Abstract
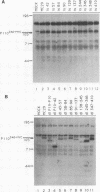
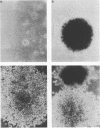
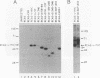
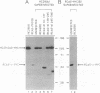
Avian myelocytomatosis virus MC29 induces a wide variety of neoplastic diseases in infected birds and transforms cells of the macrophage lineage as well as fibroblasts and epithelial cells. A biological and biochemical analysis, carried out on a series of in-frame insertion and deletion mutations within the gag-myc gene of MC29, revealed several mutations within the 5' portion of the v-myc gene that encode proteins either completely defective for transformation or compromised in their ability to transform chicken embryo fibroblasts but not macrophages. Mutations within the 3' end of the v-myc gene which disrupt sequences encoding the basic/helix-loop-helix region were defective for transformation of both fibroblasts and macrophages. Eight variants were cloned into the replication-competent avian expression vector RCAS. Analysis of cells infected with transformation-defective, replication-competent viruses confirmed the expression of functionally defective Myc proteins. Further, expression of the transformation defective variant dl91-137 in chicken fibroblasts inhibited subsequent transformation by wild-type MC29. The results reported herein support the hypothesis that Myc proteins function as regulators of transcription in a variety of cell types and clearly point out the necessity of putative regulatory domains within the amino-terminal half of the Myc protein.
Full text
PDF

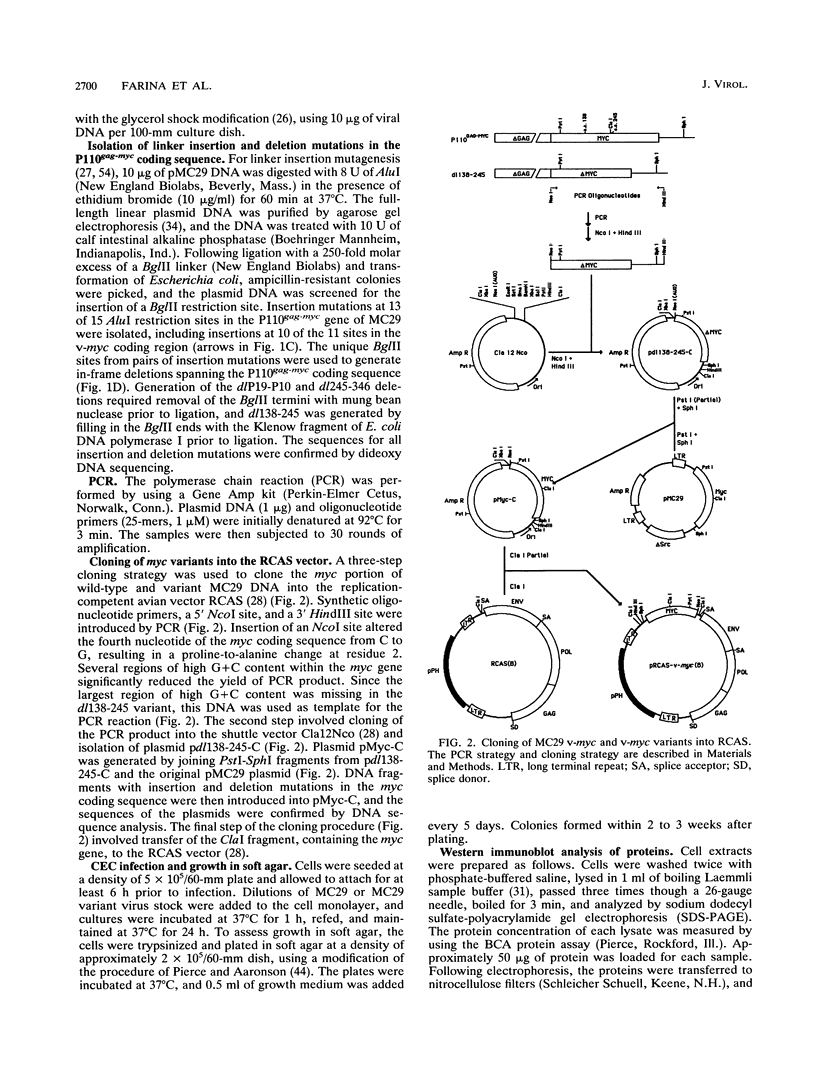

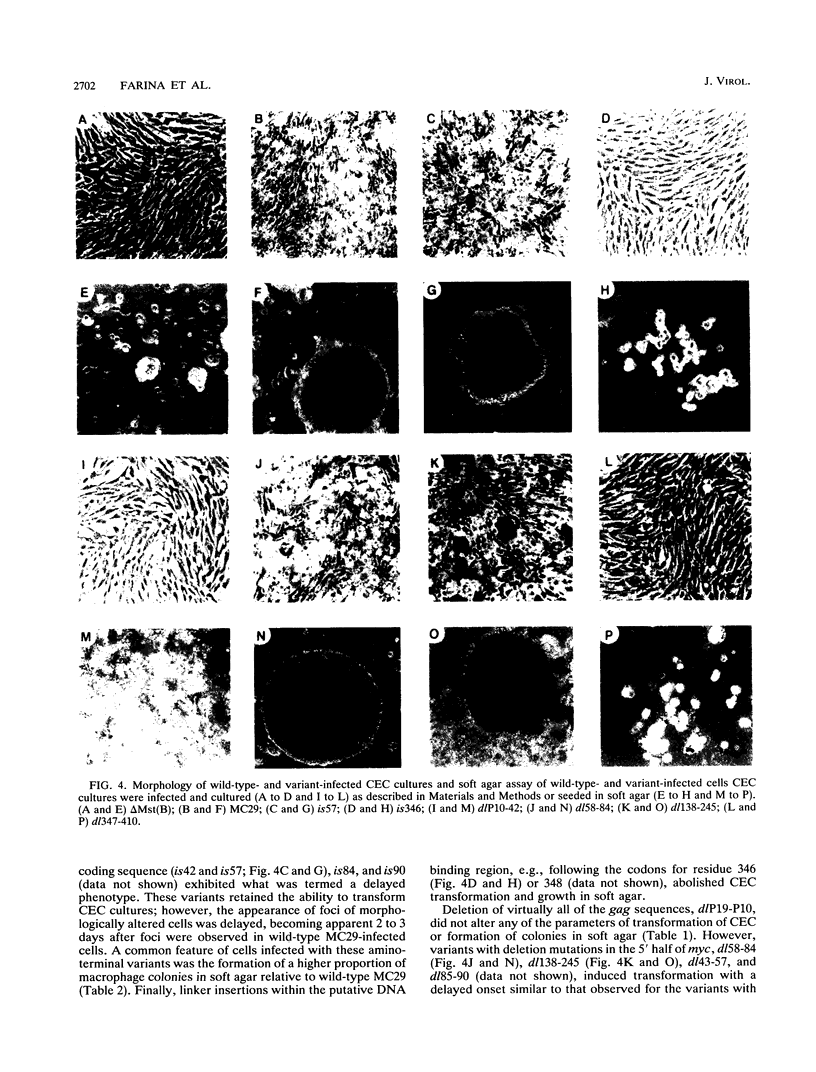
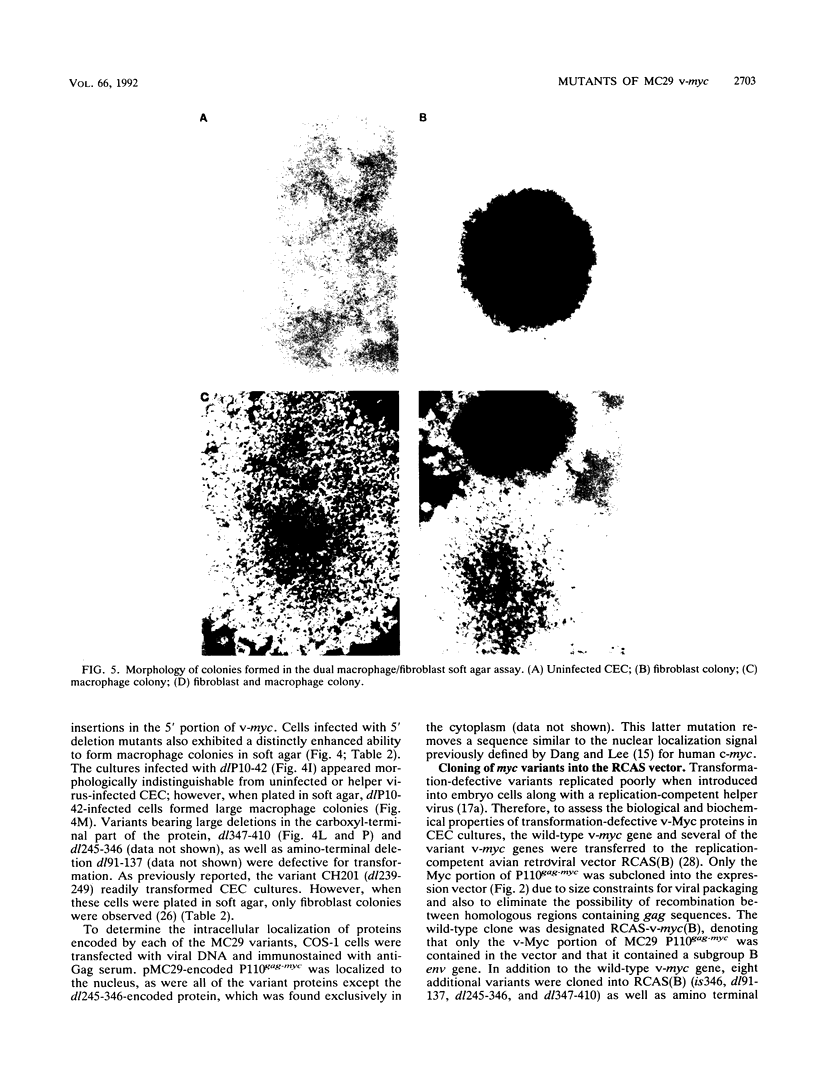

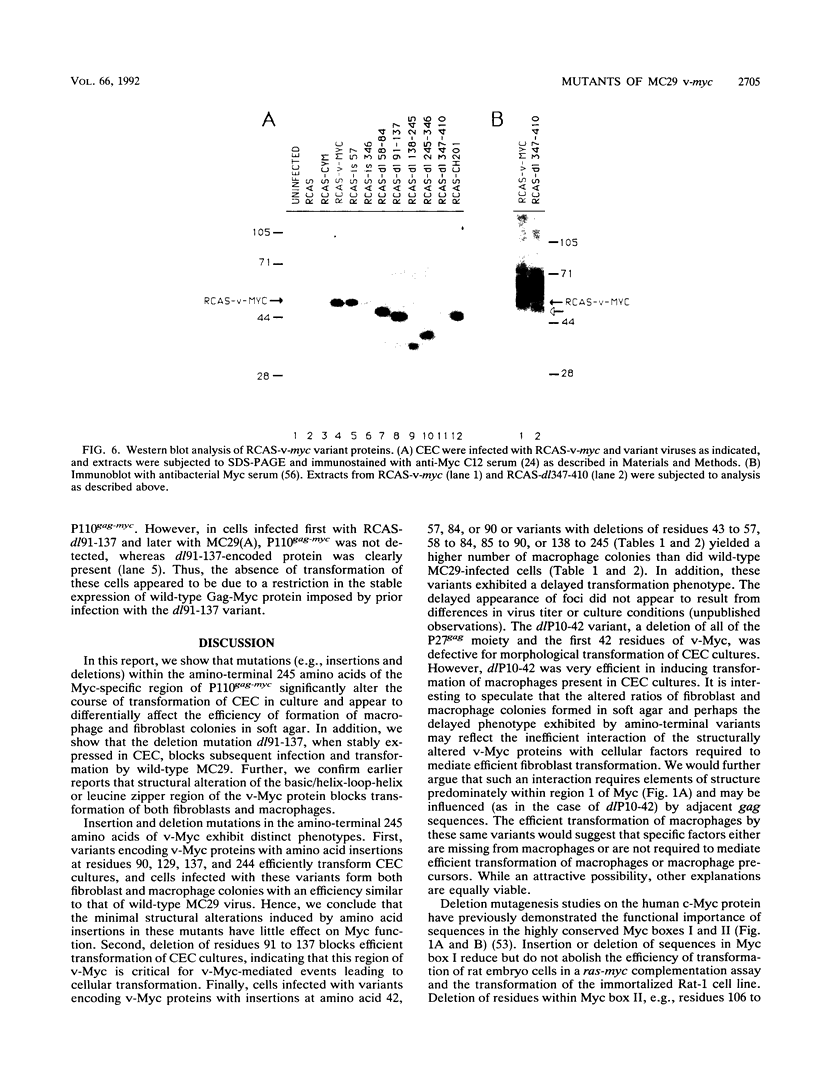
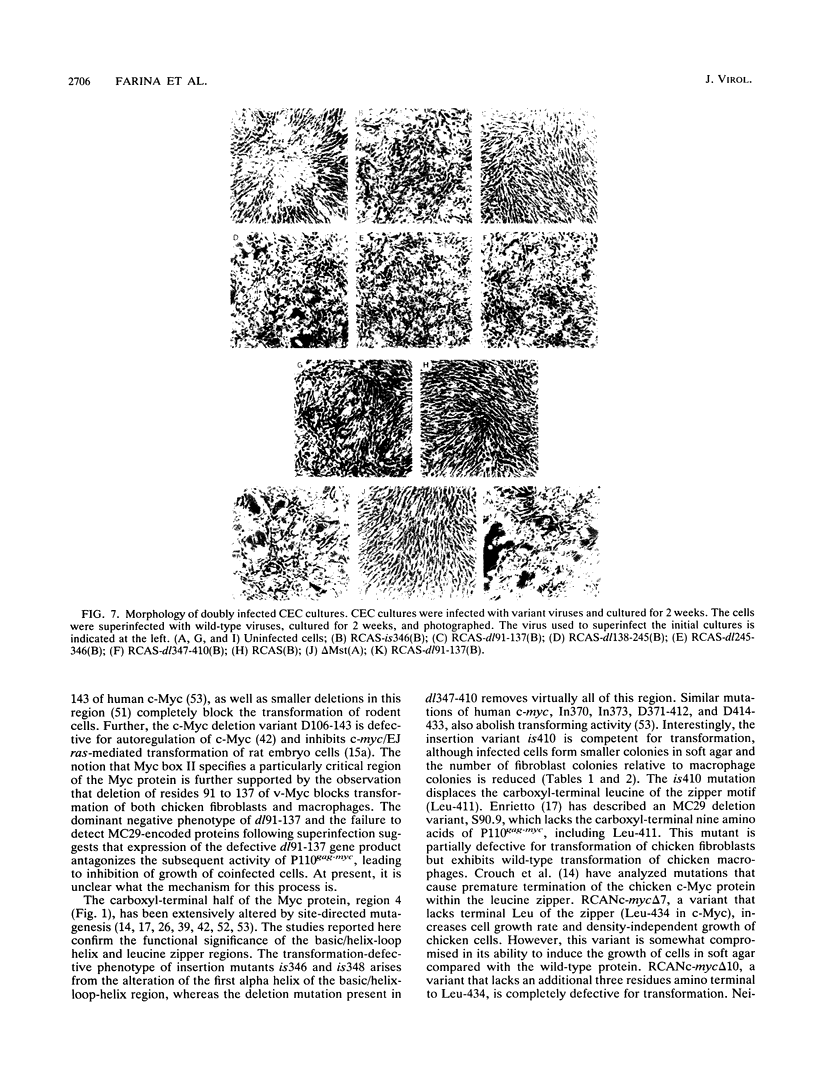


