Abstract
Background
MicroRNAs (miRNAs) represent a class of small non-coding RNAs that control gene expression by targeting mRNAs and triggering either translation repression or RNA degradation. Emerging evidence suggests the potential involvement of altered regulation of miRNA in the pathogenesis of cancers, and these genes are thought to function as both tumor suppressors and oncogenes.
Methodology/Principal Findings
Using microRNA microarrays, we identify several miRNAs aberrantly expressed in human ovarian cancer tissues and cell lines. miR-221 stands out as a highly elevated miRNA in ovarian cancer, while miR-21 and several members of the let-7 family are found downregulated. Public databases were used to reveal potential targets for the highly differentially expressed miRNAs. In order to experimentally identify transcripts whose stability may be affected by the differentially expressed miRNAs, we transfected precursor miRNAs into human cancer cell lines and used oligonucleotide microarrays to examine changes in the mRNA levels. Interestingly, there was little overlap between the predicted and the experimental targets or pathways, or between experimental targets/pathways obtained using different cell lines, highlighting the complexity of miRNA target selection.
Conclusion/Significance
Our results identify several differentially expressed miRNAs in ovarian cancer and identify potential target transcripts that may be regulated by these miRNAs. These miRNAs and their targets may have important roles in the initiation and development of ovarian cancer.
Introduction
MicroRNAs (miRNAs) are 21–23 nucleotide regulatory RNAs processed from 70–100 nucleotide hairpin pre-miRNAs [1], [2]. The miRNA are incorporated into a ribonucleoprotein complex called RNA-induced silencing complex (RISC) and guide the RISC to the target mRNA [3]. Binding of the miRNA to the target mRNA 5'UTR can downregulate gene expression through inhibition of translation or increased RNA degradation. In addition, recent evidence suggests that miRNA may also upregulate translation under certain circumstances [4]. Hundreds of miRNAs have been identified in various species, including C. elegans, humans, and the plant Arabidopsis thaliana. It is believed that mammalian miRNAs have the potential to regulate at least 20–30% of all human genes [5]. Each miRNA can target up to 200 transcripts directly or indirectly, and multiple miRNAs can target a given gene [6]. Therefore the potential regulatory circuitry afforded by miRNA is extremely complex. The expression of several miRNAs has been shown to be developmentally regulated and several studies have demonstrated that miRNA are responsible for determining cell fate. These results indicate that miRNAs play a major role in fundamental cellular processes, including the timing of cellular development, hematopoiesis, fat metabolism, organogenesis, apoptosis, cell proliferation, and differentiation. There is also strong evidence that miRNAs are implicated in the onset and progression of many diseases, including cancer [3].
Comparison between human cancers and their normal counterparts have revealed distinct miRNA expression profiles. Indeed, a number of studies have reported differentially regulated miRNAs in diverse cancer types such as breast cancer [7], lung cancer [8], chronic lymphocytic leukemia [9], colon cancer [10], thyroid carcinomas [11], pancreatic cancer [12], head and neck cancer [5], prostate cancer [13], pituitary adenomas [14], and ovarian cancer [15]–[18]. Collectively, these studies demonstrate that some human miRNAs are consistently deregulated in human cancer, suggesting a role for these genes in tumorigenesis. Specific over-expression or under expression of certain miRNAs has been shown to correlate with particular tumor types [19]. Overexpressed miRNAs could potentially target tumor suppressor genes, while downregulated miRNAs would theoretically regulate oncogenes. For example, the let-7 miRNAs, which are down regulated in lung cancer, can negatively regulate the oncogenes RAS and HMGA2, providing a mechanism for the upregulation of these oncogenes [8], [20], [21]. Thus, members of the let-7 family function biologically as tumor suppressors and their loss is predicted to promote transformation and tumor progression. Conversely, the human miRNA cluster miR-17-92 acts as an oncogene in B-cell lymphoma [22] and lung cancer [23], and can collaborate with myc [22].
Unique miRNA expression signatures have been found to be associated with bio-molecular and prognostic characteristics of human lung cancer and chronic lymphocytic leukemia [8], [24], indicating that miRNA signatures could be used to define biological or clinical features of human cancers. The development of new miRNA markers in the near future will represent one of the main goals in molecular medicine as miRNA expression profiles might better classify poorly differentiated tumors as compared with the transcript-based classifiers [19].
There is a limited number of studies in the literature on the roles of miRNAs in ovarian cancer. Zhang et al. reported high frequency genomic alterations involving miRNA genes in 227 human ovarian cancer, breast cancer and melanoma specimens [15]. In another study, 84 tissues (15 normal and 69 malignant) and 5 cell lines were analyzed for alteration in miRNA profile [16]. Finally a study analyzing 10 ovarian tumors and 10 different normal controls was recently reported [17]. In this report, we describe a series of experiments to elucidate the changes in miRNA expression in ovarian cancer and to identify possible targets of relevant candidate miRNAs.
Results
miRNA Expression patterns in ovarian tissues
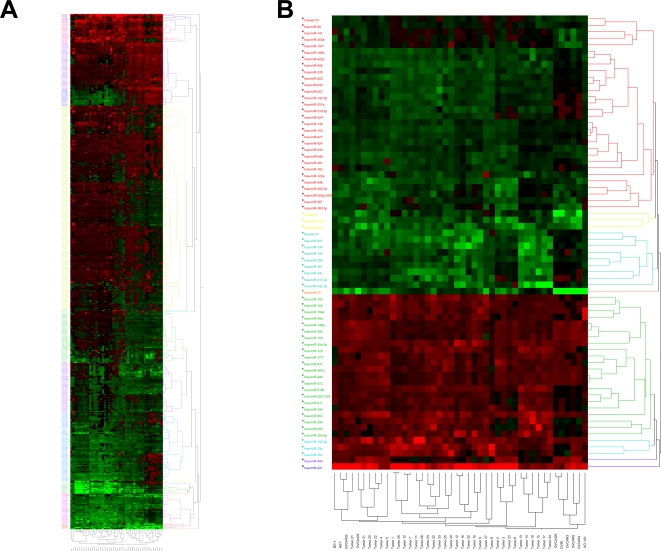
MiRNA expression profiles were determined for 34 ovarian cancer tissues ( Table 1 ) as well as 10 ovarian cancer cell lines. An immortalized human ovarian surface epithelium cell line was used as non-transformed control. The expression profiles were determined using miRCURY™ LNA miRNA Arrays. LNAs are a class of conformationally restricted nucleotide analogs that increase affinity of an oligonucleotide for its complementary RNA or DNA target. The Exiqon miRNA array is currently the most comprehensive probe set available on an array platform and allows for an in depth analysis of miRNA expression. Following RNA hybridization and array analysis, samples were clustered according to their miRNA profile using the hierarchical clustering algorithm of the JMP 6.0.0 software. Figure 1A shows hierarchical clustering analysis of tumor tissues and cell lines based on overall miRNA expression. The profiles shown are relative to ovarian surface epithelial cells (HOSE-B cells). The samples are separated into three main clusters. The middle cluster is comprised entirely of tumors (17 samples), while the left cluster contains 2 cell lines and 7 primary tumors. The right cluster is enriched in cell lines and contains 8 cell lines out of 18 members in the cluster. The hierarchical clustering was then repeated based on a much smaller number of genes and included only 70 highly differentially expressed miRNA in ovarian cancer tissues and cell lines. This clustering analysis gave rise to multiple clusters, but again, the cell lines clustered together in 2 different clusters while the primary tissues were grouped together ( Figure 1B ), suggesting specific differences in miRNA expression between ovarian cancer tissues and cell lines.
