Abstract
In this work, we report the posttranscriptional addition of poly(A)-rich sequences to mRNA in chloroplasts of higher plants. Several sites in the coding region and the mature end of spinach chloroplast psbA mRNA, which encodes the D1 protein of photosystem II, are detected as polyadenylylated sites. In eukaryotic cells, the addition of multiple adenosine residues to the 3′ end of nuclear RNA plays a key role in generating functional mRNAs and in regulating mRNA degradation. In bacteria, the adenylation of several RNAs greatly accelerates their decay. The poly(A) moiety in the chloroplast, in contrast to that in eukaryotic nuclear encoded and bacterial RNAs, is not a ribohomopolymer of adenosine residues, but clusters of adenosines bounded mostly by guanosines and rarely by cytidines and uridines; it may be as long as several hundred nucleotides. Further analysis of the initial steps of chloroplast psbA mRNA decay revealed specific endonuclease cleavage sites that perfectly matched the sites where poly(A)-rich sequences were added. Our results suggest a mechanism for the degradation of psbA mRNA in which endonucleolytic cleavages are followed by the addition of poly(A)-rich sequences to the upstream cleavage products, which target these RNAs for rapid decay.
Keywords: polyadenylylation, mRNA decay, posttranscriptional modification, psbA mRNA, spinach
The addition of multiple adenosine residues to the 3′ end of eukaryotic cell transcripts plays a key role in generating functional mRNA and in regulating mRNA decay (1–3). The poly(A) tail is formed by the addition of about 250 adenylate residues to a 3′ end generated by endonucleolytic cleavage of the precursor RNA (4). Polyadenylylation is performed by the enzyme poly(A)-polymerase and is accompanied by the complex assembly of proteins (5). More recently, poly(A) sequences were also described for bacterial RNAs (6–12). Polyadenylylation greatly accelerated the decay of several Escherichia coli RNAs, and it was therefore suggested to play a role in regulating mRNA decay (6–12).
During leaf development and plastid differentiation, the levels of many plastid mRNAs vary dramatically. RNA processing and differential stability are important factors that contribute to the developmental mRNA accumulation. In higher plant chloroplast, mRNAs are transcribed as precursor RNAs that undergo a variety of maturation events, including cis- and trans-splicing, cleavage of polycistronic messages, processing of 5′ and 3′ ends, and RNA editing (13–17). A general characteristic of the plastid protein coding region is the presence of inverted repeats sequences in the 3′ untranslated region (UTR), which form a stem–loop structure when transcribed to RNA. The 3′ ends of the chloroplast mRNAs are located several nucleotides 3′ to these stem–loop structures, which were nevertheless shown to not function as efficient transcriptional terminators (18). Instead, these structures serve as efficient RNA processing elements in vitro and are capable of stabilizing upstream RNA fragments in vivo and in vitro (18–20).
To study the degradation pathways of mRNA in the chloroplast of higher plants, an in vitro degradation system based on lysed spinach chloroplasts has been recently developed (21). It was shown that the degradation of the psbA mRNA is initiated by endonucleolytic cleavages within the amino acid coding region of the message, followed by subsequent decay that is facilitated by exonucleolytic activities (21). Nevertheless, the precise mechanism in which the stability of a specific RNA is posttranscriptionally modulated in the chloroplast during plant development and in response to physiological changes (such as light intensity and quality) is still not understood (22). Furthermore, the prokaryotic nature of the chloroplast decoding machinery, in which transcription and translation can theoretically be coupled, requires a mechanism to rapidly degrade immature RNAs, preventing translation of aberrant proteins.
The discovery that polyadenylylation of bacterial mRNA significantly affects transcript stability and may trigger rapid degradation promoted us to look for posttranscriptional polyadenylylation of mRNA in higher plant chloroplasts. The results presented in this paper show that poly(A)-rich sequences are posttranscriptionally attached to the plastid psbA mRNA at specific positions. Unlike eukaryotic and bacterial mRNAs, the sequence moiety does not consist exclusively of poly(A) but is rather composed mostly of purines, 70% adenosines and 25% guanosines. Cytidines and uridines make up the remaining 5%. Specific endonucleolytic cleavage sites that perfectly matched the sites where the poly(A)-rich sequences are added were identified. In vitro analysis of the chloroplast polyadenylylation activity revealed specificity to ATP and GTP, reflecting the composition of the poly(A)-rich tails. Furthermore, the activity is specific for the substrate structure, as unstructured RNAs are polyadenylylated with high efficiency compared with those molecules forming the stem–loop structure characteristic of the mature plastid mRNA 3′ end. Polyadenylylated RNA was rapidly degraded when incubated in chloroplast extract. The implications of these results are discussed below.
MATERIALS AND METHODS
Plant Growth, Chloroplast Isolation, and RNA Extraction.