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrams H. D., Rohrschneider L. R., Eisenman R. N. Nuclear location of the putative transforming protein of avian myelocytomatosis virus. Cell. 1982 Jun;29(2):427–439. doi: 10.1016/0092-8674(82)90159-3. [DOI] [PubMed] [Google Scholar]
- Alitalo K., Bishop J. M., Smith D. H., Chen E. Y., Colby W. W., Levinson A. D. Nucleotide sequence to the v-myc oncogene of avian retrovirus MC29. Proc Natl Acad Sci U S A. 1983 Jan;80(1):100–104. doi: 10.1073/pnas.80.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Ramsay G., Bishop J. M., Pfeifer S. O., Colby W. W., Levinson A. D. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature. 1983 Nov 17;306(5940):274–277. doi: 10.1038/306274a0. [DOI] [PubMed] [Google Scholar]
- Beard J. W., Hillman E. A., Beard D., Lapis K., Heine U. Neoplastic response of the avian liver to host infection with strain Mc29 leukosis verus. Cancer Res. 1975 Jul;35(7):1603–1627. [PubMed] [Google Scholar]
- Beug H., von Kirchbach A., Döderlein G., Conscience J. F., Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979 Oct;18(2):375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- Biegalke B. J., Heaney M. L., Bouton A., Parsons J. T., Linial M. MC29 deletion mutants which fail to transform chicken macrophages are competent for transformation of quail macrophages. J Virol. 1987 Jul;61(7):2138–2142. doi: 10.1128/jvi.61.7.2138-2142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bister K., Hayman M. J., Vogt P. K. Defectiveness of avian myelocytomatosis virus MC29: isolation of long-term nonproducer cultures and analysis of virus-specific polypeptide synthesis. Virology. 1977 Oct 15;82(2):431–448. doi: 10.1016/0042-6822(77)90017-4. [DOI] [PubMed] [Google Scholar]
