Abstract
Genomic surveys in humans identify a large amount of recent positive selection. Using the 3.9-million HapMap SNP dataset, we found that selection has accelerated greatly during the last 40,000 years. We tested the null hypothesis that the observed age distribution of recent positively selected linkage blocks is consistent with a constant rate of adaptive substitution during human evolution. We show that a constant rate high enough to explain the number of recently selected variants would predict (i) site heterozygosity at least 10-fold lower than is observed in humans, (ii) a strong relationship of heterozygosity and local recombination rate, which is not observed in humans, (iii) an implausibly high number of adaptive substitutions between humans and chimpanzees, and (iv) nearly 100 times the observed number of high-frequency linkage disequilibrium blocks. Larger populations generate more new selected mutations, and we show the consistency of the observed data with the historical pattern of human population growth. We consider human demographic growth to be linked with past changes in human cultures and ecologies. Both processes have contributed to the extraordinarily rapid recent genetic evolution of our species.
Keywords: HapMap, linkage disequilibrium, Neolithic, positive selection
Human populations have increased vastly in numbers during the past 50,000 years or more (1). In theory, more people means more new adaptive mutations (2). Hence, human population growth should have increased in the rate of adaptive substitutions: an acceleration of new positively selected alleles.
Can this idea really describe recent human evolution? There are several possible problems. Only a small fraction of all mutations are advantageous; most are neutral or deleterious. Moreover, as a population becomes more and more adapted to its current environment, new mutations should be less and less likely to increase fitness. Because species with large population sizes reach an adaptive peak, their rate of adaptive evolution over geologic time should not greatly exceed that of rare species (3).
But humans are in an exceptional demographic and ecological transient. Rapid population growth has been coupled with vast changes in cultures and ecology during the Late Pleistocene and Holocene, creating new opportunities for adaptation. The past 10,000 years have seen rapid skeletal and dental evolution in human populations and the appearance of many new genetic responses to diets and disease (4).
In such a transient, large population, size increases the rate and effectiveness of adaptive responses. For example, natural insect populations often produce effective monogenic resistance to pesticides, whereas small laboratory populations under similar selection develop less effective polygenic adaptations (5). Chemostat experiments on Escherichia coli show a continued response to selection (6), with continuous and repeatable responses in large populations but variable and episodic responses in small populations (7). These results are explained by a model in which smaller population size limits the rate of adaptive evolution (8). A population that suddenly increases in size has the potential for rapid adaptive change. The best analogy to recent human evolution may be the rapid evolution of domesticates such as maize (9, 10).
Human genetic variation appears consistent with a recent acceleration of positive selection. A new advantageous mutation that escapes genetic drift will rapidly increase in frequency, more quickly than recombination can shuffle it with other genetic variants (11). As a result, selection generates long-range blocks of linkage disequilibrium (LD) across tens or hundreds of kilobases, depending on the age of the selected variant and the local recombination rate. The expected decay of LD with distance surrounding a recently selected allele provides a powerful means of discriminating selection from other demographic causes of extended LD, such as bottlenecks and admixture (9, 12).
The important reason for this increase in discrimination is the vastly different genomic scale that LD-based approaches use compared with previous methods (scales of millions of bases rather than thousands of bases). LD methods use polymorphism distance and order information and frequency to search for selection, unlike all previous methods (9, 12). Previous methods, therefore, have difficulty defining selection unambiguously from other population architectures on the kb scale usually examined. On the megabase (Mb) scale examined by LD approaches, however, extensive modeling and simulations indicate that other demographic causes of extensive LD can be discriminated easily from those caused by adaptive selection (9). Further, current LD approaches restrict comparisons to a set of frequencies and inferred allele ages for which neutral explanations are essentially implausible.
Previously, we applied the LD decay (LDD) test to SNP data from Perlegen and the HapMap (13), finding evidence for recent selection on ≈1,800 human genes. We refer to these as ascertained selected variants (ASVs). The probabilistic LDD test searches for the expected decay of adjacent SNPs surrounding a recently selected allele. Importantly, the method is insensitive to local recombination rate, because local rate influences the extent of LD surrounding both alleles, while the method looks for LD differences between alleles. Further, the method relies only on high heterozygosity SNPs for analysis, exactly the type of data obtained for the HapMap project.