Chloroplasts were isolated on Percoll gradients from leaves of hydroponically grown spinach plants (Spinacia oleracea cv. Viroflay) under a 10.5 h light/13.5 h dark cycle as described (23). RNA was extracted from leaves or chloroplasts and depleted of DNA by DNase digestion as described (23).
Determination of Poly(A) Tail Length.
Chloroplast and total leaf RNAs (10 μg) were 3′ end-labeled with [32P]pCp and T4 RNA ligase before digestion with 25 μg RNase A and 200 units of RNase T1 for 1 h at 37°C (12). Poly(A) tails were resolved in 12% polyacrylamide sequencing gels containing 7 M urea.
PCR Amplification and Identification of Polyadenylylation Sites.
Chloroplast RNA (10 μg) was used to synthesize oligo(dT)-primed cDNA with the dT-adapter primer [5′-GACTCGAGTCGACATCGA(T)17] using avian myeloblastosis reverse transcriptase at 37°C for 10 min followed by incubation at 42°C for 2 h. This cDNA was amplified by PCR with one of the following primers: 5′-2, 5′-GATCAGGGAAACCACAG; 5′, 5′-ACTTGGGCTGATATC; or 5′-SL, CAAAACAAGAAATCGGTTATTGC, oligonucleotides extending from positions 669 to 685, 941 to 954, and 1127 to 1149 of the psbA gene, respectively (24) (see Fig. 1A), and the adapter oligonucleotide 5′-GACTCGAGTCGACATCGATT [identical to the sequence of nucleotides 3′ to the (T)17 of the dT-adapter oligonucleotide]. Amplification was carried out for 50 cycles of 1 min each at 94°C, 55°C, and 72°C, with the addition of extra enzyme after 25 cycles. PCR products were cloned, and colonies that hybridized to the dT-adapter oligonucleotide and to the psbA gene-specific probe were sequenced.
Figure 1.
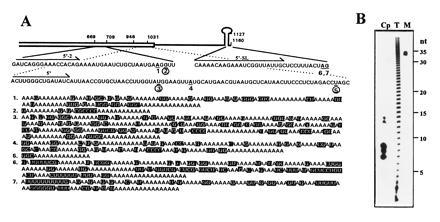
Posttranscriptional addition of poly(A)-rich sequences to spinach chloroplast psbA mRNA. (A) Nucleotide sequences and locations of the poly(A)-rich stretches that were PCR amplified from oligo(dT)-primed chloroplast cDNA. A schematic representation of the psbA RNA 3′ region is shown. The open box denotes the coding region. The single line represents the 3′ UTR in which the inverted repeats are symbolized by a stem–loop structure. The psbA gene is numbered according to Zurawski et al. (24), and the nucleotides where poly(A)-rich sequences had been added are numbered 1–7, underlined, and printed in boldface type. The poly(A)-rich addition sites coincident with endonucleolytic cleavage sites are circled (numbers 2, 3, and 5). The 3′ end of the mature psbA mRNA is located at nucleotides 1159A and 1160G, immediately following the inverted repeats that form the stem–loop structure (24); two of the poly(A)-rich addition sites, 6 and 7, were located at this position. The gene-specific primers are indicated by arrows. The poly(A)-rich stretches that are shown below, numbered 1–7, are those that were posttranscriptionally added to sites 1–7, respectively. The nucleotides that are not adenines are shaded. (B) Size of poly(A) tracts in chloroplast RNA. Total RNA from mature leaf cells (T) and purified chloroplast RNA (Cp) were labeled with [32P]pCp, followed by complete digestion of the RNAs with RNase T1 and RNase A. An end-labeled, 35-nt-long oligonucleotide was run in the same gel as a size marker (M).
Determination of the Initial Cleavage Sites in the Degradation of psbA mRNA.
Intact chloroplasts were isolated, lysed, and incubated at 25°C at a protein concentration of 10 mg/ml in 20 mM Hepes, pH 7.9/60 mM KCl/10 mM MgCl2/0.1 mM EDTA/2 mM DTT/0.5 mg/ml yeast tRNA/20% glycerol (21). Reactions were terminated as described (21), and RNA was extracted. For high resolution RNA blot analysis, the RNA was resolved in a denaturing 5% polyacrylamide gel, electroblotted to a nylon membrane, and hybridized with a 32P-labeled oligonucleotide primer complementary to positions −77 through −55 (5′-TCGCTAGAAATAGAAATTGAAAG), or to positions 1084–1105 (5′-GCCCCTTTACTTTCACTAACTC) (24). Primer extension analysis was performed as published (21). Primers used were 5′-GAAGAAGTGTAAAGTTCGAGAG and 5′-GCCCCTTTACTTTCACTAACTC (complementary to positions 801–823 and 1089–1110, respectively).
Soluble Protein Extract, in Vitro Processing, and Polyadenylylation.