Table 1. Patient data.
| Sample | General Classification | Age | Grade /Stage |
| Tumor 1 | Borderline carcinoma | 57 | - |
| Tumor 2 | Serous carcinoma low grade | 55 | Grade 2, stage 3 |
| Tumor 3 | Serous carcinoma low grade | 73 | Grade 2, stage 3 |
| Tumor 4 | Clear cell carcinoma | 50 | - |
| Tumor 5 | Clear cell carcinoma | 72 | Grade 3 |
| Tumor 6 | Serous carcinoma | 53 | Stage 4 |
| Tumor 7 | Serous carcinoma | 57 | - |
| Tumor 8 | Serous carcinoma | - | - |
| Tumor 9 | Serous carcinoma | 69 | - |
| Tumor 10 | Serous carcinoma high grade | 76 | Grade 3 |
| Tumor 11 | Serous carcinoma high grade | 74 | Grade 3 |
| Tumor 12 | Serous carcinoma high grade | 64 | Grade 3 |
| Tumor 13 | Serous carcinoma high grade | 66 | Grade 3 |
| Tumor 14 | Serous carcinoma high grade | 69 | Grade 3 |
| Tumor 15 | Serous carcinoma high grade | 48 | Grade 3 |
| Tumor 16 | Serous carcinoma high grade | 46 | Grade 3 |
| Tumor 17 | Serous carcinoma high grade | 86 | Grade 3 |
| Tumor 18 | Serous carcinoma high grade | 52 | Grade 3 |
| Tumor 19 | Serous carcinoma high grade | 64 | - |
| Tumor 20 | Serous carcinoma high grade | 61 | Grade 3, stage 3 |
| Tumor 21 | Serous carcinoma high grade | 66 | Stage 3 |
| Tumor 22 | Serous carcinoma high grade | 56 | Grade 3 |
| Tumor 23 | Serous carcinoma high grade | 76 | Grade 3, stage 3 |
| Tumor 24 | Serous carcinoma high grade | 64 | - |
| Tumor 25 | Serous carcinoma high grade | 71 | - |
| Tumor 26 | Serous carcinoma high grade | 63 | Grade 3 |
| Tumor 27 | Serous carcinoma high grade | 77 | - |
| Tumor 28 | Serous carcinoma high grade | 68 | Stage 3 |
| Tumor 29 | Serous carcinoma high grade | 80 | Grade 3 |
| Tumor 30 | Serous carcinoma high grade | 64 | Grade 3, stage 4 |
| Tumor 31 | Serous carcinoma high grade | 57 | Grade 3, stage 3 |
| Tumor 32 | Serous carcinoma high grade | 68 | - |
| Tumor 33 | Serous carcinoma high grade | 64 | Grade 4 |
| Tumor 34 | Serous carcinoma high grade | 51 | Grade 4 |
Figure 1. Cluster analysis of miRNA expression.
Tree generated by cluster analysis of ovarian cancer tissues and cell lines based on (A) all tested miRNAs in tissues and cell lines, and (B) differentially regulated miRNAs (Fold change >2.0 or <0.5 in greater than 60% of the samples) in tissues and cell lines compared to the normal control HOSE-B cells.
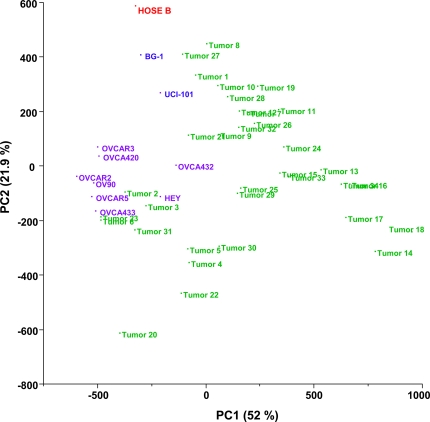
Using principal component analysis (PCA) we found that global miRNA expression could distinguish ovarian cancer tumors from ovarian cancer cell lines ( Figure 2 ). The non-tumorigenic control HOSE-B also exhibited a unique expression pattern, distinguishable from both cell lines and tissues.
Figure 2. Principal component analysis of ovarian cancer samples (PCA) based on global miRNA expression.
Two-dimensional PCA shows that global miRNA expression patterns are different in ovarian cancer cell lines (indicated in blue), ovarian cancer tissues (indicated in green), and the non-tumorigenic HOSE-B cells (in red).
Differentially expressed miRNAs
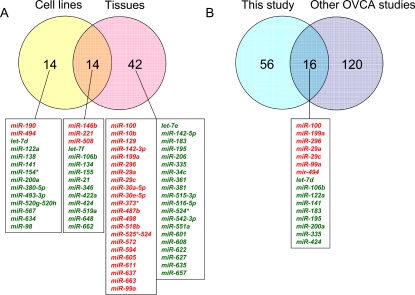
We then identified miRNAs that were differentially expressed between non-neoplastic and neoplastic samples ( Figure 3A , Table 2 ). Only miRNAs that were altered at least 2-fold in at least 60% of the samples were considered significant candidates. Using these strict criteria, we identified 25 upregulated and 31 downregulated miRNAs between control and cancer tissues. Similarly, we identified 5 upregulated and 23 downregulated miRNAs in ovarian cancer cell lines compared with non-neoplastic HOSE-B cells. 14 miRNAs were deregulated in both tissues and cell lines and are listed in Figure 3A . The number of miRNAs that showed downregulation in tumor samples (n = 31) as well as in cancer cell lines (n = 23) were higher than the number of upregulated (n = 25 in tumor tissues, n = 5 in cell lines) miRNAs. This is in agreement with previously published miRNA profiling studies, most of which have shown downregulation of miRNAs to be more common than upregulation in cancer [8], [13], [19], [25], [26].
Figure 3. Comparisons of miRNA expression in ovarian tissues.
(A) The Venn diagram shows the number miRNAs differentially expressed in ovarian cell lines, in ovarian cancer tissues and in both. For each category, the miRNAs elevated (indicated in red) and downregulated (indicated in green) are indicated below the diagram. (B) The Venn diagram shows the number of differentially expressed miRNAs identified in the current study and the number of miRNAs indentified in 3 previous ovarian cancer studies. The miRNAs in common are indicated below the diagram and color-coded (red: elevated; green: decreased).