The number of ASVs detected encompasses some 7% of human genes and is consistent with the proportion found in another survey using a related approach (12). Because LD decays quickly over time, most ASVs are quite recent (14), compared with other approaches that detect selection over longer evolutionary time scales (15, 16). Many human genes are now known to have strongly selected alleles in recent historical times, such as lactase (17, 18), CCR5 (19, 20), and FY (21). These surveys show that such genes are very common. This observation is surprising: in theory, such strongly selected variants should be rare (2, 3). The observed distribution seems to reflect an exceptionally rapid rate of adaptive evolution.
But the hypothesis that genomic data show a high recent rate of selection must overcome two principal objections: (i) The LDD test might miss older selection and (ii) a high constant rate of adaptive substitution might also explain the large number of ASVs. The first objection is addressed by recalculating the LDD test on a 3-fold larger dataset, because higher SNP density is needed to detect older selected alleles with comparable sensitivity. We test the second objection by considering a constant rate as the null hypothesis then deriving and testing genomic consequences.
Results
Finding Old Alleles.
The original Perlegen and HapMap datasets were relatively small (1.6 million and 1.0 million SNPs, respectively). The low SNP density limited the power of LD methods to detect older selection events, particularly in high-recombination areas of the genome (9). Likewise, a related study of selection (12) was biased toward newer alleles by requiring multiple adjacent SNPs to exhibit extended LD. Older selected alleles, where LDD is more rapid, would be rejected with this approach. Neither of those previous studies (9, 12) attempted to quantitate the numbers of selected events over an extended time frame, but were merely initial searches for recent extended LD at individual alleles, the most sensitive method to detect recent adaptive change. Both found abundant evidence for recent selection.
Therefore, we have now recomputed the LDD test on the newly released 3.9-million HapMap genotype dataset (13). By varying the LDD test search parameters, we can now statistically detect alleles with more rapid LDD (and hence older inferred ages) (9). For all parameters used, the detection threshold was set at an average log likelihood (ALnLH) > 2.6 SD (≥99.5th percentile) from the genome average. Again, this LDD threshold is a stringent cutoff for the detection of genomic outliers, because the high number of selective events are included in the genome average (9). The probabilistic LDD test does not require the calculation of inferred haplotypes (9), so it is not a daunting computational task to calculate ALnLH values for the HapMap 3.9 million SNPs genotyped in 270 individuals: 90 European ancestry (CEU), 90 African (Yoruba) ancestry (YRI), 45 Han Chinese (CHB), and 45 Japanese (JPT).
This analysis uncovered only 12 new SNPs (in six clusters) not originally detected in the CEU population (9) and 466 new SNPs representing 206 independent clusters in the YRI population. A total of 2,803 (CEU), 2,367 (CHB), 2,783 (JPT), and 3,486 (YRI) selection events were found. As noted (9), many inferred selected sites have faster LDD in YRI samples (with older coalescence times), resulting in lower background LD and more previously unobserved variants. The denser HapMap dataset provided better resolution of LDD (i.e., rapid decay can be reliably detected from background LD only with high density). The 3.9-million HapMap dataset discovered more ASVs, but only an incremental increase in the CEU and a (≈7%) increase in YRI values. This finding indicates that most events (defined by the LDD test) coalescing to ages up to 80,000 years ago have been detected, and any ascertainment bias against older selection is very slight within the given frequency range.
Ancient selected alleles are also more likely to be near or at fixation than recent alleles. Just as we excluded rare alleles, we also excluded high-frequency alleles (i.e., >78%) in our age distribution. But the number of such high-frequency alleles provides another test of the hypothesis that the LDD test has missed older events. We modified the LDD test to find these high-frequency “near-fixed” alleles and found only 50 candidates. Other studies have likewise found few near-fixed alleles (22, 23). These studies also show that very few ASVs are shared between HapMap samples; most are population-specific (9, 12). In our data, only 509 clusters are shared between CEU and YRI samples; many of these are likely to have been under balancing selection [supporting information (SI) Appendix]. The small number of near-fixed events and the small number of shared events are strong evidence that the LDD test has not missed a large number of ancient selected alleles.
Allele Ages.
We used a modification of described methods (24–26) to estimate an allele age (coalescence time) for each selected cluster. We focused on the HapMap populations with the largest sample sizes, which were the YRI and CEU samples. Similar results were obtained for the CHB and JPT populations (data not shown).
