). The soluble chromatin derived from DHT-treated LNCaP cells was immunoprecipitated with anti-pontin antibodies followed by release of the immune complexes and reimmunoprecipitation with anti-SUMO antibodies. In this experiment, we were able to immunoprecipitate the PSA promoter; clearly indicating that SUMO-modified pontin is indeed present on the PSA promoter under active conditions (Fig. 4B).
Fig. 4.
SUMOylation of pontin increases the proliferation and growth of prostate cancer cells. (A) SUMOylated pontin is increased in LNCaP cells in the presence of DHT. (B) Two-step ChIP assay with anti-pontin and anti-SUMO IgGs indicates that SUMO-modified pontin is present on the PSA promoter under activation condition. (C) ChIP analysis of Ubc9 on the PSA promoter with DHT treatment for 1 h in LNCaP cells. (D) AR target genes PSA and KLK2 transcripts after introduction of either shRNA against Ubc9 or nonspecific shRNA. (E) Proliferation curves of mock-, pontin-K225R-, SUMO-pontin-, or SUMO-pontin K225R-expressing LNCaP cells. Values are represented as mean ± SD of three independent experiments. (F) The anchorage-independent growth of LNCaP cells expressing pontin-, pontin-K225R-, SUMO-pontin-, or SUMO-pontin K225R in soft agar. Representative image is shown for each group.
Because Ubc9 is a single SUMO-conjugating enzyme and the interaction of Ubc9 with the AR DNA-binding domain and hinge region in yeast has been reported (27), we examined whether Ubc9 is recruited on the PSA promoter for transcriptional activation. ChIP assay revealed that Ubc9 was indeed recruited on the PSA promoter in the presence of DHT (Fig. 4C), and knockdown of Ubc9 decreased AR target transcripts such as PSA and KLK2 (Fig. 4D), suggesting that Ubc9 is required for transcriptional activation of AR target genes.
To determine whether the SUMOylation of pontin is sufficient to support PSA gene activation and, further, whether this can be extended to a physiologically relevant setting in prostate cancer cells, we examined the effects of SUMOylation status of pontin on cellular proliferation and growth of LNCaP cells. Proliferation assay measured the increase in cell number over the course of 5 days for pontin K225R-, SUMO-pontin-, and SUMO-pontin K225R-expressing LNCaP cells along with mock cells. Introduction of pontin K225R mutant suppressed growth and proliferation of LNCaP cells, whereas SUMO-pontin and SUMO-pontin K225R stimulated cell proliferation (Fig. 4E). Collectively, these results indicate that the recruitment of SUMOylated pontin by AR influences the cellular proliferation properties of androgen in prostate cancer.
To determine whether pontin SUMOylation could stimulate anchorage-independent growth, LNCaP cells stably expressing pontin, pontin K225R, SUMO-pontin, and SUMO-pontin K225R were examined for colony formation in soft agar, which is an important property of tumor cell growth (Fig. 4F). Introduction of SUMO-pontin greatly enhanced the size of colonies. Colonies with a diameter of >250 μm were not observed in mock LNCaP cells even with overexpression of pontin K225R but were present in SUMO-pontin and SUMO-pontin K225R-expressing cells. These data suggest that SUMO modification on pontin can augment the transforming potential of pontin, consistent with our in vitro AR-dependent transcriptional activation data.
Discussion
In this manuscript, we identified a signal integration pathway, SUMO modification of pontin, in the modulation of AR-dependent transcription and prostate cancer cell growth and proliferation. Given that the Ubc9 SUMO conjugating enzyme was obtained from pontin-containing complexes, we wished to explore the possible roles of SUMO modification in the regulation of pontin. We demonstrated that pontin is a substrate for Ubc9 and that SUMOylation of pontin underlies transcriptional activation of pontin in the regulation of AR target genes in prostate cancer cells. In contrast, SENP1 deSUMOylating enzyme was identified from reptin chromatin-remodeling complexes and that SUMOylation of reptin is crucial for conferring repressive function on KAI1 metastasis suppressor gene (18). In the event of SUMO modification, both SUMO conjugation and deconjugation are required steps for the coordinated regulation of this dynamic process. Pontin and reptin are closely related members of the ATPases associated with diverse cellular activities and are detected together in many types of complexes (7, 8). Given that pontin-containing complex possesses SUMO-conjugating enzyme, whereas reptin-containing complex contains the SUMO-deconjugating enzyme, it is tempting to speculate that two complex types might work coordinately and cooperatively in certain biological processes that require SUMO modification processes dynamically.
Our data show that SUMO modification of pontin is responsible for the strong transcriptional regulatory function of pontin on a subset of target genes in the nucleus through its increased nuclear retention. It is perhaps surprising that only a tiny fraction of pontin from both in vitro and in vivo SUMOylation assay was observed to be SUMOylated but that SUMO modification has such a large impact on the modulation of transcriptional activity, binding specificity, and localization of pontin.