Table 2. Differentially expressed miRNAs in ovarian cancer tissues and cell lines.
| Gene Name | Fold Change | Predicted Targets (Target Scan/PicTar) |
| Tissues: 60% samples, fold > = 2.0 or < = 0.5 | ||
| mir-221 | 9.16 | TCF12, KIT, CDKN1B, RIMS3, AIP1, NAP1L5 |
| mir-663 | 4.42 | PRRT1, IQSEC2, SCRT1 |
| mir-29a | 4.38 | PI15, COL3A1, COL4A4, COL1A1, COL4A5 var 2, COL4A5 var 3 |
| mir-142-3p | 4.27 | USP6NL, PRLR, FAM44B, FAM44B, AKT1S1, ARNTL |
| mir-296 | 3.85 | LYPLA2, IQSEC2, RNF44 |
| mir-30a-5p | 3.80 | CELSR3, FLJ35954, LYRIC |
| mir-30e-5p | 3.72 | Not available |
| mir-129 | 3.60 | TNFSF11 variant 2, TNFSF11 variant 1, SOX4 |
| mir-518b | 3.43 | MCF2L, WDR1, TSN |
| mir-29c | 3.34 | PI15, COL3A1, COL4A4, CXXC6, C1QTNF6, COL5A3 |
| mir-605 | 3.20 | VGLL3, PHACTR2, SCAMP1 |
| mir-100 | 3.16 | THAP2, KBTBD8, C4orf16, SMARCA5, BAZ2A, VLDLR |
| mir-10b | 2.94 | ARSJ, BDNF, TFAP2C, PRKWNK3 variant 2, PRKWNK3 variant 1, EPHA8 |
| mir-373* | 2.83 | KPNA4, ATXN7, SYT4 |
| mir-199a | 2.78 | ZNF17,DDR1 variant 3, DDR1 variant 1 |
| mir-637 | 2.71 | RBM9, MNT, DAGLA |
| mir-99a | 2.69 | THAP2, KBTBD8, C4orf16, SMARCA5, BAZ2A, VLDLR |
| mir-487b | 2.66 | NELF, MAP2K4, CDKN2AIP |
| mir-611 | 2.66 | RIMBP2, GTDC1, NTRK2 |
| mir-594 | 2.61 | Not available |
| mir-572 | 2.49 | C14orf101, SAP30BP, ADRBK1 |
| mir-525*-524 | 2.39 | Not available |
| mir-146b | 2.38 | Not available |
| mir-498 | 2.35 | NAP1L3, CXorf1, C9orf5 |
| mir-508 | 2.26 | Not available |
| mir-524* | 0.49 | Not available |
| let-7f | 0.49 | HMGA2, C14orf28, LIN28B, HMGA2, ARID3B, HIC2 |
| mir-424 | 0.48 | FGF2, KIF1B, TMEM16C |
| mir-627 | 0.47 | USP9X, KIAA1853, CCDC73 |
| mir-381 | 0.47 | RAB11FIP2, ARID4B, PTCH1, ARID4B, NBEA, IER5 |
| mir-183 | 0.45 | ITGB1, PFN2, SLAIN1, MAL2, PTPN4, CTDSPL |
| mir-516-5p | 0.44 | Not available |
| mir-361 | 0.43 | Not available |
| mir-551a | 0.43 | LPHN1, ERBB4, ZFP36 |
| mir-142-5p | 0.42 | ZFPM2, FAM18B, FAM91A1, RPS6KA4, AIP1̧ ATP1B1 |
| mir-635 | 0.41 | NARG1, RUNX2, UBE2D3 |
| mir-648 | 0.41 | ONECUT2, BTBD7, FUT9 |
| mir-515-3p | 0.41 | CLOCK, FAT2, FMNL3 |
| mir-542-3p | 0.40 | YPEL5, SR140, RALGPS1 |
| mir-155 | 0.40 | BACH1, ZNF652, RAB11FIP2, ZNF537, BACH1 variant 2 |
| mir-335 | 0.39 | P18SRP, HLF, CALU, KIAA0256, CALU, N-PAC |
| mir-206 | 0.39 | TNKS2, UST, GJA1, SERP1, TNKS2, POGK |
| mir-657 | 0.39 | PRKD3, COPS2, PTPRT |
| mir-195 | 0.38 | FGF2, KIF1B, TMEM16C, ARL2, LUZP1, FGF2 |
| mir-422a | 0.38 | LOC389834, HECTD2, PRKAR2A |
| mir-21 | 0.37 | YOD1, LOC150786, GPR64, PLAG1, RP2, ADNP |
| mir-622 | 0.35 | INSIG2, NFYA, ATF2 |
| mir-34c | 0.35 | DLL1, VEZATIN, FLOT2 |
| mir-106b | 0.35 | EIF5A2, ZNFX1, PKD2, KIAA1404, KIAA1196, ITGB8 |
| mir-346 | 0.33 | BCL6, ACVR2B, NFIB, KIAA0140, BCL6, ROR1 |
| mir-608 | 0.31 | DAGLA, EPHA8, SPRY4 |
| mir-519a | 0.31 | FLJ31818, TGFBR2, SLAIN1 |
| let-7e | 0.31 | HMGA2, C14orf28, LIN28B, HMGA2, ARID3B, HIC2 |
| mir-662 | 0.30 | C6orf49, NEGR1, MKX |
| mir-134 | 0.27 | NIPA1, EML4, PPP1R7, KRAS2 variant b, KRAS2 variant a, ZFPM2 |
| mir-601 | 0.26 | GOLGA8A, GOLGA8B, GOLGA8E |
| Cell lines: 60% samples, fold > = 2.0 or < = 0.5 | ||
| mir-221 | 7.23 | TCF12, KIT, CDKN1B, RIMS3, AIP1, NAP1L5 |
| mir-494 | 2.83 | SOCS6, CNR1, ARHGAP5 |
| mir-190 | 2.59 | OTUD4, NEUROD1, PHF20L1, HSHIN1, STK24, EPC2 |
| mir-508 | 2.45 | Not available |
| mir-146b | 2.4 | Not available |
| mir-424 | 0.5 | FGF2, KIF1B, TMEM16C |
| mir-98 | 0.5 | HMGA2, C14orf28, LIN28B, HMGA2, ARID3B, HIC2 |
| mir-138 | 0.5 | FOXC1, RMND5A, SLC35F1, LPHN3, FLJ13910, CLK3 |
| mir-380-5p | 0.48 | Not available |
| mir-134 | 0.48 | NIPA1, EML4, PPP1R7, KRAS2 variant b, KRAS2 variant a, ZFPM2 |
| mir-106b | 0.47 | EIF5A2, ZNFX1, PKD2, KIAA1404, KIAA1196, ITGB8 |
| mir-200a | 0.46 | ZEB2, KLF12, ZFR, PPM1E, CUL3, SEC10L1 |
| mir-422a | 0.46 | LOC389834, HECTD2, PRKAR2A |
| mir-155 | 0.46 | BACH1, ZNF652, RAB11FIP2, ZNF537, BACH1 variant 2 |
| mir-122a | 0.44 | GYS1, SLC1A5, 8D6A |
| let-7d | 0.43 | HMGA2, C14orf28, LIN28B, HMGA2, HIC2, KIAA1196 |
| mir-141 | 0.43 | ZEB2, KLF12, ZFR, PPM1E, IRS2, KIAA1078 |
| mir-567 | 0.43 | SGMS1, ACLY, GK |
| mir-634 | 0.4 | PDIK1L, NRXN3, HIPK1 |
| mir-648 | 0.39 | ONECUT2, BTBD7, FUT9 |
| mir-154* | 0.38 | ZNF281, PRPF3, C10orf86 |
| mir-346 | 0.37 | BCL6, ACVR2B, NFIB, KIAA0140, BCL6, ROR1 |
| mir-493-3p | 0.36 | Not available |
| mir-520g-520h | 0.36 | TRIM13, LOC153222, KIAA1826 |
| mir-662 | 0.3 | C6orf49, NEGR1, MKX |
| mir-519a | 0.26 | FLJ31818, TGFBR2, SLAIN1 |
| let-7f | 0.24 | HMGA2, C14orf28, LIN28B, HMGA2, ARID3B, HIC2 |
| mir-21 | 0.11 | YOD1, LOC150786, GPR64, PLAG1, RP2, ADNP |
Listed, are mRNAs with fold changes >2.0 or <0.5 in at elast 60% of the samples compared to HOSE B cells. HMGA2, BACH1, BCL6, FAM44B, FGF2, CALU, and TNKS2 were predicted by both Pictar and Target Scan for their respective miRNAs.