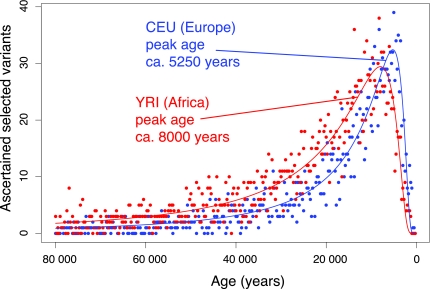
Fig. 1 presents histograms of these age estimates. The YRI sample shows a modal (peak) age of ≈8,000 years ago, assuming 25-year generations; the CEU sample shows a peak age of ≈5,250 years ago, both values consistent with earlier work (9, 12). The difference in peak age likely explains why weaker tests have found stronger evidence of selection in European ancestry samples (27, 28), unlike the current study.
Fig. 1.
Age distribution of ascertained selected alleles. Each point represents the number of variants dated to a single 10-generation bin. Fitted curves are the number of ascertained variants predicted by Eq. 2 under a constant population size and constant s̄ = 0.022 for YRI and s̄ = 0.034 for CEU. The distribution drops to zero approaching the present, because all alleles have frequencies >22% today. The 2,965 (YRI) and 2,246 (CEU) selection ages shown have had 509 alleles removed that are likely examples of ongoing balanced selection (SI Appendix). Including these alleles in the analysis does not change the overall conclusion of acceleration of selection.
Rate Estimation.
Using the diffusion model of positive selection (29), we estimated the adaptive substitution rate consistent with the observed age distribution of ASVs. For the YRI data, this estimate is 0.53 substitutions per year. For the CEU data, this estimate is 0.59 substitutions per year. The average fitness advantage of new variants (assuming dominant effects) is estimated as 0.022 for the YRI distribution and 0.034 for the CEU distribution. Curves obtained by using these estimated values fit the observed data well (Fig. 1). The higher estimated rate for Europeans emerges from the more recent modal age of variants. For further analyses, we used the lower rate estimated from the YRI sample as a conservative value.
Predictions of Constant Rate.
We can derive four predictions from the rate of adaptive substitution, each of which refutes the null hypothesis of constant rate:
The null hypothesis predicts that the average nucleotide diversity across the genome should be vastly lower than observed. Recurrent selected substitutions greatly reduce the diversity of linked neutral alleles by hitchhiking or pseudohitchhiking (30, 31). Using an approximation for site heterozygosity under pseudohitchhiking (30, 32) we estimated the expected site heterozygosity under the null hypothesis as 3.5 × 10−5 (SI Appendix). This value is less than one-tenth the observed site heterozygosity, which is between 4.0 and 6.0 × 10−4 in human populations (13, 33, 34).
Hitchhiking is more important in regions of low recombination, so the null hypothesis predicts a strong relationship between nucleotide diversity and local recombination rate. The null hypothesis predicts a 10-fold increase in diversity across the range of local recombination rates represented by human gene regions. Empirically, diversity is slightly correlated with local recombination rate, but the relationship is weak and may be partly explained by mutation rate (13, 35).
The annual rate of 0.53 adaptive substitutions consistent with the YRI data predicts an implausible 6.4 million adaptive substitutions between humans and chimpanzees. In contrast, there are only ≈40,000-aa substitutions separating these species, and only ≈18 million total substitutions (36). This amount of selection, amounting to >1/3 of all substitutions, or 100 times the observed number of amino acid substitutions, is implausible.
The null hypothesis predicts that many selected alleles should be found between 78% and 100% frequency. Positively selected alleles follow a logistic growth curve, which proceeds very rapidly through intermediate frequencies. Because selected alleles spend relatively little time in the ascertainment range, the ascertained blocks should be the “tip of the iceberg” of a larger number of recently selected blocks at or near fixation. For example, the ASVs in the YRI dataset have a modal age of ≈8,000 years ago. Based on the diffusion model for selection on an additive gene, ascertained variants should account for only 18% of the total number of selected variants still segregating. In contrast, 41% of segregating variants should be >78%. Dominant alleles (which have a higher fixation probability) progress even more slowly (>78%), so that additivity is the more conservative assumption. Empirically, few such near-fixed variants with high LD scores have been found in the human genome (13). Modifying the LDD algorithm to specifically search for high-frequency “fixed” alleles found only 50 potential sites, in contrast to the >5,000 predicted by the constant rate model. Although it is possible that the rapid LDD expected for older selected alleles near fixation may not be detected as efficiently by the LDD test, two other surveys have also found small numbers of such events (22, 23). This difference of two orders of magnitude is a strong refutation of the null hypothesis.
