Abstract
Ovarian carcinoma (OC) is a leading cause of death among women throughout the world. A number of cancer-associated genes have been shown to be inactivated by hypermethylation of CpG islands during tumorigenesis. We tested the hypothesis that methylation status of MGMT, CDH1, RAR-β and SYK could be important in the ovarian tumorigenic process and can lead to the gene(s) inactivation. Therefore, we assessed the promoter hypermethylation of MGMT, CDH1, RAR-β and SYK in 43 ovarian granulosa cell tumours (GCTs) (adult type) using methylation-specific PCR. These tumours are relatively rare, accounting for approximately 3% of all ovarian cancers. Hypermethylation of MGMT (in 14 tumours), CDH1 (in nine tumours), RAR-β (in eight tumours) and SYK (in seven tumours) have been found. Selective loss of RAR-β and RAR-β2 mRNA has been found in seven patients, while that of MGMT and SYK in three patients who also show aberrant methylation in promoter region of RAR-β in addition to MGMT, SYK and CDH1 genes. Promoter CpG hypermethylation may be an alternative to mutation(s) to inactivate tumour suppressor genes such as MGMT, CDH1, RAR-β and SYK, and this can also be an early event in the pathogenesis of OCs. Moreover, hypermethylation of the MGMT and CDH1, MGMT and RAR-β and CDH1 and RAR-β promoters occurred concordantly (P< 0.001, 0.0421 and 0.0005 respectively; Fischer's exact test). In addition to this, monosomy 22 and trisomy 14 have also been found in 10 tumours. It is clear from the results that hypermethylation of the promoter region of these tumour suppressor genes, monosomy 22 and trisomy 14, may be critical steps in the tumorigenesis, which consequently play a permissive role for tumour aggressiveness. All these events might play an important role in the early clinical diagnosis of the disease. Our results, therefore, suggest a potential role for epigenetic modification of these critical tumour suppressor genes in pathways relevant to the transformation and differentiation of rare type of ovarian cancer (GCTs).
Keywords: ovarian carcinoma, hypermethylation of MGMT, CDH1, RAR-β and SYK gene
Ovarian tumours are relatively common, and are the most lethal tumour of the female genital tract (Evan et al, 1999). Among ovarian cancers, granulosa cell tumours (GCTs) are relatively rare, accounting for approximately 3% of all ovarian cancers. The aetiology of these GCTs remains unclear. It is now accepted that accumulation of multiple genetic aberrations can lead to the development of most carcinomas. DNA replication errors (RER) have been detected in epithelial ovarian cancer, as well as in other human cancer types, and a DNA mismatch repair deficiency may be involved in their development and or progression (Suzuki et al, 2000). Genetic or epigenetic alterations in a variety of genes are fundamental to the processes of growth, cell proliferation, differentiation and programmed cell death and removal. Each alteration may be mediated through gross chromosomal changes and, hence, has the potential to be detected cytogenetically (Sandberg, 1991). It is generally thought that the events causing the activation of certain oncogenes and/or inactivation of tumour suppressor genes lead to tumour development and progression, which is caused by genetic alterations such as chromosome deletions and or loss of function mutations in the coding regions (in CpG sites) of the tumour suppressor genes.
O6-methylguanine-DNA methyltransferase (MGMT; 10q26; GenBank Accession No. U95038) is a DNA repair gene, which removes methyl groups as well as larger adducts at O6 of guanine. The alkylation of DNA at O6 position of guanine is associated with the formation of DNA mutations in cancers. CpG islands within −249 to +259 relative to transcriptional start site were chosen to study the role of promoter methylation on gene silencing. Methylation of MGMT gene has been reported in various carcinomas such as gliomas, non-small-cell lung carcinoma, lymphoma, head and neck carcinoma and colorectal cancers (Esteller et al, 1999b, 2000; Rosas et al, 2001). E-cadherin is an Mr 120 000 transmembrane glycoprotein (16q22.1; GenBank Accession No. D49685) expressed on the surface of epithelial cells and is essential for the maintenance of normal tissue homeostasis and architecture (Takeichi, 1995). Reduced E-cadherin expression has been associated with an unfavourable prognosis of cancers originating from breast, lung, nasopharynx, bladder and gastric mucosa (Zheng et al, 1999; Garcia del Muro et al, 2000; Heimann et al, 2000), and the important mechanism for loss of expression is methylation of the 5′ CpG islands within the promoter, which results in transcriptional repression of the gene (Yoshiura et al, 1995). CpG island in exon 1 of E-cadherin gene (−126 to +144) relative to transcription start site was selected to detect the promoter methylation effects on gene silencing. Retinoic acid receptor beta (RAR-β;3p24.3-24.2) induces local chromatin changes at levels of target genes containing retinoic acid responsive elements (RAREs) by recruiting multiprotein complexes with histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities that dynamically alter chromatin structure and regulate gene expression (Chambon, 1996). A RARE is also located in the RAR-β promoter region and mediates RAR-β induction in response to RA in several cell lines and tissues (de The et al, 1990). Auto regulation of the RAR-β gene may play a critical role in amplifying the RA response. Lack of RAR-β expression has been observed in a number of malignant tumours, including carcinomas of the lung (Gebert et al, 1991), head and neck (Lotan et al, 1995) and breast (Seewaldt et al, 1995). The overexpression of RAR-β induced growth arrest and apoptosis in oral cancer cell lines, and could function as a tumour suppressor (Hayashi et al, 2001). For RAR-β gene, methylation primers were designed as per the gene sequence (position 773–1007; transcription start site at position 844) to investigate the role of promoter hypermethylation in gene silencing. SYK gene (9q22; GenBank Accession No. Z29630) encodes a protein tyrosine kinase, Syk, which is highly expressed in haematopoietic cell, especially in T-cell and B-cell development and activation (Chu et al, 1998). Loss of Syk expression seems to be associated with malignant phenotype, such as increased motility and invasion (Coopman et al, 2000). Exon 1 (107 bp) and exon 2 (448 bp) are separated by ∼4.1 kb intron and the transcription start site is located in exon 2. CpG island spans from 350 bp 5′ of exon 1 to 150 bp of intron 1. A 600 bp region around exon 1 contains 62 CpG sites. Therefore, the high density of CpG sites in the 5′-regulatory region suggested that promoter methylation may influence the transcriptional regulation of SYK expression. Methylation-specific primers were chosen to cover nine CpG numbered 17–21 (forward) and 47–50 (reverse), whereas unmethylation-specific primers covered eight CpGs numbered 18–22 (forward) and 35–37 (reverse). Methylation of CpG sites in the promoter region has become recognised as an alternative mechanism for the inactivation of certain tumour suppressor genes, which is thought to be the most common mechanism of gene inactivation in various tumours.
