Abstract
Cold acclimation in plants is associated with the expression of COR (cold-regulated) genes that encode polypeptides of unknown function. It has been widely speculated that products of these genes might have roles in freezing tolerance. Here we provide direct evidence in support of this hypothesis. We show that constitutive expression of COR15a, a cold-regulated gene of Arabidopsis thaliana that encodes a chloroplast-targeted polypeptide, enhances the in vivo freezing tolerance of chloroplasts in nonacclimated plants by almost 2°C, nearly one-third of the increase that occurs upon cold acclimation of wild-type plants. Significantly, constitutive expression of COR15a also affects the in vitro freezing tolerance of protoplasts. At temperatures between −5 and −8°C, the survival of protoplasts isolated from leaves of nonacclimated transgenic plants expressing COR15a was greater than that of protoplasts isolated from leaves of nonacclimated wild-type plants. At temperatures between −2 and −4°C, constitutive expression of COR15a had a slight negative effect on survival. The implications of these data regarding possible modes of COR15a action are discussed.
Keywords: cold acclimation, COR genes, cryoprotective proteins
In 1985, Guy et al. (1) established that changes in gene expression occur in plants during cold acclimation, a developmental process that results in increased freezing tolerance (2, 3). Since then, it has repeatedly been speculated that certain COR (cold-regulated) genes might have roles in freezing tolerance. To test this notion, investigators have turned to isolating and characterizing genes that are expressed in response to low temperature. These efforts have led to the identification of a number of novel genes such as the COR15a (4), KIN1 (5), and LTI78 (6) genes of Arabidopsis thaliana; the MsaciA gene family of alfalfa (7); and the pt59 and pao86 genes of barley (8). Each of these genes encodes a polypeptide of unknown function with little or no amino acid sequence identity with previously described proteins. However, a number of findings have encouraged the notion that these genes might be involved in freezing tolerance. Some of the strongest evidence in this regard is the report of Mohapatra et al. (9) indicating that the expression level of certain cold-regulated CAS (cold acclimation-specific) genes of alfalfa correlates positively with the freezing tolerance of different alfalfa cultivars. In addition, studies have shown that the synthesis of certain COR proteins coincides closely with the development of freezing tolerance (10, 11). It is also of interest that many COR genes are induced in response to drought (4–8). The relevance of this to cold acclimation is that freezing injury is primarily a consequence of freeze-induced dehydration (3); tolerance to freezing must include tolerance to dehydration stress. Therefore, freezing and drought tolerance might be expected to have certain mechanisms in common, including the expression of specific genes. Indeed, the freezing tolerance of a number of plants has been shown to increase in response to dehydration stress (11–13).
One of the more intriguing attributes of the COR genes is that many encode polypeptides that appear to have biochemical similarities with the putative “cryoprotective proteins” described by Volger and Heber (14). Some 20 years ago, these investigators reported that the leaves of cold-acclimated cabbage and spinach, but not nonacclimated plants, contain proteins that are effective in protecting isolated thylakoid membranes against in vitro freeze–thaw damage. Subsequently, Hincha et al. (15) reported that the “cryoprotective proteins” act by reducing membrane permeability during freezing and increasing membrane expandability during thawing. Unfortunately, specific cabbage and spinach proteins with cryoprotective activity have not yet been conclusively identified and the genes encoding them have not been isolated, situations that have hampered progress to determine whether the proteins have roles in cold acclimation. What is intriguing, however, is that from the procedures used to obtain highly enriched fractions of the cryoprotective proteins, and the biochemical properties of these fractions, it would appear that the cryoprotective proteins have a number of properties in common with the polypeptides encoded by many of the novel COR genes. These include being synthesized in response to low temperature, remaining soluble upon boiling in aqueous solution, being hydrophilic, and having very low absorbance at 280 nm. Given the distinctive nature of these combined properties, we speculated that certain COR genes, like COR15a of A. thaliana, might encode homologs or analogs of the cabbage and spinach “cryoprotective proteins” and have roles in freezing tolerance (4, 16). The goal of this study was to test this hypothesis.
The cold- and drought-regulated COR15a gene of A. thaliana encodes a 15-kDa polypeptide, COR15a, that is targeted to the stromal compartment of chloroplasts (ref. 4; S.J.G. and N.N.A., unpublished data). During import, COR15a is processed to a mature 9.4-kDa polypeptide, COR15am, that is hydrophilic, remains soluble upon boiling, has a simple amino acid composition (it is rich in both alanine and lysine and devoid of proline, methionine, tryptophan, cysteine, glutamine, arginine, and histidine), is composed largely of a 13-amino acid motif that is repeated four times, and is predicted to form an amphipathic α-helix (4, 17). The relatively simple amino acid composition of COR15am together with its predicted secondary structure suggests that the polypeptide might have a nonenzymatic function. We reasoned that if COR15a encoded a homolog or analog of the putative cryoprotective proteins, constitutive expression of the gene might increase the freezing tolerance of chloroplasts in otherwise nonacclimated plants. Here we demonstrate that this is indeed the case. Moreover, we show that constitutive expression of COR15a also affects the freezing tolerance of protoplasts isolated from leaves of nonacclimated plants. Taken together, these results provide compelling evidence for COR15a having a role in A. thaliana freezing tolerance, but raise fundamental questions regarding the mode of COR15a action.