Population Growth.
The rate of adaptive evolution in human populations has indeed accelerated within the past 80,000 years. The results above demonstrate the extent of acceleration: the recent rate must be one to two orders of magnitude higher than the long-term rate to explain the genomewide pattern.
Population growth itself predicts an acceleration effect, because the number of new mutations increases as a linear product of the number of individuals (2), and exponential growth increases the fixation probability of new adaptive mutations (37). We considered the hypothesis that the magnitude of human population growth might explain a large fraction of the recent acceleration of new adaptive alleles. To test this hypothesis, we constructed a model of historic and prehistoric population growth, based on historical and archaeological estimates of population size (1, 38, 39).
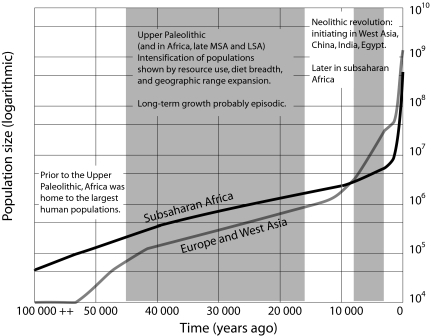
Population growth in the Upper Paleolithic and Late Middle Stone Age began by 50,000 years ago. Several archaeological indicators show long-term increases in population density, including more small-game exploitation, greater pressure on easily collected prey species like tortoises and shellfish, more intense hunting of dangerous prey species, and occupation of previously uninhabited islands and circumarctic regions (40). Demographic growth intensified during the Holocene, as domestication centers in the Near East, Egypt, and China underwent expansions commencing by 10,000 to 8,000 years ago (41, 42). From these centers, population growth spread into Europe, North Africa, South Asia, Southeast Asia, and Australasia during the succeeding 6,000 years (42, 43). Sub-Saharan Africa bears special consideration, because of its initial large population size and influence on earlier human dispersals (44). Despite the possible early appearance of annual cereal collection and cattle husbandry in North Africa, sub-Saharan Africa has no archaeological evidence for agriculture before 4,000 years ago (42). West Asian agricultural plants like wheat did poorly in tropical sun and rainfall regimes, while animals faced a series of diseases that posed barriers to entry (45). As a consequence, some 2,500 years ago the population of sub-Saharan Africa was likely <7 million people, compared with European, West Asian, East Asian, and South Asian populations approaching or in excess of 30 million each (1). At that time, the sub-Saharan population grew at a high rate, with the dispersal of Bantu populations from West Africa and the spread of pastoralism and agriculture southward through East Africa (46, 47). Our model based on archaeological and historical evidence includes large long-term African population size, gradual Late Pleistocene population growth, an early Neolithic transition in West Asia and Europe, and a later rise in the rate of growth in sub-Saharan Africa coincident with agricultural dispersal (Fig. 2).
Fig. 2.
Historic and prehistoric population size estimates for human populations (SI Appendix). Key features are the larger ancestral African population size and the earlier Neolithic growth in core agricultural areas.
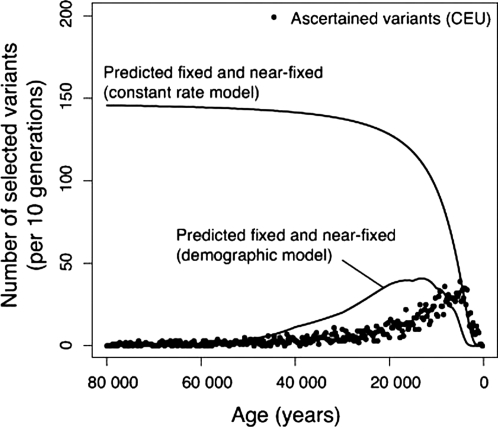
As shown in Fig. 3, the demographic model predicts the recent peak ages of the African and European distributions of selected variants, at a much lower average selection intensity than the constant population size model. In particular, the demographic model readily explains the difference in age distributions between YRI and CEU samples: the YRI sample has more variants dating to earlier times when African populations were large compared with West Asia and Europe, whereas earlier Neolithic growth in West Asia and Europe led to a pulse of recent variants in those regions. The data that falsify the constant rate model, such as the observed genomewide heterozygosity value and the probable number of human–chimpanzee adaptive substitutions, are fully consistent with the demographic model.