miR-221 was the most highly elevated miRNA in both tissues and cell lines (9-fold and 7-fold respectively), while miR-21 was significantly decreased in both sample types (3-fold and 9-fold, respectively) (Table 2). Among the different let-7 family members, let-7e and let-7f showed more than 2-fold deregulation in at least 60% of the tumor samples. The other let-7 family members (let-7g, let-7d, let-7c, let-7a-e, let-7i, let-7a, let-7b were not downregulated as consistently, but each one of them was found decreased 2-fold or more in at least 20% of the tumors. Overall, 94% of the tumors had at least one let-7 family member downregulated at least 2-fold. Cell lines exhibited downregulation of the let-7 family members as well. let-7f was downregulated more than 4-fold in cell lines, while let-7d and let-7a-e were also significantly decreased (more than 2.3-fold and 2-fold, respectively).
We then compared the differentially regulated ovarian miRNAs identified in this study with those reported in three previous studies [15]–[17]. Of a total of 136 miRNAs found deregulated in the previous studies, 16 were also identified in our study ( Figure 3B ). Of these 16 miRNAs, 9 were downregulated (let-7d, miR-106b, miR-122a, miR-141, miR-183, miR-195, miR-200a, miR-335, mir424) and 7 were upregulated (miR-100, miR-199a, miR-296, miR-29a, miR-29c, miR-99a, mir-494). Interestingly, we report 56 miRNAs that have previously not been found deregulated in ovarian cancer ( Figure 3B ).
Predicted gene targets of miRNA genes
miRNAs can regulate a large number of target genes and several databases based on various algorithms are available for predicting the targets of selected miRNAs. Target Scan 3.0 and PicTar were used to predict gene targets of the deregulated miRNAs identified in this study. Table 2 lists the top 3 predicted targets according to both Target Scan and PicTar for each differentially expressed miRNAs. There was little overlap between the targets identified with the two different databases and only 7 targets (FAM44B, BACH1, BCL6, HMGA2, CALU, FGF2, and TNKS2) for 7 miRNAs (let-7 family, miR-98, miR-155, miR-195, miR-346, miR-206, miR-335) were shared between these two databases when the top 3 targets were examined.
Experimental identification of miRNA targets
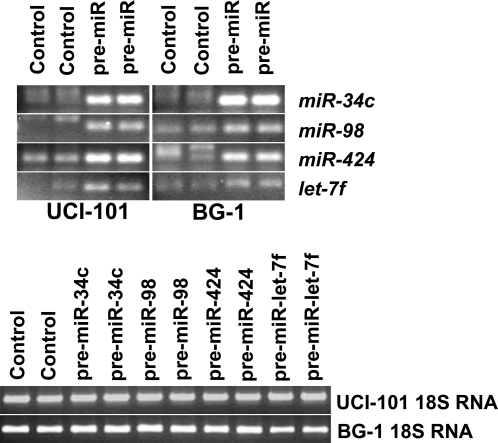
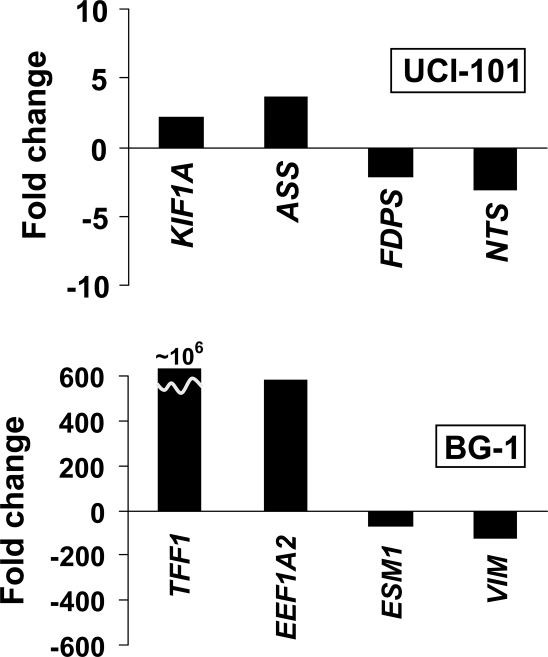
To further study the potential targets of key ovarian miRNAs, we overexpressed miR-34c, miR-98, miR-424, and let-7f in 2 ovarian cancer cell lines (BG-1 and UCI-101). miR-424 and let-7f were downregulated in both tissues and cell lines ( Figure 3A ), while miR-34c and miR-98 were found downregulated in ovarian cancer tissues and in cell lines, respectively. Figure 4 shows the successful over-expression of the miRNAs 72 hours following transfection. We then used Illumina microarrays to investigate mRNA level changes following the overexpression of these candidates. Tables 3 and 4 list the transcripts that were most significantly altered following over-expression of the miRNAs in BG-1 and UCI-101 cell lines, respectively. Interestingly, these tables include genes such as GPX3, MCM7, MSN that have previously been implicated in ovarian cancer. Among the significantly altered genes (absolute Z-ratio>1.5), there are 10 known cancer genes (MSN, PIM1, CBL, COL1A1, COX6C, EVI1, EXT1, HLXB9, PTPN11, TCEA1), 4 tumor suppressor genes (HIF1A, CAV1, GADD45A, PTTG1IP), 26 cell cycle genes (ACAT2, CDK5, FDPS, ID1, LLGL1, MAD2L1, ANLN, ATAD2, C12orf48, CD9, ECT2, GADD45A, GSTO1, HSPD1, IPO7, KNTC2, KRT18, MRPS17, NUDT1, PFN1, PRKCA, SFRP1, SKP2, SNRPA1, VAMP8, WEE1), and 6 genes involved in chromatin remodeling (ASF1A, GCN5L2, ATAD2, CBX2, CBX4, NCOA3). Very little overlap was found between the predicted (Pictar and Target Scan) and experimental targets. Interestingly, the experimental targets varied according to the cell line used, suggesting a significant influence of the molecular background on miRNA target selection. In order to validate the Illumina data, RT-PCR was performed on 8 genes found to be targets of let-7f (KIF1A, ASS, FDPS, NTS in UCI-101 cells, and TFF1, EEF1A2, ESM1, VIM, in BG-1 cells) ( Figure 5 ). We found that, while the absolute values were different, the trends were the same. KIF1A and ASS were found elevated in UCI-101, while FDPS and NTS were downregulated in these cells. TFF1 and EEF1A2 were confirmed to be elevated in BG-1, while ESM1 and VIM were downregulated.