MATERIALS AND METHODS
Plant Growth.
A. thaliana plants used to determine chloroplast freezing tolerance were grown in pots containing a 1:1:1 mixture of Baccto planting mix (Michigan Peat, Houston)/perlite/coarse vermiculite as described (18). Nonacclimated plants were grown in controlled environment chambers at 22°C on an 18/6 h day/night cycle at a light intensity of 100 μE m−2·s−1 provided by cool white fluorescent lamps. Plants were cold-acclimated for 4 days at 3°C under continuous fluorescent illumination at 50 μE m−2·s−1. A. thaliana plants used to determine protoplast freezing tolerance were grown in Terra-Lite Metro-Mix (A. H. Hummert Seed, St. Louis) in a controlled environmental chamber at 23°C for 14 days under continuous illumination at 150 μE m−2·s−1. Leaves were excised at soil level and immediately used for protoplast isolation.
Transgenic Plants.
Plants that constitutively synthesized COR15am were created by placing the coding sequence for COR15a under control of the cauliflower mosaic virus (CaMV) 35S promoter and transforming the gene into A. thaliana. Specifically, the ScaI DNA fragment from cDNA clone pLCT10A (4) that encodes the COR15a polypeptide was purified by agarose gel electrophoresis, its ends were filled in using Escherichia coli DNA polymerase I, and BamHI linkers were added using standard methods (19). The resulting fragment was digested with BamHI and cloned into the BamHI site downstream of the CaMV 35S promoter in the pCIB710 (20) plasmid vector. A recombinant plasmid, pLCT61, containing the COR15a coding sequence in the “sense” orientation, was isolated, digested with XbaI and KpnI, and the XbaI/KpnI DNA fragment containing the 35S–COR15a gene fusion was ligated into the corresponding sites of the pCIB10 binary transformation vector (20). The resulting plasmid, pLCT71, was introduced into Agrobacterium tumefaciens strain LBA4404 and used to transform A. thaliana ecotype RLD using the root explant method developed by Valvekens et al. (21). Two lines, T8 and T9, that were phenotypically uniform for kanamycin resistance and constitutive production of COR15am were chosen for further study. The 35S–β-glucuronidase (GUS) transgenic line used in this study was described previously (22). It is kanamycin resistant and carries the GUS reporter gene (23) under control of the CaMV 35S promoter.
Segregation Analysis.
The transgenic T8 line was back-crossed to RLD using T8 as the pollen donor. After selfing, F3 lines that were either uniform for kanamycin resistance and constitutive for production of COR15am or uniform for kanamycin sensitivity and did not constitutively produce COR15am, were selected by screening first for kanamycin resistance and then directly testing for COR15am production by immunoblot analysis. For some experiments, F3 populations were selfed and the F4 seed was used (the plants from these seeds were tested directly for kanamycin resistance and constitutive production of COR15am).
Immunoblot Analysis.
Total soluble protein was obtained by pulverizing leaf material (about 100 mg) in liquid N2 using a mortar and pestle, adding 0.45 ml extraction buffer containing 50 mM Tris (pH 7.6), 5 mM EDTA, 1 mM 4-(2-aminoethyl)-benzenesulfonylfluoride (Calbiochem), 10−6 M pepstatin A, and 2.5% (wt/vol) polyvinyl-polypyrrolidone and, after further grinding, removing insoluble material by centrifugation (14,000 × g for 20 min). The supernatant was collected and soluble protein was precipitated by adding 4 volumes of acetone. Soluble and insoluble protein were prepared from purified chloroplasts as described (4). Protein samples were fractionated by tricine SDS/PAGE (24) and transferred to 0.2 micron nitrocellulose membranes by electroblotting (25), and COR15am was detected using antiserum raised to purified COR15am (ref. 4; C.L., unpublished work) and protein A conjugated alkaline phosphatase (Sigma) as described (26).
Chloroplast Freezing Tolerance.