A soluble protein extract capable of 3′ end processing of chloroplast RNAs was prepared from isolated intact chloroplasts as described (25). The plasmids used for in vitro transcription of 3′ UTR mRNAs of spinach chloroplast genes psbA and petD have been described (23, 25). RNAs resembling the mature 3′ end processing products were obtained by in vitro processing of precursors RNA in scaled-up reactions and reisolation of the product. The plasmid for transcription of petD 3′ end RNA was linearized by DraI to generate the RNA that 3′ ended in the stop codon of the amino acid coding region (23). RNAs were transcribed using T7 RNA polymerase and were radioactively labeled with [α-32P]UTP to a specific activity of 8 × 103 cpm/fmol (25). The full-length transcription products were then purified on 5% denaturing PAGE gels. In vitro RNA processing experiments were carried out as described (25). Briefly, in vitro synthesized RNA (2 fmol) was incubated in the chloroplast soluble protein extract (1 mg/ml) for 1 h or for the times indicated in the figure legends. After incubation, the RNA was isolated and analyzed by gel electrophoresis and autoradiography. In vitro polyadenylylation experiments were performed as the in vitro processing assays but with the addition of 0.5 mM ATP (or the respective nucleotides) to the reaction mixture. Polyadenylylation reactions using yeast poly(A) polymerase (obtained from United States Biochemical) were performed according to the manufacturer’s instructions.
RESULTS
Detection of Polyadenylylated RNA in Chloroplast and Identification of Polyadenylylated Sites.
To detect polyadenylylated RNA in the chloroplast, intact chloroplasts from spinach were purified on Percoll gradients and washed several times to reduce the contamination of cytoplasmic polyadenylylated RNA to a minimum. RNA was extracted from these chloroplast preparations and analyzed by Northern blotting using an oligo(dT) probe. A low hybridization signal was reproducibly detected in chloroplast RNA preparations obtained from mature leaves that were illuminated before the purification of chloroplasts. In another experiment, psbA RNA was purified by hybridization of chloroplast RNA to a DNA fragment corresponding to the psbA gene. This hybrid-selected psbA RNA showed a detectable hybridization signal with oligo(dT) probe, whereas the control in vitro synthesized psbA RNA did not (not shown). Therefore, illuminated mature spinach leaves were used for further studies that focused on polyadenylylation of psbA RNA. Detection and analysis of polyadenylylated psbA mRNA was performed by cloning and sequencing of the respective cDNAs. Chloroplast RNA isolated from light-adapted mature leaves was used as a template to synthesize cDNA primed with oligo(dT)17-adapter oligonucleotide (see Materials and Methods). To precisely determine the site of the poly(A) addition, as well as the posttranscriptionally added sequences, the psbA–poly(A) junctions were amplified by PCR, cloned, and sequenced. Our experimental procedure included gel purification and cloning in bulk of the major PCR fragments. Positive colonies were selected by hybridization to both psbA gene-specific and oligo(dT) probes. With use of this strategy, about 50 colonies out of several hundred were obtained when primers 5′-2 or 5′ were used (Fig. 1A). From these, 15 were sequenced, and about equal numbers of sequences 1–5 presented in Fig. 1A were obtained. For the primer 5′-SL, which is located 12 nucleotides in front of the 3′ end of the RNA, only 2 colonies out of several hundred were found to hybridize to the psbA and oligo(dT) probes (numbers 6 and 7 in Fig. 1). Sequences cloned are part of the psbA mRNA that continues at different points in the 3′ direction as adenine-rich sequences that were not found in the DNA sequence of the gene (Fig. 1A). The psbA–poly(A) junctions are located at several positions within the amino acid coding sequence (numbers 1–5 in Fig. 1A) and at the 3′ end of mature psbA mRNA (numbers 6 and 7 in Fig. 1A). The 3′ UTRs of most chloroplast transcription units contain inverted repeats that can fold into stem–loop structures (18). The mature 3′ end of the mRNA, which is formed by processing of a longer precursor RNA, is located immediately at the 3′ end of the stem–loop structure (Fig. 1A) and, like bacteria mRNAs, is mostly not polyadenylylated in its steady-state form. Therefore, the detection of only two PCR-amplified poly(A)-rich sequences added to the mature 3′ end of the psbA mRNA, compared with about 50 clones added to nucleotides in the psbA coding region, indicated that it is polyadenylylated at a low frequency.
Analysis of the Posttranscriptionally Added Sequences.