Therefore, all these above findings led us to investigate the role of promoter hypermethylation of MGMT, CDH1, RAR-β and SYK genes, and mRNA expression of RAR-β and RAR-β2 genes in GCTs of ovaries.
MATERIAL AND METHODS
The subjects were 43 patients affected with GCTs of ovaries without a positive family history. All of them were untreated at the time of the study. The tumour grading is as follows: 19 FIGO stage IA, 10 FIGO stage IB and 14 FIGO stage IC. All these diagnoses were reviewed by a gynaecologic pathologist, and the tumours were assessed using standard criteria (Russel and Bannatyne, 1989). An informed consent was taken from all the subjects prior to the study. This study was approved by the Health Research Ethics Board of the Faculty of Natural Sciences. In all, 25 G-banded metaphases were obtained after a 5-day-old culture of an overnight collagenase disaggregated specimen of the tumour.
Methylation-Specific PCR (MSP)
DNA methylation patterns in CpG islands of tumour suppressor genes MGMT, CDH1, RAR-β and SYK were determined by chemical modification with sodium bisulphite. Briefly, 1 μg DNA was denatured by NaOH (50 μl, final concentration, 0.2 M) for 10 min at 37°C; 1 μg of salmon sperm DNA (Sigma) was added as carrier before modification. Freshly prepared 30 μl of hydroquinone (10 mM, Sigma) and 520 μl of sodium bisulphite (3 M, pH 5.0, Sigma) were mixed and the samples were incubated under mineral oil at 55°C for 16 h. The DNA samples were desalted through Wizard columns (Promega, Madison, WI, USA), then desulphonated by NaOH (final concentration, 0.3 M) treatment for 5 min at room temperature, followed by ethanol precipitation. DNA was resuspended in water and used immediately or stored at −20°C. PCR primers that distinguish between these methylated and unmethylated DNA sequences were used. Primer sequences of all genes for both the methylated and the unmethylated form, annealing temperatures and the expected PCR product sizes are summarised in Table 1 . For PCR amplification, 2 μl of bisulphite-modified DNA was added in a final volume of 25 μl of PCR mixture containing 1 × PCR buffer, MgCl2, deoxynucleotide triphosphates, and primers (100 pmol each per reaction) and 1 U of AmpiTaq Gold (Applied Biosystems, NJ, USA). Amplification was carried out in a 9700 Perkin–Elmer thermal cycler under the following conditions: 95°C for 10 min; 35 cycles of 95°C for 45 s, the specific annealing temperature for each gene for 1 min, and 72°C for 60 s; followed by a final 10 min extension at 72°C. Following sequencing, the primer pair was used to amplify bisulphite-modified DNA-containing gene for E-cadherin : (forward) 5′-GTT TAG TTT TGG GGA GGG GTT-3′ and (reverse) 5′-ACT ACT ACT CCA AAA ACC CAT AAC TAA-3′. The PCR cycle consisted of initial denaturation at 95°C for 5 min, 30 cycles (95°C × 30 s, 50°C × 30 s and 72°C × 30 s) followed by a final extension at 72°C for 5 min. The PCR product was then diluted 1 : 50 with sterile distilled water and 2 μl of this 1 : 50 diluted product was then used for nested PCR using methylated and unmethylated primer pairs. Similarly, the following sequencing primer pair was used to amplify bisulphite-modified DNA-containing gene for SYK: (forward) 5′-GAT TAA GAT ATA TTT TAG GGA ATA TG-3′ and (reverse) 5′-CAC CTA TAT TTT ATT CAC ATA ATT TC-3′. The PCR cycle consisted of hot start at 95°C × 5 min and 35 cycles (95°C × 45 s, 58°C × 50 s and 72°C × 45 s) followed by a final extension at 72°C for 5 min. The PCR product was then diluted 1 : 50 with sterile distilled water and 2 μl of this 1 : 50 diluted product was then used for nested PCR using methylated and unmethylated primer pairs. Each PCR product (10 μl) was directly loaded onto 6% nondenaturing polyacrylamide gels, stained with ethidium bromide and directly visualised under UV illumination. The MSP for all samples was repeated twice to confirm their methylation status.
Table 1. Summary of primer sequences, annealing temperature and PCR product sizes used for MSP.
| S. no. | Name and type | Primer sequence | PCR conditions | Cycles | bp |
|---|---|---|---|---|---|
| 1 | MGMT-Met: F | 5′-TTT CGA CGT TCG TAG GTT TTC GC-3′ | 95°C × 30s; 61°C × 45s; 72°C × 45 s | 35 | 81 |
| R | 5′-GCA CTC TTC CGA AAA CGA AAC G-3′ | ||||
| 2 | MGMT-UnM: F | 5′-TTT GTG TTT TGA TGT TTG TAG GTT TTT GT-3′ | 95°C × 30s; 61°C × 45s; 72°C × 45 s | 35 | 93 |
| R | 5′-AAC TCC ACA CTC TTC CAA AAA CAA AAC A-3′ | ||||
| 3 | CDH1-Met: F | 5′-TGT AGT TAC GTA TTT ATT TTT AGT GGC GTC-3′ | 95°C × 30s; 64°C × 30s; 72°C × 30 s | 40 | 112 |
| R | 5′-CGA ATA CGA TCG AAT CGA ACC G-3′ | ||||
| 4 | CDH1-UnM: F | 5′-TGG TTG TAG TTA TGT ATT TAT TTT TAG TGG CGT C-3′ | 95°C × 30s; 62°C × 30s; 72°C × 30 s | 40 | 120 |
| R | 5′-ACA CCA AAT ACA ATC AAA TCA AAC CAA A-3′ | ||||
| 5 | RARβ-Met: F | 5′-GGT TAG TAG TTC GGG TAG GTT TTA TC-3′ | 95°C × 30s; 58°C × 30s; 72°C × 30 s | 35 | 235 |
| R | 5′-CCG AAT CCT ACC CCG ACG-3′ | ||||
| 6 | RARβ-UnM: F | 5′-TTA GTA GTT TGG GTA GGG TTT ATT-3′ | 95°C × 30s; 58°C × 30s; 72°C × 30 s | 35 | 233 |
| R | 5′-CCA AAT CCT ACC CCA ACA-3′ | ||||
| 7 | SYK-Met: F | 5′-CGA TTT CGC GGG TTT CGT TC-3′ | 95°C × 30s; 67°C × 30s; 72°C × 30 s | 35 | 243 |
| R | 5′-AAA ACG AAC GCA ACG CGA AAC-3′ | ||||
| 8 | SYK-UnM: F | 5′-ATT TTT GTG GGT TTT GTT TGG TG-3′ | 95°C × 30s; 67°C × 30s; 72°C × 30 s | 35 | 140 |
| R | 5′-ACT TCC TTA ACA CAC CCA AAC-3′ |
Statistical differences were assessed with Fisher's exact test by InStat 3 Windows software. Two-sided test were used to determine the significance. P-values less than 0.05 were regarded as statistically significant.