Fully expanded leaves from plants that had not yet started to bolt were detached, wrapped in moistened cheese cloth, and inserted into a test tube that was then placed in a controlled temperature bath at −2°C. Upon temperature equilibration, freezing was induced by adding ice crystals. After 2 hr, the temperature of the bath was lowered at a rate of 1°C per 30 min. After a 30-min period at the indicated subzero temperature, the leaves were slowly thawed at 2°C as described (18). In the initial experiments (see Fig. 2), room temperature fluorescence of the treated leaves was measured using a Morgan CF-1000 chlorophyll fluorescence measurement system (P. K. Morgan Instruments, Andover, MA). Leaf samples were dark-adapted in cuvettes for 15 min before determining fluorescence in response to continuous irradiation of 900 μE m−2·s−1. Fv (variable fluorescence) was calculated as Fm (maximum fluorescence) − Fo (minimal fluorescence). In later experiments (see Fig. 3 and Table 1), room temperature fluorescence was measured using an OS-500 pulse amplitude modulation fluorometer (Opti-Sciences, Tewksbury, MA). In these experiments, leaves were dark-adapted in cuvettes for 5 min. Fo was determined by a weak modulated light of 0.12 μE m−2·s−1 and Fm was determined by a 0.8 sec light pulse of 2000 μE m−2·s−1. Fv was again calculated as Fm − Fo.
Figure 2.
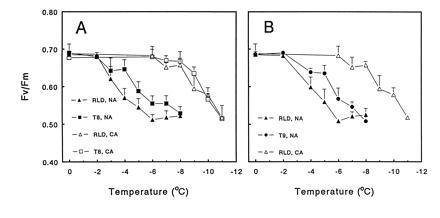
Freezing tolerance of chloroplasts in A. thaliana RLD, T8, and T9 plants. Leaves from nonacclimated (NA) and cold-acclimated (CA) plants were subjected to a freeze–thaw cycle to various subzero temperatures and damage to chloroplasts was assessed by determining Fv/Fm. (A) Nonacclimated (n = 6) and cold-acclimated (n = 2) RLD and T8 plants. (B) Nonacclimated (n = 4) RLD and T9 plants and cold-acclimated (n = 2) RLD plants. Each experiment (n) included at least three replicate samples per point.
Figure 3.
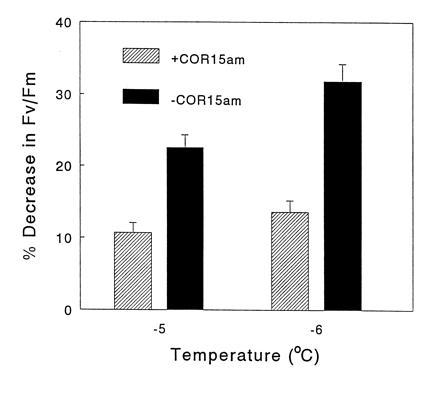
Freeze-induced damage to chloroplasts in F3 lines of nonacclimated A. thaliana plants that either did (diagonal bars) or did not (solid bars) constitutively express COR15a. Leaves from nonacclimated plants were frozen to either −5°C or −6°C, thawed, and damage to chloroplasts was assessed by determining Fv/Fm. The percentage decrease in Fv/Fm was determined by dividing the mean Fv/Fm values for the frozen leaves (values presented in Table 1) by the mean Fv/Fm values for nonfrozen leaves (and multiplying by 100). The values for the nonfrozen leaf samples for COR15am positive and negative lines (six lines for each; three leaves for each line) were 0.808 ± 0.005 and 0.804 ± 0.004, respectively.
Table 1.
Freezing tolerance of chloroplasts in F3 families of non-acclimated A. thaliana plants
| COR15am-positive
|
COR15am-negative
|
||
|---|---|---|---|
| Line | Fv/Fm (±SE) | Line | Fv/Fm (±SE) |
| Experiment 1: Leaves frozen to −5°C | |||
| 1-5 | 0.747 ± 0.013 | 1-10 | 0.671 ± 0.006 |
| 2-11 | 0.697 ± 0.041 | 1-11 | 0.603 ± 0.030 |
| 5-2 | 0.707 ± 0.006 | 2-8 | 0.614 ± 0.014 |
| 5-8 | 0.737 ± 0.004 | 5-6 | 0.606 ± 0.037 |
| Mean* | 0.722 ± 0.011 | 0.623 ± 0.015 | |
| Experiment 2: Leaves frozen to −6°C | |||
| 1-5 | 0.720 ± 0.018 | 1-6 | 0.620 ± 0.017 |
| 1-4 | 0.729 ± 0.029 | 1-1 | 0.507 ± 0.022 |
| 1-15 | 0.657 ± 0.034 | 2-5 | 0.538 ± 0.035 |
| 3-3 | 0.685 ± 0.010 | 3-1 | 0.528 ± 0.053 |
| Mean* | 0.698 ± 0.013 | 0.548 ± 0.019 | |
Wild-type RLD plants were crossed with T8 plants, and F3 families that either did (COR15am-positive) or did not (COR15am-negative) constitutively produce COR15am were selected. Leaves from nonacclimated plants were frozen to either −5°C or −6°C and thawed; damage to chloroplasts was assessed by determining Fv/Fm. Each of the Fv/Fm values listed was based on testing four leaf samples.
Mean Fv/Fm values ± SE for the 16 leaf samples tested for each set of conditions.