Figure 4. Forced overexpression of selected miRNAs in ovarian cancer cell lines.
Pre-miR-34c, Pre-miR-98, Pre-miR-424, Pre-let-7f were overexpressed in BG-1 and UCI-101. The products for each of the miRNAs is shown in duplicate for the two cell lines used. Significant overexpression of the miRNAs is confirmed. RT-PCR of 18S RNA is shown for each condition to demonstrate equal loading.
Table 3. Top 10 up-regulated and down regulated genes after over expression of mir-34c, mir-98, mir-424 and let-7f in BG-1 cell line.
| mir-34c | mir-98 | mir-424 | let-7f | ||||
| Symbol | Z-ratio | Symbol | Z-ratio | Symbol | Z-ratio | Symbol | Z-ratio |
| GFPT1 | 3.59 | HIST1H4C | 6.34 | AGRN | 4.92 | TFF1 | 8.19 |
| ALOX5AP | 3.41 | CCDC58 | 4.44 | BAT2 | 4.34 | EEF1A2 | 7.89 |
| NT5C3 | 3.36 | ABCA1 | 4.28 | GNG12 | 4.27 | S100P | 7.61 |
| SUMF2 | 3.33 | RPS7 | 4.11 | C20orf149 | 4.27 | BMP7 | 6.35 |
| SSR1 | 3.28 | ALOX5AP | 4.09 | SUMF2 | 4.27 | CRABP2 | 5.69 |
| RND3 | 3.25 | SLC38A1 | 4.04 | SLC31A1 | 4.06 | C3orf57 | 5.68 |
| TSNAX | 3.03 | DBT | 3.92 | NOMO1 | 4.03 | PRSS8 | 5.64 |
| C14orf129 | 2.96 | NRIP3 | 3.88 | PABPC3 | 3.97 | KRT19 | 5.56 |
| ASF1A | 2.96 | CUEDC1 | 3.87 | ANPEP | 3.96 | FBP1 | 5.23 |
| GTF2H3 | 2.90 | C10orf116 | 3.82 | WASF2 | 3.91 | CLDN3 | 5.21 |
| ECHDC2 | -2.57 | ZNF259 | −3.48 | GCN5L2 | −2.95 | MSN | −5.96 |
| MCM7 | −2.59 | WDR58 | −3.54 | ZNF265 | −2.96 | G0S2 | −5.99 |
| ARHGAP17 | −2.71 | MLF2 | −3.59 | CCDC14 | −3.02 | GPX1 | −6.00 |
| PIM1 | −2.71 | FLJ90652 | −3.60 | EIF2B5 | −3.04 | TM4SF1 | −6.02 |
| C8orf13 | −2.74 | GPSN2 | −3.81 | GPX3 | −3.09 | GSTP1 | −6.06 |
| FASN | −3.03 | LLGL1 | −4.13 | SLC37A4 | −3.30 | FSTL1 | −6.09 |
| LEPREL2 | −3.43 | ACTG1 | −4.81 | C16orf48 | −3.35 | IL1A | −6.49 |
| C8orf55 | −3.46 | MRCL3 | −5.08 | NPEPL1 | −3.40 | PRG1 | −6.67 |
| GPX3 | −3.53 | FKSG30 | −5.22 | MAP4K2 | −3.42 | ESM1 | −7.51 |
| NNMT | −3.54 | ACTB | −5.91 | KLF9 | −3.49 | VIM | −7.78 |
Table 4. Top 10 up-regulated and down regulated genes after over expression of mir-34c, mir-98, mir-424 and let-7f in UCI-101 cell line.
| mir-34c | mir-98 | mir-424 | let-7f | ||||
| Symbol | Z-ratio | Symbol | Z-ratio | Symbol | Z-ratio | Symbol | Z-ratio |
| FLNC | 5.03 | HIST1H4C | 5.85 | HIST1H4C | 5.27 | KIF1A | 7.89 |
| MT1A | 4.17 | PSAT1 | 4.02 | ID1 | 3.65 | ASS | 7.41 |
| AKNA | 4.1 | ATP5E | 3.93 | IRX3 | 3.52 | ASNS | 5.47 |
| ID1 | 3.73 | COX7A2 | 3.61 | ID3 | 3.4 | C6orf48 | 4.77 |
| MT2A | 3.52 | NDUFA1 | 3.58 | EBPL | 3.32 | ATF5 | 4.36 |
| EIF4EBP1 | 3.44 | SARS | 3.49 | CXXC5 | 2.88 | IGFBP5 | 4.36 |
| KRT10 | 3.39 | RPS3A | 3.47 | ATP5E | 2.83 | SFRS5 | 4.08 |
| SARS | 3.34 | RPL14 | 3.12 | ALDH9A1 | 2.8 | PHGDH | 3.99 |
| SERPINE1 | 3.32 | ERH | 3.03 | KRTCAP2 | 2.73 | SOX21 | 3.88 |
| SOST | 3.3 | ATP5O | 2.98 | COX7A2 | 2.63 | IARS | 3.73 |
| UBA2 | −2.56 | IPO4 | −2.5 | CMPK | −2.7 | ACLY | −3.56 |
| DDX39 | −2.62 | OAT | −2.54 | CLASP2 | −2.8 | C8orf52 | −3.61 |
| E2F3 | −2.68 | GTF3C3 | −2.66 | ITGAV | −2.87 | HES6 | −3.97 |
| HIF1A | −2.74 | SGK | −2.74 | C13orf23 | −2.9 | C6orf173 | −4.12 |
| LOC441087 | −2.74 | HES6 | −2.75 | TSNAX | −2.96 | DHCR7 | −4.32 |
| EVI5 | −2.78 | XRCC6BP1 | −2.76 | CAND1 | −3.02 | DHCR24 | −4.61 |
| COPS2 | −2.78 | HMBS | −2.78 | TMEM119 | −3.2 | FDFT1 | −4.91 |
| PVRL3 | −2.83 | NUDCD2 | −2.83 | MAD2L1 | −3.23 | ACAT2 | −5.53 |
| ALPP | −2.93 | CDK5 | −3.12 | PIGK | −3.28 | FDPS | −5.55 |
| FDFT1 | −2.96 | TGFBR3 | −3.25 | ARHGAP12 | −3.49 | NTS | −5.62 |
Figure 5. Validation of illumina arrays data for let-7f.