- Bister K., Ramsay G. M., Hayman M. J. Deletions within the transformation-specific RNA sequences of acute leukemia virus MC29 give rise to partially transformation-defective mutants. J Virol. 1982 Mar;41(3):754–766. doi: 10.1128/jvi.41.3.754-766.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bister K., Trachmann C., Jansen H. W., Schroeer B., Patschinsky T. Structure of mutant and wild-type MC29 v-myc alleles and biochemical properties of their protein products. Oncogene. 1987 May;1(2):97–109. [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Bryant D. L., Parsons J. T. Amino acid alterations within a highly conserved region of the Rous sarcoma virus src gene product pp60src inactivate tyrosine protein kinase activity. Mol Cell Biol. 1984 May;4(5):862–866. doi: 10.1128/mcb.4.5.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Crouch D. H., Lang C., Gillespie D. A. The leucine zipper domain of avian cMyc is required for transformation and autoregulation. Oncogene. 1990 May;5(5):683–689. [PubMed] [Google Scholar]
- Dang C. V., Lee W. M. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988 Oct;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Donner P., Greiser-Wilke I., Moelling K. Nuclear localization and DNA binding of the transforming gene product of avian myelocytomatosis virus. Nature. 1982 Mar 18;296(5854):262–269. doi: 10.1038/296262a0. [DOI] [PubMed] [Google Scholar]
- Enrietto P. J. A small deletion in the carboxy terminus of the viral myc gene renders the virus MC29 partially transformation defective in avian fibroblasts. Virology. 1989 Feb;168(2):256–266. doi: 10.1016/0042-6822(89)90265-1. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Graf T., Beug H. Avian leukemia viruses: interaction with their target cells in vivo and in vitro. Biochim Biophys Acta. 1978 Nov 17;516(3):269–299. doi: 10.1016/0304-419x(78)90011-2. [DOI] [PubMed] [Google Scholar]