RT–PCR
RAR-β and RARβ-2
Total RNA was prepared using RNeasy mini-kit (Qiagen, CA, USA) as per the manufacturer's recommendations. In brief, total RNA was extracted from the specific cancerous tissues. Cell lysates were then homogenised by passing repeatedly through a 23-gauge needle followed by spinning through a QIAshredder (Qiagen, CA, USA). RNA concentrations were determined by measuring the absorbance (260 nm UV) using a spectrophotometer (Pharmacia). Total RNA (2 μg) was used for the generation of cDNAs using Superscript reverse transcriptase (GIBCO BRL, Gaithersburg, MD, USA). The following primers were used to detect RAR-β and RAR-β2 transcripts: RAR-β forward 5′-ACC AGC TCT GAG GAA CTC GTC CCA-3′ and RAR-β reverse 5′-AGG CGG CCT TCA GCA GGG TAA TTT-3′, and for RAR-β2 forward (in exon 3), 5′-GCA TGG CAG AGT GCC CTA TC-3′; reverse (in exon 6), 5′-TCC CAG AGT CAT CCC TGC TTC AT-3′. PCR amplification was performed for 30 cycles at 95°C for 30 s, 60°C for 30 s (62°C for RAR-β2) and 72°C for 45 s (60 s for RAR-β2). Human GAPDH was used as an internal control.
MGMT gene
The following primer sequences were used for MGMT gene sense, 5′-CCTGGCT GAATGCCTATTTC-3′, and anti-sense, 5′-CAGCTTCCATAACACCTGTCTG-3′, which amplifies 116-bp PCR product. To verify the integrity of cDNA, β2-microglobulin expression was also analysed using the primer sequences sense, 5′-CATCCAGCGTACTCCAAAGA-3′, and β2-microglobulin anti-sense, 5′-GACAAGTCTGAATGCTCCAC-3′, which amplifies 165-bp PCR product. These two sets of primers span junctions between two exons, so amplification of the contaminating genomic DNA can be excluded. PCR amplification was performed for 94°C for 10 min, then 35 cycles of 94°C for 45 s, 57°C for 60 s and 72°C for 30 s, followed by a final elongation step at 72°C for 10 min.
SYK gene
The following primer sequences were used for SYK gene sense, 5′-TGTCAAGGATAAGAACATCATAG-3′, and anti-sense, 5′-CACCACGTCAT AGTAGTAATTG-3′, which amplifies 507-bp PCR product. In this PCR, β2-microglobulin was also used as an internal control. The optimised PCR program was 94°C for 5 min, then 35 cycles of 94°C for 40 s, 62°C for 40 s and 72°C for 45 s, followed by a final elongation step at 72°C for 7 min.
The PCR products were subjected to electrophoresis in 2.0% agarose gels and visualised by ethidium bromide staining.
RESULTS
Cytogenetic analysis performed on these tumours exhibited a 46,XX normal karyotype in 33 tumours. However, trisomy 14 and monosomy 22 were found in only 10 tumours (Table 2 ).
Table 2. Promoter hypermethylation in different genes in ovarian cancer.
|
Promoter hypermethylation |
Expression of |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. no. | Age (years) | Stage | MGMT | CDH1 | RAR-β | SYK | RAR-β | RAR-β2 | MGMT | SYK | Trisomy 14 | Monosomy 22 |
| 1 | 45 | IA | + | + | + | − | − | − | + | + | Yes | Yes |
| 2 | 55 | IA | − | − | − | − | + | + | + | + | No | No |
| 3 | 51 | IA | − | − | + | + | + | + | + | + | No | No |
| 4 | 65 | IA | − | − | − | + | + | + | + | − | No | No |
| 5 | 38 | IA | + | − | − | − | + | + | + | + | Yes | Yes |
| 6 | 36 | IA | + | − | − | + | + | + | + | + | No | No |
| 7 | 75 | IA | − | − | + | − | + | − | + | + | Yes | Yes |
| 8 | 81 | IA | − | − | − | − | + | + | + | + | No | No |
| 9 | 38 | IA | − | − | − | − | + | + | + | + | No | No |
| 10 | 47 | IA | − | − | − | − | + | + | + | + | No | No |
| 11 | 49 | IA | − | − | − | − | + | + | + | + | No | No |
| 12 | 64 | IA | + | + | + | − | − | − | − | + | Yes | Yes |
| 13 | 39 | IA | − | − | − | − | + | + | + | + | No | No |
| 14 | 42 | IA | − | − | − | − | + | + | + | + | No | No |
| 15 | 48 | IA | + | + | + | − | − | − | + | + | Yes | Yes |
| 16 | 53 | IA | − | − | − | − | + | + | + | + | No | No |
| 17 | 58 | IA | − | − | − | − | + | + | + | + | No | No |
| 18 | 61 | IA | + | + | + | + | − | − | − | − | Yes | Yes |
| 19 | 33 | IA | − | − | − | − | + | + | + | + | No | No |
| 20 | 52 | IB | − | − | − | − | + | + | + | + | No | No |
| 21 | 47 | IB | − | − | − | − | + | + | + | + | No | No |
| 22 | 51 | IB | − | − | − | − | + | + | + | + | No | No |
| 23 | 39 | IB | − | − | − | − | + | + | + | + | No | No |
| 24 | 49 | IB | − | − | − | − | + | + | + | + | No | No |
| 25 | 66 | IB | + | + | + | + | − | − | − | − | Yes | Yes |
| 26 | 62 | IB | − | − | − | − | + | + | + | + | No | No |
| 27 | 73 | IB | + | + | + | − | − | − | + | + | Yes | Yes |
| 28 | 58 | IB | + | + | − | − | + | + | + | + | No | No |
| 29 | 52 | IB | − | − | − | − | + | + | + | + | No | No |
| 30 | 55 | IC | − | − | − | − | + | + | + | + | No | No |
| 31 | 59 | IC | − | − | − | − | + | + | + | + | No | No |
| 32 | 62 | IC | − | − | − | + | + | + | + | + | No | No |
| 33 | 44 | IC | + | − | − | + | + | + | + | + | Yes | Yes |
| 34 | 42 | IC | − | − | − | − | + | + | + | + | No | No |
| 35 | 37 | IC | + | − | − | − | + | + | + | + | No | No |
| 36 | 66 | IC | − | − | − | − | + | + | + | + | No | No |
| 37 | 69 | IC | + | − | − | − | + | + | + | + | No | No |
| 38 | 61 | IC | − | − | − | − | + | + | + | + | No | No |
| 39 | 37 | IC | − | − | − | − | + | + | + | + | No | No |
| 40 | 45 | IC | + | + | + | + | − | − | + | + | Yes | Yes |
| 41 | 48 | IC | − | − | − | − | + | + | + | + | No | No |
| 42 | 71 | IC | − | − | − | − | + | + | + | + | No | No |
| 43 | 38 | IC | + | + | − | − | + | + | + | + | No | No |
+=hypermethylated; −=unmethylated; +=mRNA expression present; −: mRNA expression lost.