Fig. 3.
Tip of the iceberg. Both the demographic and constant-rate models can account for the age distribution of ascertained variants (CEU data shown), but they differ greatly in the expected number of variants above the ascertainment frequency (fixed or near-fixed). The demographic model predicts a low long-term substitution rate and few alleles >78%, consistent with the observed data.
Discussion
Our simple demographic model explains much of the recent pattern, but some aspects remain. Although the small number of high-frequency variants (between 78% and 100%) is much more consistent with the demographic model than a constant rate of change, it is still relatively low, even considering the rapid acceleration predicted by demography. Demographic change may be the major driver of new adaptive evolution, but the detailed pattern must involve gene functions and gene–environment interactions.
Cultural and ecological changes in human populations may explain many details of the pattern. Human migrations into Eurasia created new selective pressures on features such as skin pigmentation, adaptation to cold, and diet (25, 26, 28). Over this time span, humans both inside and outside of Africa underwent rapid skeletal evolution (48, 49). Some of the most radical new selective pressures have been associated with the transition to agriculture (4). For example, genes related to disease resistance are among the inferred functional classes most likely to show evidence of recent positive selection (9). Virulent epidemic diseases, including smallpox, malaria, yellow fever, typhus, and cholera, became important causes of mortality after the origin and spread of agriculture (50). Likewise, subsistence and dietary changes have led to selection on genes such as lactase (18).
It is sometimes claimed that the pace of human evolution should have slowed as cultural adaptation supplanted genetic adaptation. The high empirical number of recent adaptive variants would seem sufficient to refute this claim (9, 12). It is important to note that the peak ages of new selected variants in our data do not reflect the highest intensity of selection, but merely our ability to detect selection. Because of the recent acceleration, many more new adaptive mutations should exist than have yet been ascertained, occurring at a faster and faster rate during historic times. Adaptive alleles with frequencies <22% should then greatly outnumber those at higher frequencies. To the extent that new adaptive alleles continued to reflect demographic growth, the Neolithic and later periods would have experienced a rate of adaptive evolution >100 times higher than characterized most of human evolution. Cultural changes have reduced mortality rates, but variance in reproduction has continued to fuel genetic change (51). In our view, the rapid cultural evolution during the Late Pleistocene created vastly more opportunities for further genetic change, not fewer, as new avenues emerged for communication, social interactions, and creativity.
Materials and Methods
The 3.9-million HapMap release was obtained from the International HapMap Project website (www.hapmap.org). The LDD test (9) was applied to all four HapMap population datasets. Briefly, by examining individuals homozygous for a given SNP, the fraction of inferred recombinant chromosomes (FRC) at adjacent polymorphisms can be directly computed without the need to infer haplotype, a computationally daunting task on such large datasets. The test uses the expected increase with distance in FRC surrounding a selected allele to identify such alleles. Importantly, the method is insensitive to local recombination rate, because local rate will influence the extent of LD surrounding all alleles, while the method looks for LD differences between alleles. By using a large sliding window (ranging from 0.25 to 1.0 Mb in the current study), and by explicitly acknowledging the expected LD structure of selected alleles, the LDD test can distinguish selection from other population genetic/demographic mechanisms, resulting in large LD blocks (9).
A modification of the LDD test was conducted on the CEU and YRI datasets, to find selected alleles near fixation. Unlike the normal LDD test, all SNPs >78% frequency (the cutoff used for primary analysis of this data) were queried, using the same sliding windows as the normal test. Unlike the standard test, however, the requirement that the alternative allele be no more than 1 SD from the genome average was not implemented (9). Ninety-three clusters were identified in the CEU population and 85 were identified in the YRI population (with 65 overlaps), a total of 113 fixed events. Unlike normal LDD screens (9), half of these observed fixed events determined by long-range LD were in extreme centromeric or telomeric regions, which have no recombination or high recombination, respectively (13, 52). The interpretation of extended LD in these regions is ambiguous, therefore, because low recombination maintains large LD blocks (centromeres), and well documented high telomere–telomere exchange homogenizes these regions (52). Removing these centromeric and telomeric regions in which LD is likely to be the result of mechanisms different from selection yields ≈50 regions of potential fixation.