The posttranscriptionally added RNA tails detected in chloroplasts, unlike those of mRNAs encoded by nuclear and bacterial genes, are not adenosine ribohomopolymers. The tail is a mixture of mostly adenosines (70%), guanosines (25%), and cytosines and uridines (5%) (Fig. 1A). In general, sequences added posttranscriptionally to the psbA mRNA or respective fragments are mostly purines and rarely pyrimidines. However, only poly(A)-rich sequences could be amplified by using oligo(dT) to prime the cDNA synthesis. Other posttranscriptionally added sequences of nucleotides, if they exist in the chloroplast, would not have been detected. The length of the RNA tails varies, with the longest one recovered (number 3 in Fig. 1A) being 270 nucleotides. However, as an oligo(dT) primer was used for reverse transcription, it is likely that the poly(A)-rich sequence that served as a template for this clone was even longer because of the annealing position of the primer. We speculate, therefore, that the poly(A)-rich sequences added posttranscriptionally to psbA mRNA may be as much as several hundred nucleotides long.
Sensitivity of Poly(A)-Rich Tails to Ribonuclease Digestion.
The discovery of A- and G-rich instead of exclusively poly(A) sequences prompted us to examine whether this observation can be generalized for posttranscriptionally added RNA tails in spinach chloroplasts. Total leaf RNA and chloroplast RNA were labeled at the 3′ end with [32P]pCp and T4 RNA ligase and then completely digested with RNase A (cutting after C and U residues) and RNase T1 (cleaving after G residues). Analysis of the products on a 12% polyacrylamide gel revealed that the total leaf RNA was degraded into a ladder of A-containing homopolymers, as previously reported for polyadenylylated RNA (Fig. 1B, lane T) (3, 12). However, complete degradation of the chloroplast RNA by this method resulted in clusters of mainly 7, 8, 9, 13, or 14 adenosine residues (Fig. 1B, lane Cp). This result suggests that most or all chloroplast RNA tails are poly(A)-rich rather than ribohomopolymers.
Taken together, the results demonstrated that poly(A)-rich sequences are added posttranscriptionally at several points of the psbA gene in the chloroplast. Unlike the situation in nuclear-encoded and bacteria RNAs, the purine-rich sequences are not ribohomopolymers of adenosines, but mostly adenosines and guanosines.
Analysis of psbA Degradation Intermediates.
To identify stable degradation intermediates and the initial endonucleolytic cleavage sites of psbA mRNA, an in vitro assay was developed (21). We employed this assay to analyze whether polyadenylylation may occur at sites that are generated by prior endonucleolytic cleavage within the psbA mRNA. Isolated intact chloroplasts were lysed and incubated in the presence of 0.5 mg/ml tRNA, which was shown to inhibit the activity of fast proceeding exonucleases (21). Several major degradation intermediates that probably result from endonucleolytic cleavages can be observed by high-resolution RNA blot analysis (Fig. 2A). The majority of these molecules can only be detected with primers corresponding to the 3′ end of psbA mRNA, indicating a rapid degradation of the 5′ fragment. The exact positions of the cleavage sites were determined by primer extension (Fig. 2B) and are indicated in Fig. 1A by circled numbers. Three of the positions identified perfectly matched one nucleotide 3′ to the sites of poly(A)-rich addition, implying that addition of the poly(A)-rich sequence can occur following cleavage of the mRNA by an endonuclease. The 5′ moiety of the cleaved psbA mRNA, which undergoes addition of the poly(A)-rich sequence, is rapidly degraded.
Figure 2.
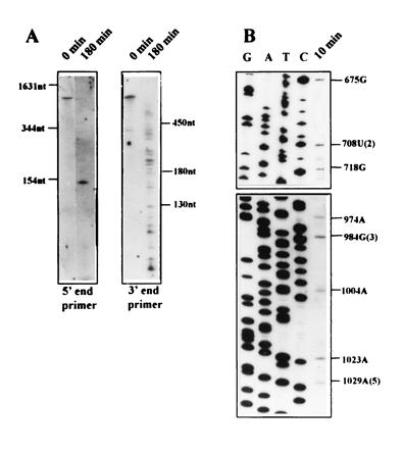
Determination of the initial cleavage sites in the degradation of psbA mRNA. (A) Characterization of psbA degradation intermediates by high-resolution RNA blot analysis. Lysed chloroplasts from spinach leaves were incubated at 25°C for 180 min, and the RNA was recovered and separated in denaturing polyacrylamide gels, which were electroblotted to nylon membranes and hybridized with a 32P-radiolabeled oligonucleotide complementary to the 5′ or 3′ end of psbA mRNA. (B) Determination of the initial psbA decay intermediate cleavage sites by primer extension analysis. The 5′ ends of the degradation intermediates were determined by primer extension analysis. Oligonucleotide primers complementary to positions 801–823 (Upper) or complementary to positions 1084–1105 (Lower) of the coding region of psbA mRNA were used. Lanes G, A, T, and C show the sequencing reactions of the corresponding cloned fragments. Nucleotide numbers of reverse transcriptase stops in the psbA gene are indicated. Positions of the three corresponding poly(A)-rich addition sites are given in parentheses.