Recently, histone demethylases such as JHDM2A and LSD1 have been reported to interact directly with AR and are recruited to AR target genes in a hormone-dependent manner (29, 30). Especially, distinct LSD1 complexes containing either coactivator or corepressor complexes have been demonstrated to exist for elegant coordinated regulation in development (31). Considering of a cohort of coactivators for transcriptional activation of AR in the presence of DHT, there may be a possible interplay between various modifying enzymes for histone or nonhistone protein substrates. Besides SUMOylation, demethylation, and acetylation, further modifying enzymes might interplay for nuclear receptor-mediated transcriptional regulation by providing another layer of transcriptional regulation.
Given that SUMO modification is involved in a variety of cellular processes, a link between SUMOylation and diseases such as tumorigenesis and cancer metastasis can be anticipated. Ubc9 was suggested to be a good candidate for a drug target because it is the only conjugating enzyme for SUMOylation process. The increased expression of Ubc9 has been reported in ovarian tumor tissues, human lung adenocarcinomas, and metastastic prostate cancer cells (18, 32). These phenomena might reflect close involvement of Ubc9 in tumorigenesis and cancer metastasis by regulating SUMOylation of various cellular targets. It is therefore tempting to explore the possibility that malignant progression of prostate cancer cells might in part prefer SUMOylated pontin and utilizes either hyperactivation of SUMO-conjugating system or conversely, inactivation of SUMO-deconjugating system.
In the present study, we provide evidence that the SUMOylation of pontin is important for maintaining and exerting a subset of nuclear receptor-dependent transcriptional activation processes, and SUMOylated pontin further led to an increase in proliferation and growth of prostate cancer cells. We speculate that the SUMOylation status of certain proteins is a crucial modulator of cancer progression, and determining the upstream signal for the SUMO modification of these proteins may shed light on the role of SUMO modification in human disease settings. The elucidation of the biological importance of protein SUMOylation and their roles in various disease states will provide tremendous information for understanding disease and developing therapeutic reagents.
Materials and Methods
Reagents.
The following antibodies were obtained from Santa Cruz Biotechnology: anti-β-catenin (E-5), Tip60 (N-17), Ubc9 (N-15), and SUMO (D-11, FL-101). Anti-p400 (ab5201) antibody was purchased from Abcam. DHT was from Sigma, and Lipofectamine 2000 reagent was obtained from Invitrogen.
Plasmids and shRNAs.
To generate the SUMO-pontin wild-type or K225R fusion constructs, SUMO (amino acids 1–96) lacking the C-terminal Gly-Gly was amplified by PCR using the primers 5′-CAAAGCTTA TGTCTGACCAGGAGGCAAAACCT-3′ and 5′-GTGCGGCCGCCCCCGTTTGTTCCTGATA AACTTC-3′ and subcloned into pCMV2. Wild-type or K225R-mutated pontin was cloned downstream of SUMO, and the resulting SUMO fusion constructs were sequenced to confirm the reading frame. shRNA sequences against pontin and Ubc9 were reported in refs. 18 and 19).
Cell Culture and Luciferase Assays.
293T cells were grown and transiently transfected by using Lipofectamine 2000 reagents (Invitrogen). For luciferase assays, 1 × 105 cells were seeded in phenol red-free DMEM supplemented with 5% charcoal-dextran-stripped FBS for 24 h. Cells were transfected with 500 ng of an ARE-luciferase reporter along with 25 ng of AR and 300 ng of each pontin expression construct. After 24 h of transfection, cells were treated with 20 nM DHT for 24 h, and luciferease activity was measured. Transfection efficiency was normalized by using β-galactosidase expression constructs, and the results were obtained from at least three independent experiments.
ChIP and Two-Step ChIP Assays.
Chromatin immunoprecipitation assays were conducted as described in ref. 19, with a sheared fragment size of ≈300 bp to 1 kb. For PCR, 1 μl from 30 μl of DNA extract and 25–30 cycles of amplification were used. For the two-step ChIP assay, components were eluted from the first immunoprecipitation reaction by incubation with 10 mM dithiothreitol (DTT) at 37°C for 30 min and diluted 1:50 with ChIP dilution buffer [20 mM Tris·HCl (pH 8.1), 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), and 1% Triton X-100] followed by reimmunoprecipitation with the secondary antibody.
In Vivo SUMOylation Assay.