- Graf T. Two types of target cells for transformation with avian myelocytomatosis virus. Virology. 1973 Aug;54(2):398–413. doi: 10.1016/0042-6822(73)90152-9. [DOI] [PubMed] [Google Scholar]
- Graf T., von Kirchbach A., Beug H. Characterization of the hematopoietic target cells of AEV, MC29 and AMV avian leukemia viruses. Exp Cell Res. 1981 Feb;131(2):331–343. doi: 10.1016/0014-4827(81)90236-6. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Abrams H. D., Rohrschneider L. R., Eisenman R. N. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983 Oct;34(3):789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney M. L., Pierce J., Parsons J. T. Site-directed mutagenesis of the gag-myc gene of avian myelocytomatosis virus 29: biological activity and intracellular localization of structurally altered proteins. J Virol. 1986 Oct;60(1):167–176. doi: 10.1128/jvi.60.1.167-176.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Greenhouse J. J., Petropoulos C. J., Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987 Oct;61(10):3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. Transcriptional regulation by dimerization: two sides to an incestuous relationship. Cell. 1990 Apr 6;61(1):9–11. doi: 10.1016/0092-8674(90)90207-u. [DOI] [PubMed] [Google Scholar]
- Kornfeld S., Beug H., Doederlein G., Graf T. Detection of avian hematopoietic cell surface antigens with monoclonal antibodies to myeloid cells. Their distribution on normal and leukemic cells of various lineages. Exp Cell Res. 1983 Feb;143(2):383–394. doi: 10.1016/0014-4827(83)90065-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981 Dec;27(2 Pt 1):279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mladenov Z., Heine U., Beard D., Beard J. W. Strain MC29 avian leukosis virus. Myelocytoma, endothelioma, and renal growths: pathomorphological and