Protoplast Freezing Tolerance.
Protoplasts were isolated from leaves according to Uemura et al. (27). Briefly, individual leaves were cut into three pieces and placed in an isotonic sorbitol solution (0.4 M) containing 1.3% (wt/vol) cellulysin (Calbiochem), 0.4% (wt/vol) macerase (Calbiochem), 1 mM CaCl2, and 10 mM Mes [2-(N-morpholino)ethanesulfonic acid] buffer (pH 5.5) for 2 hr at 28°C in the dark. Undigested leaf sections were removed by filtering the suspension through four layers of cheesecloth, and protoplasts in the filtrate were collected by centrifugation at 50 × g for 10 min at 0°C. Pelleted protoplasts were suspended in the isotonic sorbitol solution containing 1 mM CaCl2 and 10 mM Mes buffer (pH 5.5) and washed twice by repeated centrifugation and resuspension. Washed protoplasts were suspended in the isotonic sorbitol solution and kept on ice.
Freezing tolerance of the protoplasts was determined as described (28). Briefly, an aliquot of the washed protoplast suspension (0.5 ml, 105 protoplasts) was placed in a glass test tube (10 × 100 mm), which was then placed in an ethanol bath at −2°C. After 15 min, ice formation in the suspension was effected by touching the outside of the test tube with a spatula precooled in liquid N2. After an additional 30-min isothermal period, the samples were cooled to the indicated temperatures at a rate of 0.8°C per min. After 30 min at the specified temperatures, the samples were thawed at room temperature and then kept on ice. Protoplast survival was determined by staining with fluorescein diacetate (FDA) (29) at a final concentration of 0.001% (wt/vol). After the samples were incubated with FDA for 5 min at room temperature, the number of protoplasts that retained the dye was counted in a hemocytometer.
RESULTS
Construction of Transgenic Plants That Constitutively Produce COR15am.
Transgenic A. thaliana plants that constitutively produce COR15am were created by placing a cDNA copy of the COR15a gene under control of the CaMV 35S promoter and transforming the chimeric gene into A. thaliana ecotype RLD. Two of the resulting transgenic lines, T8 (Fig. 1) and T9 (not shown), constitutively produced COR15am at levels that were approximately equal to the amount of COR15am produced in cold-acclimated RLD plants. Like the COR15am protein synthesized from the endogenous COR15a gene in cold-acclimated RLD plants (4), the COR15am polypeptide produced in the nonacclimated transgenic plants was present in the chloroplasts as a soluble protein (Fig. 1; N.N.A. and S.J.G., unpublished results); it was not detected in insoluble protein fractions prepared from the chloroplasts (not shown).
Figure 1.
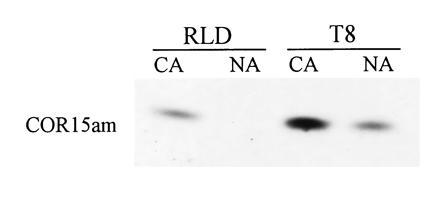
Presence of COR15am in the soluble protein fraction of chloroplasts isolated from A. thaliana RLD and T8 plants. Chloroplasts were purified from the leaves of nonacclimated (NA) and cold-acclimated (CA) T8 and RLD plants. Total soluble (25 μg) protein was fractionated by tricine SDS/PAGE. COR15am was detected by immunoblot analysis.
Enhancement of Chloroplast Freezing Tolerance.
Given the presence of COR15am in chloroplasts, our first question was whether constitutive expression of COR15a had an effect on chloroplast freezing tolerance. Specifically, the in vivo freezing tolerance of chloroplasts in wild-type and transgenic plants was assessed by freezing leaves at various temperatures and then, after thawing, measuring the chlorophyll fluorescence of the leaves. Most of the chlorophyll fluorescence that is emitted from leaves at physiological temperatures arises from the chlorophyll a associated with photosystem II. Butler and colleagues (30, 31) established the potential of using chlorophyll fluorescence to monitor the amount of absorbed light energy that is directed toward useful photochemical reactions. The ratio of variable fluorescence, Fv, to maximum fluorescence, Fm, is used to estimate the quantum yield of photosystem II photochemisty (32). Environmental stresses that decrease the efficiency of photosystem II result in a decrease of the Fv/Fm ratio (32).
A decrease in the Fv/Fm ratio was observed after freezing leaves of nonacclimated RLD, T8, and T9 plants to temperatures below −2°C (Fig. 2). The Fv/Fm “inactivation curves” for the T8 (Fig. 2A) and T9 (Fig. 2B) plants, however, were shifted nearly 2°C lower in temperature than the curves for RLD plants (Fig. 2). While there was considerable variability in the assay, a paired t test indicated that the mean Fv/Fm values for leaves of nonacclimated RLD plants were significantly different from those for nonacclimated T8 plants frozen to −4°C (P < 0.05, n = 6) and −5°C (P < 0.05, n = 7) and nonacclimated T9 plants frozen to −4°C (P < 0.05, n = 6) and −5°C (P < 0.05, n = 5) (the analysis included the data presented in Fig. 2 and additional experiments in which leaf samples were frozen to −4°C and −5°C). No difference was observed in the Fv/Fm inactivation curves for RLD and 35S–GUS, a transgenic line of A. thaliana carrying the GUS reporter gene fused to the CaMV 35S promoter, indicating that the transformation procedure per se did not affect chloroplast freezing tolerance.