Clustering.
The LDD test produces “clusters” of SNPs with the signature of selection, because of the extensive LD surrounding these alleles (9). Each cluster is likely to represent a single selection event, and hence we have attempted to minimize potential overcounting by cluster analysis. Using a simple nearest-neighbor technique, we assign a 10-kb radius to each selected SNP. Each pass through the data produces a new set of centroids, and cluster membership is reassigned to the nearest centroid. A SNP that lies >20 kb away from the nearest centroid is considered a new cluster, with it being the sole member. Using larger window sizes (up to 100 kb) reduces the number of independent clusters (by approximately half), however, at the cost of “fusing” likely independent events (data not shown). We believe the 10-kb window, therefore, is a conservative first-pass clustering of the observed selection events.
Each selected SNP identified by the LDD test was sorted and mapped to its physical location on human chromosomes (University of California Santa Cruz Human Genome 17). We iterate through the SNP list, starting with the most distal, and a SNP and its closest neighbor (within 10 kb radius) are clustered together with a new centroid (average) i computed. To be included as part of the ith cluster, the next SNP on the sorted SNP list must fall within 20 kb of the ith cluster. If it is within 20 kb of both an upstream and downstream cluster, to be integrated in the ith cluster it must have a distance to the ith centroid closer than the next closest centroid (i + 1). Otherwise, a new centroid and cluster is initiated. This task is repeated for all SNPs identified by the LDD test.
Allele Age Calculations.
Coalescence times (commonly referred to as allele ages) were calculated by methods described (24–26). Briefly, information contained in neighboring SNPs and the local recombination frequency is used to infer age. The genotyped population is binned (at the SNP under inferred selection, the target SNP) into the major and minor alleles (9). While every neighboring SNP gives information on the age of the target SNP, a single recombination event carries all of the downstream neighbors to an equal or higher FRC. Hence, our algorithm moves away (positively and negatively) from the target SNP and computes allele age only when a higher FRC level is reached in a neighboring SNP. A single neighboring SNP with no neighbors within 20 kb is not used for computation. This method is consistent with the theoretical and experimental expectations of LDD surrounding selected alleles (9).
For neighboring SNPs, allele age is computed by using:
where t = allele age (in generations), c = recombination rate (calculated at the distance to the neighboring SNP), xt = frequency in generation t, and y = frequency on ancestral chromosomes. This method is a method-of-moments estimator (24), because the estimate results from equating the observed proportion of nonrecombinant chromosomes with the proportion expected if the true value of t is the estimated value. It requires no population genetic or demographic assumptions, only the exponential decay of initially perfect LD because of recombination. Estimates are obtained until FRC reaches 0.3, to avoid allele age calculations of lower reliability. We assume the ancestral allele is always the allele with neutral or genome average LDD ALnLH scores (9). Average regional recombination rates were obtained by querying data from ref. 53 in the University of California Santa Cruz database (http://genome.ucsc.edu). Regions with <0.1 cM/Mb average recombination rate were excluded. All allele age estimates are averages of the individual calculations at the target SNP (26).
Estimating the Rate of Adaptive Substitutions.
Under the null hypothesis of a constant rate of adaptive substitution, the age distribution of ASVs can estimate the mean fitness advantage (s̄) of new selected variants. The empirical distribution of fitness effects of adaptive substitutions is not known. On theoretical grounds, this distribution is expected to approximate a negative exponential (3). Other studies have assumed this distribution or a gamma distribution with similar shape (54–56), and selected mutations in laboratory organisms appear to fit this theoretical model (57, 58). In these expressions, s is the selection coefficient favoring a new mutation, and s̄ is the mean selection coefficient among the set of all advantageous mutations. We assume that adaptive alleles are dominant in effect, which allows the highest fixation probability (59) and the most rapid increase in frequencies and is therefore conservative (less dominance requires a higher substitution rate to explain the observed distribution). The value of s̄ is not known, and we are concerned with finding the single value that creates the best fit of the population size prediction to the observed data. We assumed a negative exponential distribution of s, in which Pr[s] = e−s/s̄. The number of ascertained new adaptive variants originating in any single generation t is given by the equation:
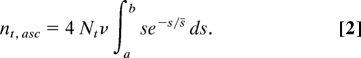 |
Here, ν is the rate of adaptive mutations per genome per generation, and Nt is the effective population size in generation t. This integral derives from the expectation of adaptive mutations in a diploid population (here, 2Nν) multiplied by the fixation probability 2s for each, again assuming dominant fitness effect. Under the null hypothesis, the population size Nt is constant across all generations, so the expected number of new adaptive mutations (ascertained and nonascertained) is likewise constant.