In Vitro Polyadenylylation of Synthetic RNAs in the Chloroplast Protein Extract.
To characterize the biochemical properties of the enzyme activity responsible for adding the poly(A)-rich sequences, we tested whether synthetic RNA can be polyadenylylated by components of the chloroplast soluble protein extract. This chloroplast protein extract allows accurate transcription and 3′ end processing of chloroplast RNA (18, 23, 25). As depicted in Fig. 3A, synthetic transcripts corresponding to the precursor of the psbA 3′ end were adenylated by an activity in this extract to the same extent as with yeast poly(A) polymerase. Accumulation of polyadenylylated RNA in the chloroplast protein extract is only transient: under our experimental conditions, polyadenylylated psbA RNA was fully degraded after 90 min of incubation (Fig. 3A). The transient accumulation of the in vitro polyadenylylated RNA is reminiscent of the low steady state concentration of polyadenylylated RNAs in vivo, which are only detected by PCR. Specificity of the in vitro polyadenylylation activity to different nucleotides was tested by replacing the ATP with each of the other nucleotides. Addition of about 200 residues of ribohomopolymer was observed with GTP or ATP, but not with UTP or CTP (Fig. 3B). This specificity of activity toward ATP and GTP is in good agreement with the poly(A)-rich sequences detected in vivo, which were 95% adenines and guanosines (Fig. 1). However, we cannot exclude the possibility that in addition to the poly-purine sequences, poly-pyrimidine stretches are also present in the chloroplast and were not detected in this work as the result of using oligo(dT) primer for the reverse transcription–PCR. In addition, 0.5 mM ATP and GTP inhibited the in vitro 3′ end processing reaction usually observed; the stable, right size, processing product of 219 nucleotides does not accumulate (Fig. 3). A similar polyadenylylation activity was obtained using a soluble protein extract isolated from chloroplasts of the green algae Chlamydomonas reinhardtii (V. Liveanu, unpublished results). In addition to the psbA 3′ end RNA, in vitro polyadenylylation activity was also observed for synthetic RNAs corresponding to rbcL (encoding the large subunit of the ribolose-1, 5-bisphosphate carboxylase), petD (encoding subunit IV of cytochrome b6f) (Fig. 3B), and rbs14 (encoding 30S ribosomal subunit protein 14) precursor 3′ ends that were tested (not shown).
Figure 3.
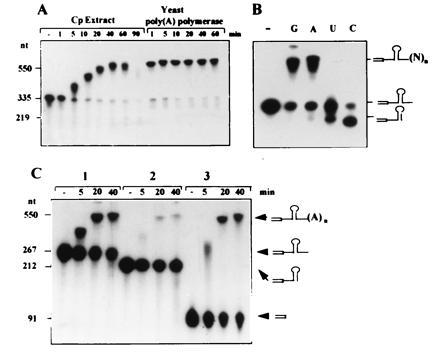
Polyadenylylation of synthetic psbA transcripts in vitro using an extract of soluble chloroplast proteins. (A) Radioactive RNA corresponding to the unprocessed 3′ UTR of the chloroplast psbA mRNA was incubated in the presence of 0.5 mM ATP with chloroplast extract (Cp Extract) or yeast poly(A) polymerase. Lane −, RNA that was incubated for 60 min without addition of protein. The lengths of the RNAs in nucleotides are shown at the left. (B) The synthetic RNA described in A was incubated for 1 h with the chloroplast protein extract in the presence of 0.5 mM ATP (lane A), CTP (lane C), GTP (lane G), or UTP (lane U). (C) RNA terminating with a stem–loop structure is poorly polyadenylylated in chloroplast protein extract. Synthetic RNAs corresponding to the unprocessed precursor, mature 3′ end and part of the coding region of the petD 3′ end RNA, were incubated for the times indicated in the figure with chloroplast extract in the presence of 0.5 mM ATP. A schematic representation of the RNA substrates is shown on the right. The open box denotes the amino acid coding region of the mRNA.
A 100-kDa RNA-binding chloroplast protein (100RNP-PNPase) has been recently described as displaying an exonuclease activity and sequence similarity to the bacteria enzyme polynucleotide phosphorylase (PNPase) (26). To determine if the activity described here reflected artificial polymerase activity of the 100RNP-PNPase, the chloroplast protein extract was depleted of this enzyme by fractionation on a single-stranded DNA cellulose or on heparin agarose columns binding the 100RNP-PNPase (26). Polyadenylylation activity was observed exclusively in the 100RNP-PNPase-depleted fractions (the unbound fraction of the columns) (data not shown). Because immunoblots using specific antibodies to the 100RNP-PNPase (26) demonstrated that the unbound fractions of the heparin-agarose and single-stranded DNA-cellulose columns were completely depleted of this protein, we concluded that the posttranscriptional addition of poly(A)-rich sequences described in this work was performed by a different enzyme.