Transcripts identified by illumina arrays to be altered following let-7f overexpression are validated by RT-PCR. Fold changes for genes KIF1A, ASS, FDPS, NTS (in UCI-101 cells), and TFF1, EEF1A2, ESM1, VIM (in BG-1 cells) are shown and confirm the changes identified by illumina arrays.
Using these experimental targets as a starting point ( Table 3 and Table 4 ), we used Ingenuity Pathway Analysis (IPA) to reveal potential diseases, molecular functions, physiological systems, and canonical pathways associated with expression of these miRNAs in BG-1 ( Table 5 ) and UCI-101 ( Table 6 ). Not surprisingly, the analysis identified “cancer” as the main disease associated with these patterns of expression in both cell lines. GI disease was also commonly identified. Interestingly, cellular movement, cell cycle, and cardiovascular functions were systems often found to be related to these expression patterns. There was little consistency in the canonical pathways identified through IPA and except for integrin signaling (found following miR34c or miR98 expression in BG-1), none of the canonical pathways was found more than once in Tables 5 and 6 . This analysis therefore suggests that while the general pathways affected by miR-34c, miR-98, miR-424, and let-7f expression in these two cell lines are related to cancer, the exact molecular pathways targeted are variable and depend on the cell line and on the miRNAs. This again points to the high level of complexity of miRNA target selection and regulation.
Table 5. Ingenuity analysis of Genes affected by transfection of miRNAs in BG-1.
| miR34c | miR98 | miR424 | Let7f | |
| Disease | Cancer | Cancer | Cancer | Cancer |
| Reproductive System | GI disease | GI disease | GI disease | |
| Molecular and Cellular Function | Cell Cycle | Protein Synthesis | Cellular Movement | Cell Growth and Proliferation |
| Lipid Metabolism | Carbohydrate metabolism | Cellular assembly and organization | Cellular Movement | |
| Physiological System | Connective tissue development and function | Hair and Skin development and function | Organism survival | Tissue development and function |
| Hematological System | Hepatic system development and function | Connective tissue development and function | Cardiovascular development and function | |
| Canonical pathways | Nicotinamide Metabolism | Integrin signaling | Tight Junction Signaling | Aryl Hydrocarbon receptor signaling |
| Integrin Signaling | Protein Ubiquitination Pathways | Interferon Signaling | IGF Signaling |
Table 6. Ingenuity analysis of Genes affected by transfection of miRNAs in UCI-101.
| miR34c | miR98 | miR424 | Let7f | |
| Disease | Organismal Injury and abnormalities | Renal and Urological Disease | Cancer | Cancer |
| Hepatic System Disease | Cancer | GI disease | GI disease | |
| Molecular and Cellular Function | Cellular Movement | Cellular signaling | Cellular Movement | Amino acid Metabolism |
| Cell Cycle | Cellular Function and Maintenance | Cell Cycle | Small Molecule Biochemistry | |
| Physiological System | Tissue Morphology | Hair and Skin development and function | Cardiovascular development and function | Nervous system development and function |
| Organismal Development | Cardiovascular development and function | Tumor Morphology | Cardiovascular development and function | |
| Canonical pathways | Aminoacyl t-RNA Biosynthesis | Oxidative Phosphorylation | Oxidative Phosphorylation | Propanoate Metabolism |
| Propanoate Metabolism | Glycine | Ubiquinone Synthesis | Aminoacyl t-RNA Biosynthesis |
Discussion
Previous work has shown that specific miRNA expression signatures in various human cancers can be associated with diagnosis, prognosis, and therapy response [27]. Moreover it has been suggested that specific miRNAs may have crucial roles in the initiation and/or progression of human cancers through their effects on various molecular pathways. A better understanding of miRNA expression in cancer may uncover novel molecular pathways, or novel mechanisms of activation for known pathways. Using the Exiqon miRNA array, the most comprehensive miRNA array available, we have obtained miRNA expression signatures in ovarian cancer and identified several miRNAs differentially expressed in ovarian cancer tissues and cell lines, as well as putative targets for several of these miRNAs.
We have identified a total of 70 miRNAs deregulated in ovarian tissues and 14 of these were also aberrantly expressed in ovarian cancer cell lines ( Figure 3A ). Overall, there were relatively few miRNAs found in common between this study and previous studies ( Figure 3B ). There are several possible explanations for this discrepancy. First, the array platform used for the identification of miRNA is different and may yield different patterns. Second, the material we utilized as normal control is different than what was used in previous studies and the choice of normal is known to influence the outcome of gene profiling analysis [28]. We used immortalized ovarian surface epithelial cells while two of the other three miRNA ovarian studies used whole ovaries. We believe that whole ovarian bulk tissue is not the most appropriate normal control as the ovary is composed of several cell types and that the epithelium only represents a small fraction of the ovarian tissue. The use of an immortalized cell strain also has drawbacks, but we believe it to be a better approximation of normal ovarian epithelial tissues than the mixture present in whole ovaries. In any case, the independent validation of expression patterns is crucial, regardless of the source of normal tissue or tumors used in the study.
We identify let-7f as highly downregulated in both cell lines and tumors. Expression of various let-7 miRNAs have been reported to be down regulated in breast cancers [7], lung cancers [29], thyroid cancer [11], prostate cancer [13], and ovarian cancer [15]–[17]. An association between let-7 downregulation and poor prognosis has been reported in human lung [29] and breast cancer [7]. The finding that the let-7 family of miRNAs regulates the expression of the RAS oncogene family provides a potential molecular basis for the role of let-7 miRNAs in human cancer [20]. let-7 is therefore considered to be a tumor suppressor miRNA and, interestingly, let-7 genes map to loci deleted in multiple types of cancers, including ovarian cancer [20]. In addition, let-7f has been reported to promote angiogenesis by targeting anti-angiogenic genes [30]. The current work as well a previous reports [15]–[17] showing that members of the let-7 family are altered in ovarian cancer suggest that let-7 may represent an important player in this disease. It will be important to identify let-7 downstream targets relevant to ovarian tumorigenesis.