We considered the range of s between value a, yielding a current mean frequency of 0.22, and value b, yielding a current mean frequency of 0.78, as derived from the diffusion approximation for dominant advantageous alleles (60). The parameter ν is constant in effect across all generations, while the number of ascertained variants originating in each generation varies with the range of s placing new alleles in the ascertainment range. We applied a hill-climbing algorithm to find the best-fit value of s̄ for the empirical distribution of block ages, allowing ν to vary freely. With an estimate for s̄, the rate of adaptive mutations, ν, can be estimated as the value that satisfies Eq. 2. This value is also sufficient to estimate the expected number of substitutions per generation, which is the value of the integral in Eq. 2 over the range 0 to infinity (in our analyses, the vast majority had 0.01 ≤ s ≤0.1). For the YRI data, assuming dominant fitness effects, the resulting estimate of adaptive substitution rate is 13.25 per generation, or 0.53 per year.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Alan Fix, Dennis O'Rourke, Kristen Hawkes, Alan Rogers, Chad Huff, Milford Wolpoff, Balaji Srinivasan, and five anonymous reviewers for comments and discussions. This work was supported by grants from the U.S. Department of Energy, the National Institute of Mental Health, and the National Institute of Aging (to R.K.M.), the Unz Foundation (to G.M.C.), the University of Utah (to H.C.H.), and the Graduate School of the University of Wisconsin (to J.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707650104/DC1.
References
- 1.Biraben J-N. Population Sociétés. 2003;394:1–4. [Google Scholar]
- 2.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon; 1930. [Google Scholar]
- 3.Orr HA. Genetics. 2003;163:1519–1526. doi: 10.1093/genetics/163.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armelagos GJ, Harper KN. Evol Anthropol. 2005;14:68–77. doi: 10.1002/evan.20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roush RT, McKenzie JA. Annu Rev Entomol. 1987;32:361–380. doi: 10.1146/annurev.en.32.010187.002045. [DOI] [PubMed] [Google Scholar]
- 6.Lenski RE, Travisano M. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wick LM, Weilenmann H, Egli T. Microbiology. 2002;148:2889–2902. doi: 10.1099/00221287-148-9-2889. [DOI] [PubMed] [Google Scholar]
- 8.Wahl LM, Krakauer DC. Genetics. 2000;156:1437–1448. doi: 10.1093/genetics/156.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang ET, Kodama G, Baldi P, Moyzis RK. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. Science. 2006;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Nielsen R. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voight BF, Kudaravalli S, Wen X, Pritchard JK. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przeworski M. Genetics. 2001;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 16.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollox EJ, Poulter M, Zvarek M, Ferak V, Krause A, Jenkins T, Saha N, Kozlov AI, Swallow DM. Am J Hum Genet. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novembre J, Galvani AP, Slatkin M. PLoS Biol. 2005;3:e339. doi: 10.1371/journal.pbio.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabeti PC, Walsh E, Schaffner SF, Varilly P, Fry B, Hutcheson HB, Cullen M, Mikkelsen TS, Roy J, Patterson N, et al. PLoS Biol. 2005;3:e378. doi: 10.1371/journal.pbio.0030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamblin MT, Thompson EE, Di Rienzo A. Am J Hum Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson S, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura R, Fujimoto A, Tokunaga K, Ohashi J. PLoS One. 2007;2:e286. doi: 10.1371/journal.pone.0000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slatkin M, Rannala B. Annu Rev Genom Hum Genet. 2000;1:225–249. doi: 10.1146/annurev.genom.1.1.225. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y-C, Chi H-C, Grady DL, Morishima A, Kidd JR, Kidd KK, Flodman P, Spence MA, Schuck S, Swanson JM, et al. Proc Natl Acad Sci USA. 2002;99:309–314. doi: 10.1073/pnas.012464099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang E, Ding Y-C, Flodman P, Kid JR, Kidd KK, Grady DL, Ryder OA, Spence MA, Swanson JM, Moyzis RK. Am J Hum Genet. 2004;74:931–944. doi: 10.1086/420854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kayser M, Brauer S, Stoneking M. Mol Biol Evol. 2003;20:893–900. doi: 10.1093/molbev/msg092. [DOI] [PubMed] [Google Scholar]