The shift in the inactivation curves for the nonacclimated transgenic plants was small in absolute terms, but amounted to nearly one-third of the shift observed upon cold acclimation: the Fv/Fm freeze-inactivation curve for leaves from cold-acclimated RLD plants was shifted about 6°C lower than that for leaves from nonacclimated RLD plants (Fig. 2A). This increase in freezing tolerance is in good agreement with previous studies (18) in which nonacclimated A. thaliana plants were killed at about −3°C, and fully cold-acclimated plants were killed at about −9°C (lethal temperatures in these experiments were estimated by measuring electrolyte leakage). No significant difference was observed between the Fv/Fm freeze-inactivation curves for leaves of cold-acclimated RLD and T8 plants (Fig. 2A). Thus, constitutive expression of COR15a at the level that occurred in T8 plants had no discernible additive effect on chloroplast freezing tolerance over that which occurred in wild-type plants with cold acclimation.
The results of a genetic segregation analysis confirmed that the in vivo freezing tolerance of chloroplasts in nonacclimated plants was greater in lines that constitutively expressed COR15a. Specifically, wild-type RLD plants were crossed with T8 plants, and F3 families that either did (seven lines), or did not (eight lines), constitutively express COR15a were selected. Leaves from these plants were then frozen to either −5°C or −6°C, thawed, and chloroplast damage was assessed by measuring Fv/Fm. The mean Fv/Fm values for COR15am positive and negative lines frozen to −5°C were 0.722 ± 0.011 and 0.623 ± 0.015, respectively, and those frozen to −6°C were 0.698 ± 0.013 and 0.548 ± 0.019, respectively (Table 1). Analysis of the data using the Student t test indicated that the differences in the Fv/Fm values for the COR15am-producing and nonproducing lines were highly significant (P < 0.005). Further analysis indicated that the freeze-induced decrease in Fv/Fm at −5°C and −6°C was only about half as much in the COR15am-producing plants as in the nonproducing plants (Fig. 3).
Effects of COR15a Expression on Protoplast Freezing Tolerance.
Constitutive expression of COR15a had a significant effect on the freezing tolerance of isolated protoplasts. Between −5 and −8°C, the survival of protoplasts isolated from leaves of nonacclimated T8 plants was greater than that of protoplasts isolated from leaves of nonacclimated RLD plants (Fig. 4A). The average difference in survival was 13% at −5.5°C, 17% at −6.0°C, 18% at −6.5°C, and 10% at −7.0°C. Though these differences were small, they were extremely reproducible, such that the results of six different experiments, conducted over an eight-month period, were virtually superimposable in the range of −5 to −8°C (Fig. 4A). The Student t test indicated that the differences were highly significant at −5.5°C (P < 0.005) and −6.0°C, −6.5°C, and −7.0°C (P < 0.0001), which is approximately the same temperature range over which the freezing tolerance of chloroplasts was enhanced by constitutive expression of COR15a.
Figure 4.
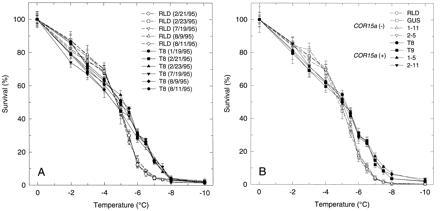
Effects of constitutive COR15a expression on the freezing tolerance of protoplasts isolated from leaves of nonacclimated A. thaliana plants. Isolated protoplasts were frozen to the indicated temperatures and thawed; survival was determined by staining with FDA (expressed as a percentage of the unfrozen control). Three hemocytometer samples were counted for each temperature in a given experiment. The results shown are the average and SD of individual experiments. (A) Transgenic T8 and wild-type RLD plants. (B) Plants that constitutively express COR15a (T8, T9, 1-5, and 2-11) and those that do not [RLD, GUS (35S–GUS), 1-11, and 2-5].
A close inspection of the data (Fig. 4A) revealed that constitutive expression of COR15a had an additional effect on protoplast freezing tolerance. After freezing at −2°C, −3°C, and −4°C, the survival of protoplasts isolated from the leaves of nonacclimated T8 plants was somewhat less than that of the protoplasts isolated from nonacclimated RLD plants. The average differences were 4% at −2°C and 8% at −3°C and −4°C. The Student t test indicated that the differences were highly significant at −3°C (P < 0.0001) and −4°C (P < 0.001).