MGMT, CDH1, RAR-β and SYK promoter hypermethylation
The methylation status of ovarian tumours (GCTs) was determined at the respective loci using MSP. DNA from 43 ovarian tumours was modified using sodium bisulphite, which converts all unmethylated cytosine residues to uracil, but leaves methylated cytosines unchanged. To confirm that the modification was successful, all samples were first amplified with primers specific for unmethylated DNA at the p16 or MLH1 loci (even tumours methylated at these loci would be expected to be positive due to contamination with normal tissue, which is known to be unmethylated). The samples were next subjected to MSP using primers specific for methylated DNA at the respective loci being studied.
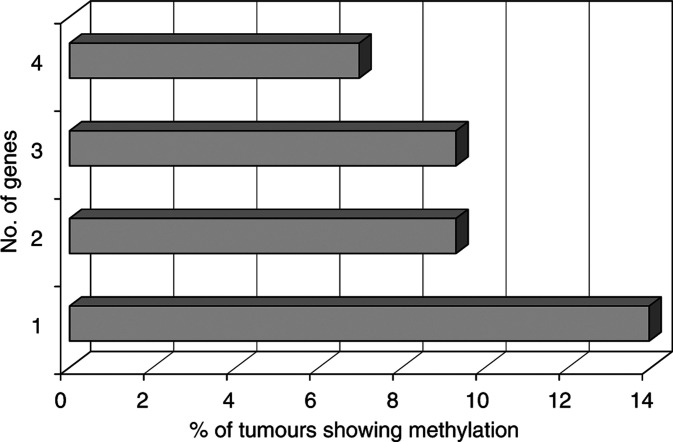
At least one of these four genes showed aberrant methylation in 17 tumour samples (17 out of 43). Methylation of only one gene was found in 13.95% (6 of 43) of tumours. The percentage of the tumours with methylation in genes 2, 3 and 4 is 9.3% (4 of 43), 9.3% (4 of 43) and 6.97% (3 of 43), respectively (Figure 1). Three tumours were found to be hypermethylated for four tumour suppressor genes (Table 2) investigated in the present study. The representative examples of MSP are shown in Figure 2.
Figure 1.
Percentage of distribution of the number of genes methylated in ovarian carcinoma.
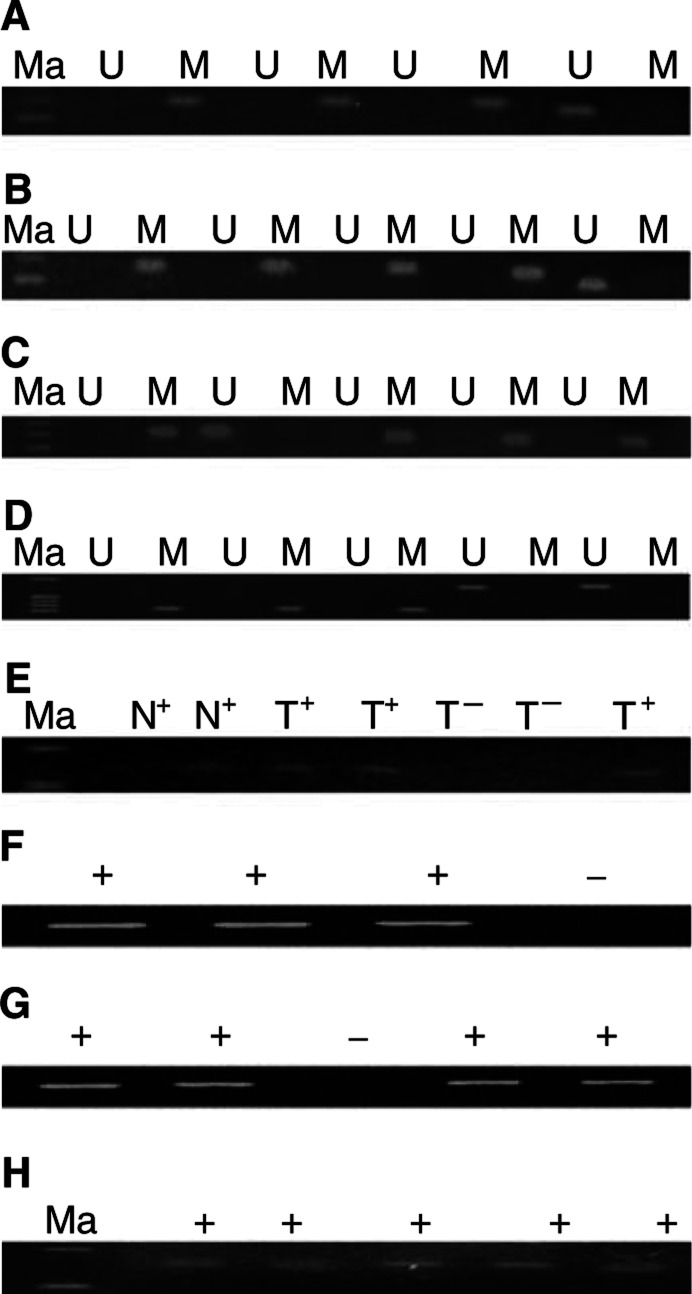
Figure 2.