- 28.Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright S. The Theory of Gene Frequencies, Evolution and the Genetics of Populations. Vol 2. Chicago: Univ Chicago Press; 1969. [Google Scholar]
- 30.Gillespie JH. Genetics. 2000;155:909–919. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y. Genetics. 2006;172:1967–1978. doi: 10.1534/genetics.105.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betancourt AJ, Kim Y, Orr HA. Genetics. 2004;168:2261–2269. doi: 10.1534/genetics.104.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Fan J, Siao C, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1081. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 34.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han J-H, et al. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 35.Hellmann I, Ebersberger I, Ptak SE, Pääbo S, Przeworski M. Am J Hum Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 37.Otto SP, Whitlock MC. Genetics. 1997;146:723–733. doi: 10.1093/genetics/146.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coale AJ. Sci Am. 1974;231:40–52. doi: 10.1038/scientificamerican0974-40. [DOI] [PubMed] [Google Scholar]
- 39.Weiss K. Hum Biol. 1984;56:637–649. [PubMed] [Google Scholar]
- 40.Stiner MC, Munro ND, Surovell TA. Curr Anthropol. 2000;41:39–73. [PubMed] [Google Scholar]
- 41.Bar-Yosef O, Belfer-Cohen A. In: Transitions to Agriculture in Prehistory. Gebauer AB, Price TD, editors. Madison, WI: Prehistory Press; 1992. pp. 21–48. [Google Scholar]
- 42.Bellwood P. First Farmers: The Origins of Agricultural Societies. Oxford: Blackwell; 2005. [Google Scholar]
- 43.Price TD, editor. Europe's First Farmers. Cambridge, UK: Cambridge Univ Press; 2000. [Google Scholar]
- 44.Relethford JH. Evol Anthropol. 1999;8:7–10. [Google Scholar]
- 45.Gifford-Gonzalez D. Afr Archaeol Rev. 2000;17:95–139. [Google Scholar]
- 46.Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege JEO. Science. 2002;296:336–339. doi: 10.1126/science.1069878. [DOI] [PubMed] [Google Scholar]
- 47.Diamond J, Bellwood P. Science. 2003;300:597–603. doi: 10.1126/science.1078208. [DOI] [PubMed] [Google Scholar]
- 48.Frayer DW. Am J Phys Anthropol. 1977;46:109–120. doi: 10.1002/ajpa.1330460114. [DOI] [PubMed] [Google Scholar]
- 49.Larsen CS. Annu Rev Anthropol. 1995;24:185–213. [Google Scholar]
- 50.McNeill W. Plagues and Peoples. Garden City, NY: Doubleday; 1976. [Google Scholar]
- 51.Crow JF. BioScience. 1966;16:863–867. [Google Scholar]
- 52.Riethman HC, Xiang Z, Paul S, Morse E, Hu X-L, Flint J, Chi H-C, Grady DL, Moyzis RK. Nature. 2001;409:948–951. doi: 10.1038/35057180. [DOI] [PubMed] [Google Scholar]
- 53.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 54.Keightley PD, Lynch M. Evolution (Lawrence, Kans) 2003;57:683–685. doi: 10.1111/j.0014-3820.2003.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 55.Shaw FH, Geyer CJ, Shaw RG. Evolution (Lawrence, Kans) 2002;56:453–463. doi: 10.1111/j.0014-3820.2002.tb01358.x. [DOI] [PubMed] [Google Scholar]
- 56.Elena SF, Ekunwe L, Hajela N, Oden SA, Lenski RE. Genetica. 1998;102/103:349–358. [PubMed] [Google Scholar]
- 57.Imhof M, Schlötterer C. Proc Natl Acad Sci USA. 2001;98:1113–1117. doi: 10.1073/pnas.98.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kassen R, Bataillon T. Nat Genet. 2006;38:484–488. doi: 10.1038/ng1751. [DOI] [PubMed] [Google Scholar]
- 59.Haldane JBS. Trans Cambridge Philos Soc. 1927;23:19–41. [Google Scholar]
- 60.Ewens WJ. Mathematical Population Genetics. Cambridge, UK: Cambridge Univ Press; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.