A comparison of additional A. thaliana lines including F3 families used in the chloroplast studies confirmed that constitutive expression of COR15a affected the freezing tolerance of isolated protoplasts. Specifically, protoplasts isolated from leaves of nonacclimated plants that constitutively expressed COR15a (T8, T9, 1-5, and 2-11) were more freezing tolerant over the temperature range of −5 to −8°C and slightly less freezing tolerant over the range of −2 to −4°C than protoplasts isolated from nonacclimated plants that did not constitutively express COR15a (RLD, 35S–GUS, 1-11, 2-5) (Fig. 4B). Again, the differences in freezing tolerance observed, though small, were highly reproducible; the Student t test indicated that the differences were highly significant at −3°C (P < 0.005) and −4°C, −5.5°C, −6.0°C, −6.5°C, and −7.0°C (P < 0.0001). Neither the increase nor decrease in protoplast freezing tolerance that occurred in the constitutive COR15a lines occurred in the 35S–GUS transgenic line (Fig. 4B), indicating that the differences were not a consequence of the transformation process per se.
DISCUSSION
Since 1985, when Guy et al. (1) first established that changes in gene expression occur during cold acclimation, considerable effort has been directed at determining whether COR genes have a functional role in freezing tolerance. Here we present direct evidence in support of this notion. Collectively, the independent studies showing that constitutive expression of COR15a affects the freezing tolerance of both chloroplasts frozen in vivo and protoplasts frozen in vitro—together with the extreme reproducibility of the protoplast studies and fact that the enhanced freezing tolerance of chloroplasts and protoplasts occurs over approximately the same temperature range—provide compelling evidence that COR15a is intimately associated with the increase in freezing tolerance that occurs during cold acclimation. Presumably, the cold-regulated COR15b gene of A. thaliana (17) and the cold-regulated BN115, BN26, and BN19 genes of Brassica napus (33), all of which are apparent homologs of COR15a, also encode polypeptides that affect freezing tolerance.
The results of the in vivo chlorophyll fluorescence experiments (Figs. 2 and 3; Table 1) indicate that constitutive expression of COR15a reduces freeze-induced damage to photosystem II in nonacclimated plants. A major goal now is to determine the mechanism by which COR15a expression brings about this effect on chloroplasts. One possibility that we have considered is that COR15am might act akin to the putative cryoprotective proteins described by Heber and colleagues (14, 15). As mentioned earlier, these polypeptides are reported to protect isolated thylakoid membranes against freeze-induced damage in vitro. The presence of COR15am in the stromal compartment of the chloroplasts is compatible with the possibility of COR15am protecting thylakoid membranes against freeze-induced damage. However, unlike the cryoprotective proteins, which are reported to reduce freeze-induced inactivation of cyclic photophosphorylation in isolated thylakoids (14), COR15am does not protect isolated thylakoid membranes against freeze-induced inactivation of either cyclic photophosphorylation or light-induced proton uptake (34). Thus, the COR15am polypeptide does not appear to function in the same manner as the cryoprotective proteins described by Volger and Heber (14).
The results of the protoplast survival studies reveal that the effect of COR15a expression extends beyond the chloroplasts—a finding that has important implications regarding the mechanism of COR15a action. In the protoplast experiments, survival was measured by FDA staining, a method that reports on retention of the semipermeable characteristics of the plasma membrane. Thus, both the observed increase in survival over the range of −5°C to −8°C and the small decrease in survival over the range of −2°C to −4°C indicate that constitutive expression of COR15a affects the cryostability of the plasma membrane. If COR15am were to act as a cryoprotectant, the most simple scenario to explain the protoplast results would be for COR15am to alter the cryostability of the plasma membrane by interacting directly with it. This, however, would not seem likely as the COR15a gene product is targeted to the chloroplasts: the COR15a-encoded polypeptide has a chloroplast import sequence (4), and cell fractionation (4) and immunolocalization (N.N.A. and S.J.G., unpublished results) studies have demonstrated that COR15am is present in the stromal compartment of chloroplasts. Thus, unless there are small amounts of COR15am present in the cytoplasm (a formal possibility that has not been ruled out), a more complex scenario would seem to be required to explain how COR15am might act as a cryoprotectant.