ultrastructural aspects. J Natl Cancer Inst. 1967 Mar;38(3):251–285. [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Ikeda M., Tsuchida N., Nishimura S., Taya Y. Inactivation of the N-myc gene product by single amino acid substitution of leucine residues located in the leucine-zipper region. Oncogene. 1989 Aug;4(8):999–1002. [PubMed] [Google Scholar]
- Parsons S. J., Wilson L. D., Ely C. M., Parsons J. T., Benjamin D. C. Monoclonal antibodies specific for the virion polypeptide, p27, of avian retroviruses. Hybridoma. 1984;3(1):25–31. doi: 10.1089/hyb.1984.3.25. [DOI] [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Littlewood T. D., Morgenstern J. P., Evan G. I., Lee W. M., Land H. Domains of human c-myc protein required for autosuppression and cooperation with ras oncogenes are overlapping. Mol Cell Biol. 1990 Sep;10(9):4961–4966. doi: 10.1128/mcb.10.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H., Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984 Aug 17;225(4663):718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- Pierce J. H., Aaronson S. A. In vitro transformation of murine pre-B lymphoid cells by Snyder-Theilen feline sarcoma virus. J Virol. 1983 Jun;46(3):993–1002. doi: 10.1128/jvi.46.3.993-1002.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Ralston R., Bishop J. M. The protein products of the myc and myb oncogenes and adenovirus E1a are structurally related. Nature. 1983 Dec 22;306(5945):803–806. doi: 10.1038/306803a0. [DOI] [PubMed] [Google Scholar]
- Ramsay G. M., Hayman M. J. Isolation and biochemical characterization of partially transformation-defective mutants of avian myelocytomatosis virus strain MC29: localization of the mutation to the myc domain of the 110,000-dalton gag-myc polyprotein. J Virol. 1982 Mar;41(3):745–753. doi: 10.1128/jvi.41.3.745-753.