293T cells were transfected with pcDNA-pontin and His6-SUMO. Thirty-six hours after transfection, the cells were lysed in lysis buffer [150 mM NaCl, 25 mM Tris·HCl (pH 7.8), 0.1% Nonidet P-40, and 1 mM EDTA] supplemented with complex protease inhibitor mixture (Roche Molecular Biochemicals) and 0.2% SDS, sonicated briefly, and centrifuged. The clarified extracts were incubated with 15 μl of Ni-NTA agarose beads (Qiagen) for 2 h. The slurry was washed with lysis buffer. After denaturation, proteins were separated by SDS/PAGE, and the existence of pontin was confirmed by immunoblotting.
In Vitro SUMOylation Assay.
In vitro SUMOylation assays were carried out in 10-μl reaction volumes containing 1 μg of recombinant GST-SUMO, 150 ng of purified GST-E1 (SAE1/SAE2), 10 ng of purified His-E2 (Ubc9), and ATP regeneration system [50 mM Tris·HCl (pH 7.6), 5 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate, 3.5 units/ml creatine kinase, and 0.6 units/ml inorganic pyrophosphatase] with 1 μl of [35S]methionine-labeled in vitro-translated pontin prepared by using a TNT T7 Quick-coupled reticulocyte lysate kit (Promega). The reaction products were analyzed by autoradiography.
Cell Transformation Assay.
Anchorage-independent growth of mock-, pontin-, pontin-K225R-, SUMO-pontin-, and SUMO-pontin K225R-expressing LNCaP cells was determined by analyzing cellular growth in semisolid medium. Cells (105) were placed in Iscove's media containing 0.4% noble agar containing 10% FCS. Cells were allowed to grow for 3 weeks in 5% CO2, and the formation of colonies containing >50 cells was analyzed.
Supplementary Material
ACKNOWLEDGMENTS.
We thank I. S. Kim for technical assistance. This work was supported by grants from the National R & D Program for Cancer Control of the Ministry of Health and Welfare and the Molecular and Cellular BioDiscovery Research Program (to S.H.B.); grants from the Korea Research Foundation and the Science Research Center program of the Ministry of Science and Technology and Korea Science and Engineering Foundation (to S.H.B. and K.I.K.); a grant from the Ubiquitome Research Program (to C.H.C. and K.I.K.); Brain Korea 21 fellowships (to J.H.K., J.M.L., H.J.N., H.J.C., J.W.Y, and J.S.L.); and a Seoul Science Fellowship (to J.M.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710343105/DC1.
References
- 1.Baek SH, Rosenfeld MG. Biochem Biophys Res Commun. 2004;319:707–714. doi: 10.1016/j.bbrc.2004.04.169. [DOI] [PubMed] [Google Scholar]
- 2.Feldman BJ, Feldman D. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 3.Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AAK, Miner JN, Diamond MI. Proc Natl Acad Sci USA. 2005;102:9802–9807. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Proc Natl Acad Sci USA. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger SL. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Bauer A, Chauvet S, Huber O, Usseglio F, Rothbächer U, Aragnol D, Kemler R, Pradel J. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 9.Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M. Cell. 2002;111:661–672. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- 10.Kerscher O, Felberbaum R, Hochstrasser M. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 11.Kim KI, Baek SH, Chung CH. J Cell Physiol. 2002;191:257–268. doi: 10.1002/jcp.10100. [DOI] [PubMed] [Google Scholar]
- 12.Kim KI, Baek SH. Mol Cells. 2006;22:247–253. [PubMed] [Google Scholar]
- 13.Bylebyl GR, Belichenko I, Johnson ES. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 14.Lee MH, Lee SW, Lee EJ, Choi SJ, Chung SS, Lee JI, Cho JM, Seol JH, Baek SH, Kim KI, et al. Nat Cell Biol. 2006;8:1424–1431. doi: 10.1038/ncb1512. [DOI] [PubMed] [Google Scholar]
- 15.Mikolajczyk J, Drag M, Békés M, Cao JT, Ronai Z, Salvesen GS. J Biol Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 16.Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M. Mol Cell Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS, Lee MH, Choi SJ, Kim KI, Kim SI, et al. Nat Cell Biol. 2006;8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, et al. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 21.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 22.Johnson ES. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 23.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 24.Ross S, Best JL, Zon LI, Gill G. Mol Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 25.Chalkiadaki A, Talianidis I. Mol Cell Biol. 2005;25:5095–5105. doi: 10.1128/MCB.25.12.5095-5105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang Y, Myers M, Brown M. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 27.Poukka H, Aarnisalo P, Karvonen U, Palvimo JJ, Janne OA. J Biol Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 28.Kokontis JM, Hay N, Liao S. Mol Endocrinol. 1998;12:941–953. doi: 10.1210/mend.12.7.0136. [DOI] [PubMed] [Google Scholar]
- 29.Metzger E, Wissman M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 30.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 32.Mo YY, Yu Y, Theodosiou E, Ee PLR, Beck WT. Oncogene. 2005;24:2677–2683. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.