RNAs Terminating with a Stem–Loop Structure Are Poorly Polyadenylylated in Chloroplast Protein Extract.
To investigate whether in vitro polyadenylylation activity discriminates between RNAs of different structure, three RNAs corresponding to the petD 3′ end were incubated in the chloroplast protein extract in the presence of ATP. The first RNA (267 nt) included sequences of the 3′ UTR and extended 55 nt 3′ to the stem–loop structure. This RNA represented the precursor of the 3′ end processing reaction. The second RNA (212 nt) terminated right in the 3′ end of the stem–loop and resembled the mature 3′ end of the petD mRNA in the chloroplast. The third RNA (91 nt) terminated in the end of the gene’s amino-acid coding region. Results of this experiment demonstrate that the efficiency of polyadenylylation of the RNA terminated by the stem–loop structure was very low (Fig. 3C). Nevertheless, this is the 3′ end that is present in the chloroplast at high concentrations. In comparison, the two other RNAs were highly polyadenylylated (Fig. 3C). Similar results were observed with psbA, rbcL, and rps14 3′ end RNAs (data not shown). It was therefore concluded that RNAs terminated with a stem–loop structure, resembling the mature 3′ ends of chloroplast RNAs, are poorly adenylated in the chloroplast protein extract. This result is again in good agreement with the data obtained in vivo. The poly(A)-rich sequences’ addition sites at the mature 3′ end of psbA mRNA, as detected in vivo by reverse transcription–PCR and cloning (numbers 6 and 7 in Fig. 1), were much less frequently obtained than the other clones. These data show that the structured mature 3′ end is an inefficient substrate for polyadenylylation activity in vivo and in vitro.
Polyadenylylated RNA Is Specifically Degraded in the Chloroplast Protein Extract.
To determine whether polyadenylylation affects the stability of RNA when incubated in the chloroplast protein extract, a psbA precursor 3′ end RNA was polyadenylylated in vitro, re-isolated, and analyzed in the standard in vitro 3′ end processing assay. Fig. 4A shows that nonpolyadenylylated psbA 3′ end RNA is processed and a stable product of correct size accumulates, as previously described (18, 23, 25). When the polyadenylylated precursor 3′ end RNA was incubated with the chloroplast protein extract, however, it was degraded without the accumulation of a stable product, and after 2 h of incubation, most of it could no longer be detected (Fig. 4A). To verify that destabilization of the psbA 3′ RNA did not occur as a result of the addition of 200 nucleotides to the 3′ end of the RNA, an RNA of 550 nucleotides extended into the vector sequences was transcribed using the plasmid of the psbA 3′ end DNA sequence digested with PvuII instead of HindIII (25). When this RNA was incubated with chloroplast protein extract, it was not degraded, as compared with the polyadenylylated RNA (data not shown). Previous studies have shown that RNA that does not carry a stem–loop structure in the 3′ end is rapidly degraded in the chloroplast protein extract (27). Detection of the poly(A)-rich sequences at the 3′ end of the unstable endonucleolytic cleavage products suggests that polyadenylylation may target the RNA to rapid degradation. To test whether the RNA degrading enzymes of the chloroplast extract have a higher affinity for polyadenylylated RNA, synthetic RNA corresponding to a truncated mutant of the petD 3′ end, which does not end with a stem–loop structure (23), was polyadenylylated. When polyadenylylated and nonpolyadenylylated RNAs were added together to the chloroplast extract, the polyadenylylated RNA was degraded much more rapidly and the nonpolyadenylylated RNA was stabilized (Fig. 4B). Addition of poly(G), in contrast, had no such effect. An increase in protein concentration of the chloroplast extract resulted in a more rapid degradation of the polyadenylylated RNA as well as slower degradation of the nonpolyadenylylated RNA (not shown). Therefore, the addition of poly(A) sequences to RNA resulted in destabilization and prevented accumulation of a product of correct size in the case of precursor 3′ end RNA.
Figure 4.
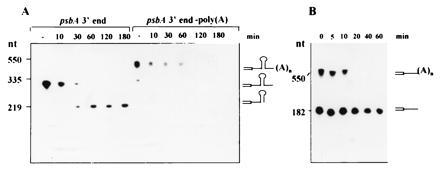
Effect of polyadenylylation on the 3′ end processing and degradation of RNAs in vitro. In vitro-synthesized [32P]RNA corresponding to the 3′ end of the psbA precursor RNA (A) or the petD amino acid coding region and part of the 3′ UTR (B), and the same RNAs that were first polyadenylylated in vitro, were incubated in the chloroplast protein extract without the addition of ATP, either each alone (A) or as a mixture (B). Samples were taken at the times indicated in the figure and analyzed by denaturing gel electrophoresis and autoradiography.