Representative examples of MSP analysis of MGMT, CDH1, RAR-β and SYK genes in GCTs of ovarian origin. (A) Unmethylated and methylated MGMT PCR products were detected in one and three cases. (B) Unmethylated and methylated CDH1 PCR products were detected in one and four cases. (C) Unmethylated and methylated RAR-β products were detected in one and four cases. (D) Unmethylated and methylated SYK PCR products were detected in two and three cases, respectively. (E) Gene expression of RAR-β in GCTs of ovaries (F) loss of gene expression of MGMT gene in one patient (G) loss of gene expression of SYK gene in one patient and (H) GADPH as internal control (+) for expression studies: U, unmethylated PCR products; M, methylated PCR products; Ma, Molecular weight marker; N+, gene expression in lymphocytes; T+, gene expression in cancerous tissue; T−, loss of expression in cancerous tissues.
We compared hypermethylation of MGMT, CDH1, RAR-β and SYK promoters in ovarian carcinoma (OC). As shown in Table 3 , we found concordant hypermethylation of MGMT and CDH1 promoters (0.001), and MGMT and RAR-β promoters (0.0421). Similarly, hypermethylation of CDH1 and RAR-β promoters (0.0005) was also found. However, we found no correlation between the methylation status of MGMT and SYK promoters, CDH1 and SYK promoters or RAR-β and SYK promoters. There was no significant correlation between the DNA methylation status and clinical/pathological parameters with respect to age, tumour size and pathological grading.
Table 3. Correlation of promoter methylation of MGMT, CDH1, RARb and SYK in ovarian carcinomas.
| Methylated | Unmethylated | P-value | |
|---|---|---|---|
| MGMT methylation status | |||
| CDH1 methylation status | |||
| Methylated | 9 | 0 | 0.001* |
| Unmethylated | 5 | 11 | |
| RAR-β methylation status | |||
| Methylated | 7 | 1 | 0.0421* |
| Unmethylated | 7 | 10 | |
| SYK methylation status | |||
| Methylated | 4 | 2 | 0.6609 |
| Unmethylated | 10 | 9 | |
| CDH1 methylation status | |||
| RAR-β methylation status | |||
| Methylated | 7 | 1 | 0.0005* |
| Unmethylated | 2 | 15 | |
| SYK methylation status | |||
| Methylated | 3 | 4 | 1 |
| Unmethylated | 6 | 11 | |
| RAR-β methylation status | |||
| SYK methylation status | |||
| Methylated | 2 | 5 | 1 |
| Unmethylated | 6 | 12 | |
Significant at P< 0.05.
RT–PCR for RAR-β, RAR-β2, MGMT and SYK gene expression study
To examine the biological role of promoter hypermethylation in GCTs of ovaries, we assessed the levels of gene expression by semiquantitative RT–PCR in tumours with known methylation status in RAR-β gene. Both RAR-β and RAR-β2 were found to have negative expression of mRNA in seven patients with OC showing promoter hypermethylation in RAR-β gene. Another patient was essentially negative for RAR-β2 mRNA, although it shows aberrant methylation in the RAR-β promoter region. All other patients of OC patients showed normal expression of both RAR-β and RAR-β2 and were not methylated (Table 2). The data suggest that promoter hypermethylation leading to gene silencing may affect a variety of key pathways in germ cell tumorigenesis. Aberrant promoter methylation changes that occur in cancer are associated with transcriptional repression and loss of function of the gene by interrupting the binding of proteins involved in the transcription activator complex. Our gene expression analysis by RT–PCR demonstrated that all tumours that showed methylation of RAR-β genes also showed downregulation of mRNA levels in the methylated tumours. Thus in these cases, promoter hypermethylation is one mechanism whereby gene expression can be deregulated in GCTs. We also assessed the levels of gene expression in MGMT and SYK genes by semiquantitative RT–PCRs in these patients. Only three patients showed negative expression of mRNA in these genes. Two patients showed promoter hypermethylation in all four tumour suppressor genes and negative expression of mRNA in RARβ, MGMT and SYK.
DISCUSSION
Monosomy 22 and trisomy 14 appear to be emerging as nonrandom chromosome abnormalities in this type of tumours, although it is also associated with other complex chromosome abnormalities. In light of the data, the present finding of monosomy 22 as the sole chromosome change in these tumours suggests that this karyotypic change, possibly followed by the acquisition of an extra chromosome 14 (trisomy), may be nonrandom but an early, nonobligatory event of the tumorigenesis of GCTs of the ovary (Dal Cin et al, 1997; Speleman et al, 1997, Van den Berghe et al, 1999). The loss of fidelity in the inherent order of replication of allelic pairs during cell division provides a source for generating mutations involving genetic (aneuploidy) and epigenetic (gene silencing or allelic inactivation) events required for the generation and establishment of malignant phenotype (Duesberg et al, 2000). As reported in previous studies and in the present findings, monosomy of chromosome 22 and trisomy 14 provide a prognostic factor for the disease (Speleman et al, 1997). This emphasizes the association between MGMT, CDH1, SYK and RARβ methylation and aneuploidy. Genome-wide hypomethylation due to reduced levels of DNA methyl transferase, Dnmt 1 may lead to the loss or gain of certain chromosomes, that is, monosomy 22 and/or trisomy 14 as seen in the present study. Therefore, altered methylation results in chromosomal instability and hence increased risk of developing cancers. Instability of chromosomes is of such fundamental importance to the development of human or animal cancers that possibly there may be a relationship between DNA methylation status and chromosome loss/gain in somatic cells (Eden et al, 2003, Gandet et al, 2003).