Is there, in fact, any direct evidence for COR15am acting as a cryoprotectant? At present, the data are equivocal on this point. For instance, Lin and Thomashow (35) reported that the unprocessed COR15a polypeptide is effective in decreasing the incidence of freeze-induced inactivation of lactate dehydrogenase in vitro, but subsequent studies indicate that the cryoprotective effect of COR15am on lactate dehydrogenase is no greater than that of bovine serum albumin (S.J.G., unpublished data). Uemura et al. (36) have found that COR15am decreases the incidence of freeze-induced fusion of liposomes frozen in vitro. However, this effect only occurs under unusual conditions—when the liposomes are frozen in dilute buffer in the absence of any solutes. Webb et al. (37) found that unlike sucrose, a well-known cryoprotectant, COR15am has no effect on the dehydration-induced increase in the liquid crystalline-to-gel phase transition temperature (Tm) of either dipalmitoylphosphatidylcholine or dioleoylphosphatidyl-choline. Similarly, COR15am does not alter the osmotic pressure (hydration) at which multilamellar vesicles composed of a mixture of dioleoylphosphatidylethanolamine and dioleoylphosphatidylcholine undergo the lamellar-to-hexagonal II phase transition. However, when the vesicles are dehydrated in the presence of COR15am at osmotic pressures greater than 39 MPa, the resultant lipid aggregates frequently have a polyhedral shape with angular corners and the surface of the lamellae have a distinctive striated appearance. These effects on morphology suggest that the COR15am polypeptide interacts with lipid bilayers, but how this relates to potential mechanisms of cryoprotection is not known at this time.
If the COR15am polypeptide does not act as a cryoprotectant, how then might COR15a expression affect freezing tolerance? One possibility is that COR15am could mediate biochemical or physiological changes (i.e., alter gene expression, lipid composition, etc.) that affect freezing tolerance at both the organelle and cellular level. More detailed comparisons of the wild-type and COR15am-producing transgenic plants in regard to differences in cryobehavior and biochemical composition should provide further insight into the mechanism of COR15a action.
One of the more intriguing aspects of the results presented is that constitutive expression of COR15a increased the freezing tolerance of both chloroplasts and protoplasts frozen at temperatures between −5°C and −8°C, but did not significantly affect the LT50 values of the protoplast survival curves and resulted in a slight decrease in protoplast survival over the range of −2°C to −4°C. Although these results might be considered paradoxical, they can be explained when considered from a perspective of specific freeze-induced lesions in the plasma membrane rather than survival per se. Survival of isolated protoplasts, as determined by FDA staining, is limited by different lesions that occur at different subfreezing temperatures (3, 27). Over the range of −2°C to −4°C, the predominant form of injury is expansion-induced lysis, which is a consequence of the osmotic excursion incurred during a freeze–thaw cycle. Over the range of −4°C to −8°C, the predominant form of injury is freeze-induced lamellar-to-hexagonal II phase transitions involving the plasma membrane and various endomembranes—most frequently the chloroplast envelope—that are brought in close apposition as a consequence of freeze-induced removal of water from the surfaces of cellular membranes. Thus, it is possible that constitutive expression of COR15a might decrease the incidence of freeze-induced formation of hexagonal II phase lipids, but have little or even possibly a negative effect on the incidence of expansion-induced lysis. Preliminary studies (M.U. and P.L.S., unpublished results) are consistent with this explanation.
In recent years, there has been considerable interest (and hope) that biotechnology might offer new strategies to improve the freezing tolerance of crop plants. This is due, in part, to the fact that classical plant breeding approaches have met with limited success in improving the freezing tolerance of agronomic plants (38). The freezing tolerance of the best wheat varieties today, for instance, is essentially the same as the most freezing-tolerant varieties developed in the early part of this century. One idea has been that if COR genes have a functional role in cold acclimation, they might provide new tools for improving freezing tolerance. Here we show for the first time that expression of a single COR gene can affect freezing tolerance. The effects observed were relatively small—a finding that is not unexpected given that plant freezing tolerance is a (polygenic) quantitative genetic trait (38). However, an increase in freezing tolerance on the order of 2°C would be significant for a number of crop and horticultural plant species. Whether individual cold-regulated genes such as COR15a will be useful for altering freezing tolerance remains to be determined. Perhaps a more promising approach will be to alter expression of the entire battery of COR genes. The recent cloning of a transcription factor that binds to a cis-acting cold-regulatory element present in a number of COR genes (E. Stockinger, S.J.G., and M.F.T., unpublished data) may make such an approach possible in the near future.
Acknowledgments
We are grateful to Don Ort and Neil Baker for helpful discussions and advice regarding plant chlorophyll fluorescence; Rebecca Grumet and Richard Lenski for help with statistical analysis of the data; and Don Ort, Rebecca Grumet, and Barbara Sears for constructive comments on an early version of the manuscript. This research was supported by grants from the National Science Foundation (IBN-9307348) and the Michigan Agricultural Experiment Station to M.F.T., and a grant from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (93–37100-8835) to P.L.S.
Footnotes
Abbreviations: CaMV, cauliflower mosaic virus; COR, cold-regulated; FDA, fluorescein diacetate; Fv, variable fluorescence; Fm, maximum fluorescence; Fo, minimal fluorescence; GUS, β-glucuronidase.