DISCUSSION
In this work, the addition of poly(A)-rich tails to chloroplast-encoded mRNAs was examined. Poly(A)-rich tracts could be detected at the 3′ end of mRNA fragments cleaved endonucleolytically, as well as at the 3′ end of mature mRNAs. Poly(A)-rich tailed RNAs are highly unstable; therefore, they could only be detected by the sensitive technique of reverse transcription–PCR. We suggest that degradation of chloroplast psbA mRNA is initiated by endonucleolytic cleavages, followed by the addition of a poly(A)-rich sequence that may target this RNA for rapid degradation, possibly to prevent translation of aberrant RNAs.
Despite clear indications that polyadenylylation of mRNA exists in bacteria, for about 20 years it was considered to be exclusively one of the unique properties of nuclear-encoded mRNA in eukaryotic cells (1–7). It is believed to function in both mRNA turnover and in facilitating translation (1–4). However, polyadenylylation of several RNAs has been recently reported in bacteria (6–12). In these cases, it was suggested that this posttranscriptional modification is part of a mechanism that targets the corresponding RNA for rapid degradation (6, 9–12). Polyadenylylation of RNA in the mitochondria was described a long time ago (34). In addition, cDNAs of mitochondria-encoded genes harboring poly(A) stretches in the 3′ end could be isolated from oligo(dT)-primed cDNA libraries, suggesting that polyadenylylation of mRNA occurs in that organelle (28). Surprisingly, the detection of poly(A)-containing RNA from plastids of maize was described 20 years ago (29). The rediscovery of posttranscriptional addition of poly(A)-rich sequences in the chloroplast demonstrates that mRNA polyadenylylation is a universal posttranscriptional modification, occurring in all major genetic systems.
In higher plant chloroplasts, psbA mRNA stability is modulated during leaf development and in response to physiological changes such as dark–light transitions (13–16, 30–32). The results of this work suggest that targeting of mRNA for rapid degradation could be achieved by the addition of poly(A)-rich sequence to an endonucleolytic cleavage product, full-length transcribed molecule, or possibly an unprocessed transcribed precursor RNA. Our in vitro experiments imply that precursor RNA to which poly(A)-rich sequence is added will be degraded and does not accumulate as a 3′ end processed stable product of correct size (Fig. 4). In other words, following transcription, the precursor psbA RNA in the chloroplast can either undergo 3′ end processing to generate a stable product or the addition of poly(A)-rich sequences and subsequent rapid degradation. In vitro, the respective pathway depends on the concentration of ATP present in the reaction. Processing occurs at an ATP concentration of less than 0.5 mM, and polyadenylylation followed by rapid decay occurs at high concentration (Fig. 3). In the chloroplast, however, other factors such as redox potential, photosynthetic electron flow, posttranslational modifications of the enzymes involved, or specific regulatory proteins may determine which way the precursor RNA will go.
Our experiments showed that mRNAs terminated with a stem–loop structure at their 3′ end are polyadenylylated in vitro and in vivo at very low efficiency. Therefore, the 3′ end processing of precursor RNA will generate mature RNA that is stable, translatable, and mostly unable to be elongated in its 3′ end by the addition of poly(A)-rich sequence. In this case, what will be the initial process to target this mRNA for degradation at an indicated time? A possible scenario is endonucleolytic cleavage removing the stem–loop 3′ end, allowing the addition of poly(A)-rich sequences and targeting for degradation. An endonucleolytic cleavage has been shown to be the initial step in the degradation of many bacterial and nuclear-encoded RNAs (1–3).
Recent work in bacteria suggests that polyadenylylated RNAs are degraded by the coordinated activity of the endonuclease RNase E and the exonuclease polynucleotide phosphorylase (PNPase), which have been found in the same multiprotein complex in E. coli cells (33). An analogous high-molecular-weight enzymatic complex that contains a PNPase-like exoribonuclease and a site-specific endoribonuclease has been recently described in spinach chloroplasts (26). This suggests a similarity between bacteria and chloroplast in the regulation of RNA degradation by the addition of poly(A)-rich sequences to endonucleolytic cleavage sites. For example, when the endonuclease of the high-molecular-weight complex, a 67-kDa protein that crossreacts to antibodies raised against bacterial endonuclease RNase E, was analyzed in vitro, it cleaved synthetic RNA corresponding to the 3′ end of the petD mRNA in the amino acid coding region (26). Ongoing experiments are exploring the possibility that the chloroplast 67-kDa endonuclease can mediate degradation of mature RNA by generating truncated RNA, which is elongated by poly(A)-rich sequences and subsequently degraded.