DNA methylation is an inheritable epigenetic change in human cancers and the transcriptional silencing by hypermethylation of CpG islands in the promoter region is becoming more recognised as a common mechanism for the inactivation of various tumour suppressor genes and also affects a number of molecular pathways in human cancer. About 50% of human genes have unmethylated CpG clusters at their 5′-regulatory sequences. Aberrant methylation or hypermethylation of CpG islands in the promoters is associated with transcriptional inactivation and silencing of tumour suppressor genes. DNA replication errors have been found in ovarian GCTs (Suzuki et al, 2000). These findings further suggest that DNA mismatch repair deficiency may also contribute to the pathogenesis of ovarian cancer, and this deficiency may be an early event in the development and progression of the disease. Studies on methylation of specific genes known to play important roles in tumour development have contributed greatly to our current understanding of ovarian cancer. We found hypermethylation in MGMT gene in 32.5% cases of OC (14 out of 43). There are reports in the literature regarding the promoter hypermethylation and gene silencing of MGMT in many cancers (Esteller et al, 2000; Rosas et al, 2001; Bae et al, 2002). We found hypermethylation in E-cadherin gene (CDH1) in 20.9% OCs (9 out of 43). Hypermethylation in CDH1 gene and its reduced expression in gut, liver, prostate and breast cancers, which could be due to the disruption of intercellular adhesion and impairment of β-catenin mediated transactivation of cadherin–catenin complex (Graff et al, 1995; Sadot et al, 1998; Tamura et al, 2000; Corn et al, 2001; Oue et al, 2002), has been reported. DNA methylation in the RAR-β promoter region has been found in 18.6% ovarian tumours (8 out of 43). Methylation in the promoter region of RAR-β gene is frequently found to be associated with downregulation/loss of its expression in tissues affected with gastric carcinoma and in gastric cell lines, head and neck cancers, breast cancer and non-small-cell lung carcinoma (Xu et al, 1994, 1997; Hayashi et al, 2001; Oue et al, 2002). DNA hypermethylation in SYK gene has been found in 16.3% ovarian tumours (7 out of 43). DNA methylation and loss of SYK expression has been reported in breast cancer (Coopman et al, 2000; Yuan et al, 2001). It could be associated with DNA hypermethylation mediated by methyl-CpG binding proteins that bind to methylated cytosines and forms a repressive and inactive complex of proteins that helps in repressing the transcription including that of HDACs and are gene silencing (Wade et al, 1999). Similar type of results have been reported for hMLH1 gene in primary gastric cancers and endometrial cancers (Fleisher et al, 1999; Leung et al, 1999). Several genes including tumour suppressor genes and DNA repair genes such as hMLH1, RB1, VHL, p15, p16, RASSF1A, MGMT and BRCA1 in human cancers (Herman et al, 1994, 1996, 1998; Merlo et al, 1995; Esteller et al, 1999a; Baldwin et al, 2000; Gras et al, 2001; Nakayama et al, 2001; Yoon et al, 2001) were shown to be epigenetically inactivated by DNA methylation in tumours. Hypermethylation of CpG islands of another tumour suppressor gene RASSF1A has been reported in ovarian and renal cell carcinoma (Yoon et al, 2001). One of the major mechanisms of tumour progression is thought to be the inactivation of tumour suppressor genes. This inactivation can be induced by mechanisms such as chromosomal deletion and loss of function mutation in the coding region of genes or by epigenetic alteration in the form of methylation of promoter regions. We found that 15 of the MSI-positive tumours had hypermethylation of either p16, BRCA1 or RASSF1A gene, whereas none of the MSI-negative tumours (MSS) demonstrated the promoter hypermethylation and this could be due to the epigenetic inactivation of either p16, BRCA1 or RASSF1A genes (unpublished data). While it is possible that most of the methylation events we observed contribute in some way to the carcinogenic process, it is unlikely that all the methylated loci play a direct role in ovarian tumorigenesis. Rather, methylation of only certain classes of genes, such as tumour suppressor genes may be crucial to cancer progression.
We found concordant hypermethylation in promoters of MGMT and CDH1, MGMT and RAR-β, CDH1 and RAR-β. Retinoic acid (RA) has profound regulatory effects in the control of many biological processes such as development, immunomodulation, differentiation, proliferation, reproduction and apoptosis (Lotan, 1980). The regulation of cell growth and differentiation of normal, premalignant and malignant cells by retinoids is thought to result from the direct and indirect effects of retinoids on gene expression. These effects are mediated by the nuclear receptors, including RAR-β2 located at 3p24. Retinoic acid receptor beta has four alternative splicing forms and the β-2 form appears to possess tumour-antagonizing activity at least in some cancers. There are no reports of the mutations in the RAR-β gene, but it undergoes epigenetic inactivation by promoter methylation in tumours of different origin. Retinoic acid receptor beta underwent DNA promoter region methylation in significant percent in several other tumours such as lung (Virmani et al, 2000), cervical (Yang et al, 2001) bladder (Maruyama et al, 2001), and prostate (Maruyama et al, 2002). Squamous cell carcinomas and premalignant dysplasias demonstrate a specific downmodulation of RAR-β that can be restored by systemic retinoid therapy (Lotan, 1995). It has also been reported that RA induces upregulation of CDH1 expression and the morphological change of fibroblastoid to epithelioid growth of human pancreatic cancer cell line SUIT-2 (Jimi et al, 1998). Promoter hypermethylation in three genes MGMT, CDH1 and RAR-β together in seven cases of OC, which also show loss of expression of both RAR-β and RAR-β2, may be due to the involvement of the signalling pathway mediated by β-catenin and other factors in the tumorigenic process in these OCs. It has already been reported that the reduced expression of E-cadherin gene and its hypermethylation (Tamura et al, 2000), and the reduced expression of RAR-β due to the promoter hypermethylation may be associated with the reduced expression of CDH1. E-cadherin is involved in a signalling pathway mediated by β-catenin and lymphocyte enhancer factor and T cell transcription factors and can, therefore, explain the concordant hypermethylation in the promoter regions of these three genes. Promoter hypermethylation of RAR-β2 may block or interfere with the retinoid signalling pathways in OC. Deregulation of this pathway results in constant activation of β-catenin, lymphocyte enhancer factor and T cell factor target genes, including c-myc and cyclin D1. For the cyclin-dependent kinase inhibitor p16, the silencing of the gene mediated by promoter hypermethylation seems to be an early event in the development and progression of the tumorigenic process in ovarian cancers. CpG methylation patterns are replicated together with DNA replication during the S phase (Stein et al, 1982), and the altered transcriptional regulation via aberrant promoter methylation could play an important and significant role in the tumorigenic process of these ovarian cancers. Our results suggest promoter methylation of tumour suppressor genes such as MGMT, CDH1 and SYK. We also analysed the relationship between promoter methylation status and basal expression levels of RAR-β, RAR-β2, MGMT and SYK in GCTs of ovaries. Our data clearly show that methylation in the promoter region of RAR-β gene is correlated with downregulation/loss of RAR-β2 gene expression of GCTs of ovaries. Therefore, it might be the only mechanism responsible for loss of or downregulation of RAR-β gene expression in this type of cancer and plays a critical role in the tumour growth and development, which can cause disruption in the retinoid signalling pathway, the metastasis-related process and DNA repair processes in OC. Similarly, out of 14 patients who show promoter hypermethylation in MGMT, three show loss of expression while out of 7 patients who show promoter hypermethylation in SYK gene, only three were found to have loss of expression. It is clear from these results that it is not always true for all the tumour suppressor genes to show a positive correlation between promoter hypermethylation and loss of expression in cancers.