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G., Graf T., Hayman M. J. Mutants of avian myelocytomatosis virus with smaller gag gene-related proteins have an altered transforming ability. Nature. 1980 Nov 13;288(5787):170–172. doi: 10.1038/288170a0. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Reynolds R. K., Watson D. K., Schultz R. A., Lautenberger J., Papas T. S. Nucleotide sequence analysis of the proviral genome of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1983 May;80(9):2500–2504. doi: 10.1073/pnas.80.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid J., Halazonetis T. D., Murphy W., Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. J., Charron-Prochownik D. C., Prochownik E. V. The leucine zipper of c-Myc is required for full inhibition of erythroleukemia differentiation. Mol Cell Biol. 1990 Oct;10(10):5333–5339. doi: 10.1128/mcb.10.10.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. C., Atkinson T., Smith M., Pawson T. Identification of functional regions in the transforming protein of Fujinami sarcoma virus by in-phase insertion mutagenesis. Cell. 1984 Jun;37(2):549–558. doi: 10.1016/0092-8674(84)90385-4. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D. K., Reddy E. P., Duesberg P. H., Papas T. S. Nucleotide sequence analysis of the chicken c-myc gene reveals homologous and unique coding regions by comparison with the transforming gene of avian myelocytomatosis virus MC29, delta gag-myc. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2146–2150. doi: 10.1073/pnas.80.8.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R. A., Shatzman A. R., Rosenberg M. Expression and characterization of the human c-myc DNA-binding protein. Mol Cell Biol. 1985 Mar;5(3):448–456. doi: 10.1128/mcb.5.3.448. [DOI] [PMC free article] [PubMed] [Google Scholar]