We have demonstrated that poly(A)-rich sequences are posttranscriptionally added to chloroplast mRNA at 3′ termini generated by endonuclease(s) cleavages and exonucleases (the mature 3′ end). In this respect, chloroplast poly(A) polymerase(s) are similar to eukaryotic enzymes that polyadenylylate mRNA at processing sites (4, 5). The chloroplast poly(A) polymerase(s) may have affinity either for the RNA processing complexes as in mammalian cells (4, 5) or for other features of mRNA sequence or structure. The results presented here suggest that poly(A)-rich tails play a major role in the rapid degradation of intermediary products of mRNA decay as well as precursor and mature mRNA.
Note Added in Proof.
Polyadenylylation of chloroplast mRNA has also been described in a recent paper by Kudla et al. (35).
Acknowledgments
We thank Drs. David Stern, Eliezer Lifshitz, Haim Manor, Benjamin Horwitz, and T. Baumstark for critical reviews of the manuscript. We thank Prof. D. Riesner for continuous support and E. Reinartz for excellent technical assistance. This research was supported by the Israel Science Foundation administrated by the Israel Academy of Sciences and Humanities (to G.S.) and by the Deutsche Forschungsgemeinschaft (to P.K.).
Footnotes
Abbreviation: UTR, untranslated region.
References
- 1.Jackson R J, Standart N. Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 2.Baker E J. In: Control of mRNA Stability. Belasco J G, Brawerman G, editors. New York: Academic; 1993. pp. 367–415. [Google Scholar]
- 3.Sachs A B. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 4.Wahle E, Keller W. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 5.Manley J L. Curr Opin Genet Dev. 1995;5:222–228. doi: 10.1016/0959-437x(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S N. Cell. 1995;80:829–832. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 7.Manley J L. Proc Natl Acad Sci USA. 1995;92:1800–1801. doi: 10.1073/pnas.92.6.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao G J, Sarkar N. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu F, Lio-Chao S, Cohen S N. Proc Natl Acad Sci USA. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu F, Cohen S N. Nature (London) 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 11.Hajnsdorf E, Braun F, Haugel-Nielsen J, Regnier P. Proc Natl Acad Sci USA. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Hara E, Chekanova J A, Ingle C A, Kushner Z R, Peters E, Kushner S R. Proc Natl Acad Sci USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullet J E. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:475–502. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- 14.Mullet J E. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruissem W. Cell. 1989;56:161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- 16.Gruissem W, Schuster G. In: Control of mRNA Stability. Belasco J G, Brawerman G, editors. New York: Academic; 1993. pp. 329–365. [Google Scholar]
- 17.Rochaix J-D. Annu Rev Cell Biol. 1992;8:1–28. doi: 10.1146/annurev.cb.08.110192.000245. [DOI] [PubMed] [Google Scholar]
- 18.Stern D B, Gruissem W. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- 19.Stern D B, Radwanski E R, Kindle K L. Plant Cell. 1991;3:285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blowers A D, Klein U, Ellmore G S, Bogorad L. Mol Gen Genet. 1993;238:339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- 21.Klaff P. Nucleic Acids Res. 1995;23:4885–4892. doi: 10.1093/nar/23.23.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruissem W, Tonkyn J C. Crit Rev Plant Sci. 1993;12:19–55. [Google Scholar]
- 23.Lisitsky I, Liveanu V, Schuster G. Plant Physiol. 1995;7:933–941. doi: 10.1104/pp.107.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zurawski G, Bohnert H J, Whitfeld P R, Bottomley W. Proc Natl Acad Sci USA. 1982;79:7699–7703. doi: 10.1073/pnas.79.24.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster G, Gruissem W. EMBO J. 1991;10:1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W. EMBO J. 1996;15:1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 27.Stern D B, Jones H, Gruissem W. J Biol Chem. 1989;264:18742–18750. [PubMed] [Google Scholar]
- 28.Sprecher H, Barr H M, Slotky J I, Tzukerman M, Eytan G D, Assaraf Y G. J Biol Chem. 1995;270:20668–20676. doi: 10.1074/jbc.270.35.20668. [DOI] [PubMed] [Google Scholar]
- 29.Haff L A, Bogorad L. Biochemistry. 1976;15:4110–4141. doi: 10.1021/bi00663a030. [DOI] [PubMed] [Google Scholar]
- 30.Klaff P, Gruissem W. Plant Cell. 1991;3:517–529. doi: 10.1105/tpc.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M, Christopher D A, Mullet J E. Plant Mol Biol. 1993;22:447–463. doi: 10.1007/BF00015975. [DOI] [PubMed] [Google Scholar]
- 32.Rapp J C, Baumgatner B J, Mullet J E. J Biol Chem. 1992;267:21404–21411. [PubMed] [Google Scholar]
- 33.Carpousis A J, Houwe G V, Ehretsmann C, Krisch H M. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 34.Ojala D, Attardi G. Proc Natl Acad Sci USA. 1974;71:563–566. doi: 10.1073/pnas.71.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudla, J., Hayes, R. & Gruissem, W. (1996) EMBO J., in press. [PMC free article] [PubMed]