There exist two different pathways that can contribute to the development of cancers; genome-wide hypomethylation may lead to the loss of chromosomes thus leading to chromosomal instability, whereas promoter methylation in certain tumour suppressor genes, which is responsible for gene silencing, can lead to the development of cancers in somatic cells. Therefore, the balance in DNA methylation is very important, and alteration in these may be protective in one pathway but deleterious in the other. It can therefore be concluded that promoter hypermethylation of MGMT, CDH1, RAR-β and SYK genes, and loss of expression of RAR-β and RAR-β2 in ovarian cancers (GCTs) is relatively common and this may also be useful as a tumour marker for early diagnosis and subsequent disease monitoring. Hence, these epigenetic signatures could play a decisive role in designing treatment options for this category of ovarian cancer.
References
- Bae SI, Lee HS, Kim SH, Kim WH (2002) Inactivation of O6-methylguanine-DNA methyltransferase by promoter CpG island hypermethylation in gastric cancers. Br J Cancer Res 86: 1888–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin RL, Nemeth E, Tran H, Shvartsman H, Cass I, Narod S, Karlan BY (2000) BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res 60: 5329–5333 [PubMed] [Google Scholar]
- Chambon P (1996) A decade of molecular biology of retinoic acid receptors. FASEM J 10: 940–954 [PubMed] [Google Scholar]
- Chu DH, Morita CT, Weiss A (1998) The Syk family of protein tyrosine kinases in T-cell activation and development. Immunol Rev 165: 167–180 [DOI] [PubMed] [Google Scholar]
- Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, Basyuk E, Blancato JK, Vezza PR, McLeskey SW, Mangeat PH, Mueller SC (2000) The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature (London) 406: 742–747 [DOI] [PubMed] [Google Scholar]
- Corn PG, Heath EI, Heitmiller R, Fogt F, Forastiere AA, Herman JG, Wu TT (2001) Frequent hypermethylation of the 5′ CpG island of E-Cadherin in esophageal adenocarcinoma. Clin Cancer Res 7: 2765–2769 [PubMed] [Google Scholar]
- Dal Cin P, Qi H, Pauwels P, Backx C, Van den Berghe H (1997) Monosomy 22 in a fibrothecoma. Cancer Genet Cytogenet 99: 129–131 [DOI] [PubMed] [Google Scholar]
- de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A (1990) Identification of a retinoic acid response element in the retinoic acid receptor b gene. Nature (London) 343: 177–180 [DOI] [PubMed] [Google Scholar]
- Duesberg P, Li R, Rasmick D, Rausch C, Willer A, Kraemer A, Yerganian G, Hehlman R (2000) Aneuploidy precedes and segregates with chemical carcinogenesis. Cancer Genet Cytogenet 119: 83–93 [DOI] [PubMed] [Google Scholar]
- Eden A, Gandet F, Waghmare A, Jaenisch R (2003) Chromosomal instability and tumour promoted by DNA hypomethylation. Science 300: 455. [DOI] [PubMed] [Google Scholar]
- Esteller M, Catasus L, Matias-Guiu X, Mutter GL, Prat J, Baylin SB, Herman JG (1999a) hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol 155: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG (1999b) Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 59: 793–797 [PubMed] [Google Scholar]
- Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG (2000) Inactivation of the DNA repair gene O6-Methylguanine-DNA Methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res 60: 2368–2371 [PubMed] [Google Scholar]
- Evan MF, McDicken IW, Herrington CS (1999) Numerical abnormalities of chromosomes 1, 11, 17, and X are associated with stromal invasion in serous and mucinous epithelial ovarian tumours. J Pathol 189: 53–59 [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN, Shi YQ, Rhyu MG, Powell SM, James SP, Wilson KT, Herman JG, Meltzer SJ (1999) Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res 59: 1090–1095 [PubMed] [Google Scholar]
- Gandet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R (2003) Induction of tumours in mice by genomic hypomethylation. Science 300: 489–492 [DOI] [PubMed] [Google Scholar]
- Garcia del Muro X, Torregrosa A, Munoz J, Castellsague X, Condom E, Vigues F, Arance A, Fabra A, Germa JR (2000) Prognostic value of the expression of E-cadherin and β-catenin in bladder cancer. Eur J Cancer 36: 357–362 [DOI] [PubMed] [Google Scholar]
- Gebert JF, Moghal N, Frangioni JV, Sugerbaker DJ, Neel BG (1991) High frequency of retinoic acid receptor β abnormalities in human lung cancer. Oncogene 6: 1859–1868 [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB (1995) E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res 55: 5195–5199 [PubMed] [Google Scholar]
- Gras E, Catasus L, Arguelles R, Moreno-Bueno G, Palacios J, Gamallo C, Matias-Guiu X, Prat J (2001) Microsatellite instability, MLH-1 promoter hypermethylation, and frame shift mutations at coding mononucleotide repeat microsatellites in ovarian tumours. Cancer 92: 2829–2836 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yokozaki H, Naka K, Yasui W, Lotan R, Tahara E (2001) Overexpression of retinoic acid receptor induces growth arrest and apoptosis in oral cancer cell lines. Jpn J Cancer Res 92: 42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann R, Lan F, McBride R, Hellman S (2000) Separating favourable from unfavourable prognostic markers in breast cancer: the role of E-cadherin. Cancer Res 60: 298–304 [PubMed] [Google Scholar]
- Herman JG, Jen J, Merlo A, Baylin SB (1996) Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res 56: 722–727 [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman LI, Zbar B, Lin S, Samid D, Duan DS, Gnarra JR, Linechan WM (1994) Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA 91: 9700–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB (1998) Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95: 6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi S, Shono T, Tanaka M, Kono A, Yamada Y, Sudo K, Kuwano M (1998) Effect of retinoic acid on morphological changes of human pancreatic cancer cells on collagen gels: a possible association with the metastatic potentials. Oncol Res 10: 7–14 [PubMed] [Google Scholar]
- Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC (1999) hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res 59: 159–164 [PubMed] [Google Scholar]
- Lotan R (1980) Effects of vitamin A and its analogs (retinoids) on normal and neoplastic cells. Biochim Biophys Acta 605: 33–91 [DOI] [PubMed] [Google Scholar]
- Lotan R (1995) Retinoids and apoptosis: implications for cancer chemoprevention and therapy. J Natl Cancer Inst 87: 1655–1657 [DOI] [PubMed] [Google Scholar]
- Lotan R, Xu C, Lippman SM, Ro JY, Lee JS, Lee JJ, Hong WK (1995) Suppression of retinoic acid receptor β in premalignant oral lesions and its up-regulation by isotretinoin. N Engl J Med 332: 1405–1410 [DOI] [PubMed] [Google Scholar]
- Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zochbauer-Muller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A, Czerniak B, Gazdar AF (2001) Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res 61: 8659–8663 [PubMed] [Google Scholar]
- Maruyama R, Toyooka S, Toyooka KO, Virmani AK, Zochbauer-Muller S, Farinas AJ, Minna JD, McConnell J, Frenkel EP, Gazdar AF (2002) Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res 8: 514–519 [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabreilson E, Burger PC, Baylin SB, Sidransky D (1995) 5′-CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers [see comments]. Nat Med 1: 686–692 [DOI] [PubMed] [Google Scholar]
- Nakayama K, Takebayashi Y, Namiki T, Tamahashi N, Nakayama S, Uchida T, Miyazaki K, Fukumoto M (2001) Comprehensive allelotype study of ovarian tumours of low malignant potential: potential differences in pathways between tumours with and without genetic predisposition to invasive carcinoma. Int J Cancer 94: 605–609 [DOI] [PubMed] [Google Scholar]
- Oue N, Motoshita J, Yokozaki H, Hayashi A, Tahara E, Taniyama K, Matsusaki K, Yasui W (2002) Distinct promoter hypermethylation of p16INK4a, CDH1 and RAR-beta in intestinal, diffuse-adherant, and diffuse-scattered type gastric carcinomas. J Pathol 198: 55–59 [DOI] [PubMed] [Google Scholar]
- Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, Jen J, Sidransky G (2001) Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res 61: 939–942 [PubMed] [Google Scholar]
- Russel P, Bannatyne P (1989) Surgical Pathology of Ovaries. London: Churchill Livingstone [Google Scholar]
- Sadot E, Simcha I, Shtutman M, Ben-Ze'ev A, Geiger B (1998) Inhibition of β-catenin-mediated transactivation by cadherin derivatives. Proc Natl Acad Sci USA 95: 15339–15344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg AA (1991) Chromosome abnormalities in human cancer and leukaemia. Mutat Res 247: 231–240 [DOI] [PubMed] [Google Scholar]
- Seewaldt VL, Johnson BS, Parker MB, Collins SJ, Swisshelm K (1995) Expression of retinoic acid receptor-b mediates retinoic acid-induced growth arrest and apoptosis in breast cancer cells. Cell Growth Differ 6: 1077–1088 [PubMed] [Google Scholar]
- Speleman F, Dermaut B, De Potter CR, Van Gele M, Van Roy N, De Paepe A, Laureys G (1997) Monosomy 22 in a mixed germ cell-sex cord-stromal tumor of the ovary. Genes Chromosomes Cancer 19: 192–194 [PubMed] [Google Scholar]
- Stein R, Gruenbaum Y, Pollack Y, Rajin A, Cedar H (1982) Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci USA 79: 61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ohwada M, Saga Y, Ochiai K, Sato I (2000) DNA replication error is frequent in ovarian granulosa cell tumors. Cancer Genet Cytogenet 122: 55–58 [DOI] [PubMed] [Google Scholar]
- Takeichi M (1995) Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 7: 619–627 [DOI] [PubMed] [Google Scholar]
- Tamura G, Yin J, Wang S, Fleisher AS, Zou T, Abraham JM, Kong D, Smolinski KN, Wilson KT, James SP, Silverberg SG, Nishizuka S, Terashima M, Motoyama T, Meltzer SJ (2000) E-cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst (Bethesda) 92: 569–573 [DOI] [PubMed] [Google Scholar]
- Van den Berghe I, Dal Cin P, De Groef K, Michielssen P, Van den Berghe H (1999) Monosomy 22 and trisomy 14 may be early events in the tumorigenesis of adult granulosa cell tumor. Cancer Genet Cytogenet 112: 46–48 [DOI] [PubMed] [Google Scholar]
- Virmani AK, Rathi A, Zochbauer-Muller S, Sacchi N, Fukuyama Y, Bryant D, Maitra A, Heda S, Fong KM, Thunnissen F, Minna JD, Gazdar AF (2000) Promoter methylation and silencing of the retinoic acid receptor-beta gene in lung carcinomas. J Natl Cancer Inst 92: 1303–1307 [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballester E, Aubry F, Wolffe AP (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 23: 62–66 [DOI] [PubMed] [Google Scholar]
- Xu X-C, Ro JY, Lee JS, Shin DM, Hong YK, Lotan R (1994) Differential expression of nuclear retinoid receptors receptors in normal, premalignant and malignant head and neck tissues. Cancer Res 54: 3580–3587 [PubMed] [Google Scholar]
- Xu X-C, Sneige N, Liu X, Nandagiri R, Lee JJ, Lukmanji F, Hortobagyi G, Lippman SM, Dhingra K, Lotan R (1997) Progressive decrease in nuclear retinoic acid receptor b messenger RNA levels during breast carcinogenesis. Cancer Res 57: 4992–4996 [PubMed] [Google Scholar]
- Yang Q, Mori I, Shan L, Nakamura M, Nakamura Y, Utsunomiya H, Yoshimura G, Suzuma T, Tamaki T, Umemura T, Sakurai T, Kakudo K (2001) Biallelic inactivation of retinoic acid receptor beta2 gene by epigenetic change in breast cancer. Am J Pathol 158: 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Dammann R, Pfeifer GP (2001) Hypermethylation of the CpG islands of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer 94: 212–217 [DOI] [PubMed] [Google Scholar]
- Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S (1995) Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA 92: 7416–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Mendez R, Sahin A, Dai JL (2001) Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res 61: 5558–5561 [PubMed] [Google Scholar]
- Zheng Z, Pan J, Chu B, wong YC, Cheung AL, Tsao SW (1999) Down-regulation and abnormal expression of E-cadherin and β-catenin in nasopharyngeal carcinoma: close association with advanced disease stage and lymph node metastasis. Hum Pathol 30: 458–466 [DOI] [PubMed] [Google Scholar]