References
- 1.Guy C L, Niemi K L, Brambl R. Proc Natl Acad Sci USA. 1985;82:3673–3677. doi: 10.1073/pnas.82.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guy C L. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:187–223. [Google Scholar]
- 3.Steponkus P L, Uemura M, Webb M S. In: Advances in Low Temperature Biology. Steponkus P L, editor. Vol. 2. London: JAI; 1993. pp. 211–312. [Google Scholar]
- 4.Lin C, Thomashow M F. Plant Physiol. 1992;99:519–525. doi: 10.1104/pp.99.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurkela S, Franck M. Plant Mol Biol. 1990;15:137–144. doi: 10.1007/BF00017731. [DOI] [PubMed] [Google Scholar]
- 6.Nordin K, Vahala T, Palva E T. Plant Mol Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- 7.Laberge S, Castonguay Y, Vézina L P. Plant Physiol. 1993;101:1411–1412. doi: 10.1104/pp.101.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cattivelli L, Bartels D. Plant Physiol. 1990;93:1504–1510. doi: 10.1104/pp.93.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohapatra S S, Wolfraim L, Poole R J, Dhindsa R S. Plant Physiol. 1989;89:375–380. doi: 10.1104/pp.89.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houde M, Dhindsa R S, Sarhan F. Mol Gen Genet. 1992;234:43–48. doi: 10.1007/BF00272343. [DOI] [PubMed] [Google Scholar]
- 11.Guy C, Haskell D, Neven L, Klein P, Smelser C. Planta. 1992;188:265–270. doi: 10.1007/BF00216823. [DOI] [PubMed] [Google Scholar]
- 12.Siminovitch D, Cloutier Y. Cryobiology. 1983;20:487–503. doi: 10.1016/0011-2240(83)90037-8. [DOI] [PubMed] [Google Scholar]
- 13.Cox W, Levitt J. Plant Physiol. 1976;57:553–555. doi: 10.1104/pp.57.4.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volger H G, Heber U. Biochim Biophys Acta. 1975;412:335–349. doi: 10.1016/0005-2795(75)90048-3. [DOI] [PubMed] [Google Scholar]
- 15.Hincha D K, Heber U, Schmitt J M. Planta. 1990;180:416–419. doi: 10.1007/BF00198794. [DOI] [PubMed] [Google Scholar]
- 16.Lin C, Guo W W, Everson E, Thomashow M F. Plant Physiol. 1990;94:1078–1083. doi: 10.1104/pp.94.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilhelm K S, Thomashow M F. Plant Mol Biol. 1992;23:1073–1077. doi: 10.1007/BF00021822. [DOI] [PubMed] [Google Scholar]
- 18.Gilmour S J, Hajela R K, Thomashow M F. Plant Physiol. 1988;87:745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Rothstein S J, Lahners N K, Lotstein R J, Carozzi N B, Jayne S M, Rice D A. Gene. 1987;53:153–161. doi: 10.1016/0378-1119(87)90003-5. [DOI] [PubMed] [Google Scholar]
- 21.Valvekens D, Van Montagu M, Van Lijsebettern M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker S S, Wilhelm K S, Thomashow M F. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 23.Jefferson R A. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 24.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 25.Towbin H, Staehlelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blake M S, Johnston K H, Russell-Jones G T, Gotschlich E C. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 27.Uemura M, Joseph R A, Steponkus P L. Plant Physiol. 1995;109:15–30. doi: 10.1104/pp.109.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uemura M, Steponkus P L. Plant Physiol. 1989;91:1131–1137. doi: 10.1104/pp.91.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widholm J M. Stain Technol. 1972;47:189–194. doi: 10.3109/10520297209116483. [DOI] [PubMed] [Google Scholar]
- 30.Butler W L. In: Encyclopedia of Plant Physiology. Trebst A, Avron M, editors. Vol. 5. Berlin: Springer; 1977. pp. 149–167. [Google Scholar]
- 31.Butler W L. Annu Rev Plant Physiol. 1978;29:345–378. [Google Scholar]
- 32.Krause G H, Weis E. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:313–349. [Google Scholar]
- 33.Weretilnyk E, Orr W, White T C, Iu B, Singh J. Plant Physiol. 1993;101:171–177. doi: 10.1104/pp.101.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uemura M, Gilmour S J, Thomashow M F, Steponkus P L. Cryobiology. 1994;31:558. (abstr.). [Google Scholar]
- 35.Lin C, Thomashow M F. Biochem Biophys Res Commun. 1992;183:1103–1108. doi: 10.1016/s0006-291x(05)80304-3. [DOI] [PubMed] [Google Scholar]
- 36.Uemura M, Gilmour S J, Thomashow M F, Steponkus P L. Plant Physiol. 1996;111:313–327. doi: 10.1104/pp.111.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb M S, Gilmour S J, Thomashow M F, Steponkus P L. Plant Physiol. 1996;111:301–312. doi: 10.1104/pp.111.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomashow M F. Adv Genet. 1990;28:99–131. [Google Scholar]