The most consistently and highly upregulated miRNA in both tissues and ovarian cancer cell lines was miR-221 ( Table 2 ). miR-221 has been shown to be highly upregulated in pancreatic cancers [12], in glioblastoma [31] and was implicated in thyroid cancer [32]. miR-221 has been shown to target the oncogene KIT [33] as well as the tumor suppressor p27kip1 [34].
While miR-21 overexpression has been observed in several cancers, including glioblastoma [31], breast cancer [7], [35], lung cancer [8], pancreatic cancer [36], human malignant cholangiocyte cell lines [37] and colon cancer [38], we found this gene highly downregulated in our samples. As a matter of fact, miR-21 was the most significant downregulated miRNA in ovarian cancer when our data was averaged across all samples and cell lines. Our finding is counter-intuitive since it has been found that miR-21 downregulation was associated with increased apoptosis and decreased cell proliferation [35]. miR-21 was suggested to function as an oncogene modulating tumorigenesis through regulation of genes such as BCL2 [35], PTEN [37] and PDCD4 [39]. We are currently investigating the possible targets of miR-21 in our cells as well as the possible reasons behind its downregulation in our system. It is possible that the targets of certain miRNAs could be tissue-specific, which would explain differences in the roles of various miRNA in different tissues.
Other miRNAs found differentially expressed in both tissues and cell lines are miR-146b, miR-508, miR-106b, miR-134, miR-155, miR-346, miR-422a, miR-424, miR-519a, miR-648, miR-662. Several of these miRNAs have been implicated in the other malignancies or in the control of growth and apoptosis. For example, miR-134 downregulated cell growth, and miR-155 increased cell growth in lung carcinoma cells, A549 [40]. The contribution of each of these players to ovarian tumorigenesis remains to be determined.
miR-34c has recently been shown to be a transcriptional target of p53 [41], [42] and can suppress proliferation and colony formation in soft agar in neoplastic epithelial ovarian cells [42]. Interestingly, we found miR-34c deregulated in ovarian cancer tissues and this finding suggests that miR-34c may play a role in ovarian tumorigenesis through its role in the p53 pathway.
Because of the tissue specificity of the miRNAs, different sets of miRNAs are likely to be upregulated or downregulated in cancers of different cellular origins, although it has been reported that miRNA signatures of different cancer types especially epithelial would share some individual miRNAs [38]. Of the miRNAs that were reported here to be differentially expressed in ovarian cancer, several have been similarly deregulated in other cancers. Overall, 31 miRNAs identified in the current study have also been reported de-regulated in other cancer (data not shown). Moreover, 16 miRNAs identified here have previously been reported to be altered in ovarian cancer ( Figure 3b ). More interestingly, we identify 56 miRNAs that have not previously been found differentially expressed in ovarian cancer. These miRNA may play a role in ovarian tumorigenesis and are currently being investigated.
In order to identify potential targets of the differentially expressed miRNAs in ovarian cancer we searched two major databases, PicTar and Target Scan. We found that there was very little overlap between the predicted targets of each algorithm. Indeed, only 7 (FAM44B, BACH1, BCL6, HMGA2, CALU, FGF2, and TNKS2) predicted targets were shared between these two databases. This result illustrates the well-documented difficulties of predicting targets for miRNA [43]. Moreover, it is possible that targets may depend on the cellular environment, (relative ratios of different potential targets may affect the ones most likely to be downregulated) adding another layer of complexity in miRNA target selection. In order to experimentally investigate the changes in mRNA levels, we over-expressed 4 different miRNA candidates (miR-98, miR-424, miR-34c and let-7f) in 2 cell lines and assessed transcript levels using an Illumina oligonucleotide array. Clearly, this approach can only identify miRNA targets that have altered mRNA levels (as opposed to translation inhibition targets), but it has previously been shown that miRNAs can downregulate the levels of a large number of transcripts [44]. While some of the transcripts altered may not represent direct targets of the corresponding miRNA, further analysis of individual candidate mRNAs can provide evidence as to whether they represent direct or secondary targets. Ingenuity Pathway Analysis of the targets revealed a number of pathways and system potentially regulated by the overexpressed miRNAs ( Table 5 and Table 6 ). Interestingly, the predicted pathways were not consistent between the 2 cell lines and again suggested that miRNA target selection and therefore, miRNA function is highly tissue dependent.
When the predicted targets (obtained with PicTar and Target Scan) were compared with the experimental targets obtained by Illumina microarray, we observed no overlap. This could be due to a number of reasons. First, the algorithms for the identification of miRNA targets may preferentially identify targets that result in translational repression, which obviously would not be identified by our microarray experiment. Moreover, as discussed previously, it is likely that the miRNA targets are tissue-specific. In agreement with this possibility is the fact that the targets we identified experimentally were completely different in the two cell lines we used. Interestingly, most of the highly up regulated and down regulated (top 10) genes have already been reported to have roles in cancer or other cellular activities related to cancer ( Table 3 and 4 ), such as GSTP1 [45], [46], HIF-1 [47], [48], and SGK [49].
In this study we have identified miRNAs differentially expressed in ovarian cancer. As a first step in determining their roles in ovarian tumorigenesis, we have started to identify targets of these miRNAs. We show that the predicted targets are different from the experimental and that experimental targets vary according to the cellular background in which miRNAs are expressed. It will therefore be important to methodically study each miRNAs in several models in order to clarify their roles in ovarian cancer. In addition, a combination of overexpression/targeting events by miRNA along with protein array technology will be crucial in identifying the miRNA:mRNA players of ovarian cancer. The picture that is likely to emerge is a highly complex set of interactions between miRNAs and target mRNAs. A better understanding of these pathways will bring us yet closer to an understanding of the molecular mechanism underlying this disease and hopefully to novel approaches for detection and therapy.
Materials and Methods
Cell Lines and Tissue Samples
Ovarian cancer lines BG-1, UCI-101, HEY, OVCA420, OVCA432, OVCA433, OVCAR2, OVCAR3 and OVCAR5 cells were cultured in McCoy's 5A growth medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 units/ml penicillin and 100 µg/ml streptomycin). Ovarian cancer cell line OV90 was cultivated in MCBD 105 medium (Sigma)+Medium 199 (Invitrogen) supplemented with 15% FBS and antibiotics. HOSE-B, an ovarian surface epithelial cell line immortalized with E6 and E7 [50], was cultivated in RPMI1640 supplemented with 10% FBS, antibiotics and 5 ng/ml EGF. Eighteen frozen primary ovarian cancers were obtained through the Collaborative Human Tissue Network Gynecologic Oncology Group (Children's Hospital, Columbus, OH). In addition, nine frozen primary ovarian cancer samples and seven RNA samples (from primary ovarian cancer tissues) were obtained from the Department of Pathology of The Johns Hopkins Medical Institutions (Baltimore, MD). Histological classification of all the tissues is presented in Table 1 and all samples were found to contain greater that 80% cancer cells.
RNA extraction & quantification
Total RNA was obtained using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA was quantified and assessed using the RNA 6000 Nano Kit and 2100 Bioanalyzer (Agilent Technologies UK Ltd, West Lothian, UK).
miRNA microarray hybridization
miRCURY™ LNA miRNA Arrays (Exiqon), consisting of control probes, mismatch probes and 1458 capture probes, perfectly matched probes for all miRNA in all organisms as annotated in miRBase Release 8.1, July 2006 (human miRBase 8.2 is also covered) were used in this study. The capture probes cover 92.3% of miRNAs annotated in miRBAse 9.0. The control probes includes 10 spike-in control probes to assure optimal labeling and hybridization, eight negative capture probes and twelve capture probes that hybridize to small nuclear RNAs. RNA labeling and hybridization was completed according to the manufacturer's instructions. Briefly, RNA from each sample was labeled with Cy3 using miRCURY™ LNA miRNA Array labeling kit. After labeling, the samples were loaded onto the microarray slide and incubated 16–18 hrs at 60°C. After hybridization the slides were washed, dried by centrifugation, and scanned using the Agilent Microarray Scanner (model G2565B).
miRNA microarray analysis
Average values of the replicate spots of each miRNA on the microarray were normalized using global normalization. The correction factor was calculated by dividing the sum of intensities of each sample by the average of all the samples. The normalized values were calculated by multiplying average intensities of each miRNA with the correction factor. The expression levels for each tumor sample were then computed relative to the expression observed in the non-tumorigenic HOSE-B cells. The JMP 6.0.0 software (SAS Institute Inc, Cary, NC) was used for hierarchical clustering, generation of heat maps, and principal component analysis (PCA). Agglomerative hierarchical clustering was applied using the complete linkage method to investigate whether there was evidence for natural groupings of tumor samples based on correlations between gene-expression profiles. The miRNA array data is MIAME compliant has been submitted to the NCBI Gene Expression Omnibus (GEO) database (Accession: GSE10150).
miRNA over-expression in cancer cell lines
Cell lines were tranfected with Pre-miR™ miRNA precursor (Ambion) and control siRNA (Dharmacon) in 24 well plates using siPORT Neo-FX™ (Ambion) according to the manufacturer's protocol. Three days post-transfection, RNA was prepared using RNeasy kit (Qiagen) and changes in RNA levels analyzed using illumina microarrays (below). The changes in miRNA levels were monitored using N-code cDNA synthesis and qPCR.
Illumina Microarray analysis and validation
Biotinylated cRNA was prepared using the Illumina RNA Amplification Kit (Ambion, Inc., Austin, TX) according to the manufacturer's directions starting with approximately 500 ng total RNA. Samples were purified using the RNeasy kit (Qiagen, Valencia, CA). Hybridization to the Sentrix HumanRef-8 Expression BeadChip (Illumina, Inc., San Diego, CA), washing and scanning were performed according to the Illumina BeadStation 500× manual (revision C). Array data processing and analysis was performed using Illumina Bead Studio software. The illumina microarray data is MIAME compliant has been submitted to the NCBI Gene Expression Omnibus (GEO) database.
Validation of the expression patterns of the following genes KIF1A, ASS, FDPS, NTS (in UCI-101), and TFF1, EEF1A2, ESM1 VIM (in BG-1) was performed using RT-PCR as previously described [51]. Primer sequences are available from the authors. Gnes diffentially expressed following miRNA expression were analyzed using Ingenuity Pathway Analysis (Redwood City, California).
Acknowledgments
We thank the members of our laboratory for useful comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007 doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 5.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 6.Lai EC. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 8.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 10.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 11.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007 doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 12.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. Jama. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 13.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 14.Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol. 2007;210:370–377. doi: 10.1002/jcp.20832. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 18.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2007;7 doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 24.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 25.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 26.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 28.Zorn KK, Jazaeri AA, Awtrey CS, Gardner GJ, Mok SC, et al. Choice of normal ovarian control influences determination of differentially expressed genes in ovarian cancer expression profiling studies. Clin Cancer Res. 2003;9:4811–4818. [PubMed] [Google Scholar]
- 29.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 30.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 31.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 32.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 33.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 34.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si ML, Zhu S, Wu H, Lu Z, Wu F, et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 36.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 37.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2007 doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 40.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He L, He X, Lim LP, de Stanchina E, Xuan Z, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 43.John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima T, Elovaara E, Anttila S, Hirvonen A, Camus AM, et al. Expression and polymorphism of glutathione S-transferase in human lungs: risk factors in smoking-related lung cancer. Carcinogenesis. 1995;16:707–711. doi: 10.1093/carcin/16.4.707. [DOI] [PubMed] [Google Scholar]
- 46.Joshi MB, Shirota Y, Danenberg KD, Conlon DH, Salonga DS, et al. High gene expression of TS1, GSTP1, and ERCC1 are risk factors for survival in patients treated with trimodality therapy for esophageal cancer. Clin Cancer Res. 2005;11:2215–2221. doi: 10.1158/1078-0432.CCR-04-1387. [DOI] [PubMed] [Google Scholar]
- 47.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 48.Kitada T, Seki S, Sakaguchi H, Sawada T, Hirakawa K, et al. Clinicopathological significance of hypoxia-inducible factor-1alpha expression in human pancreatic carcinoma. Histopathology. 2003;43:550–555. doi: 10.1111/j.1365-2559.2003.01733.x. [DOI] [PubMed] [Google Scholar]
- 49.Wu W, Chaudhuri S, Brickley DR, Pang D, Karrison T, et al. Microarray analysis reveals glucocorticoid-regulated survival genes that are associated with inhibition of apoptosis in breast epithelial cells. Cancer Res. 2004;64:1757–1764. doi: 10.1158/0008-5472.can-03-2546. [DOI] [PubMed] [Google Scholar]
- 50.Gregoire L, Munkarah A, Rabah R, Morris RT, Lancaster WD. Organotypic culture of human ovarian surface epithelial cells: a potential model for ovarian carcinogenesis. In Vitro Cell Dev Biol Anim. 1998;34:636–639. doi: 10.1007/s11626-996-0012-z. [DOI] [PubMed] [Google Scholar]
- 51.Rangel LBA, Agarwal R, D'Souza T, Pizer ES, Alò PL, et al. Tight Junction Proteins Claudin-3 and Claudin-4 Are Frequently Overexpressed in Ovarian Cancer but Not in Ovarian Cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]