Abstract
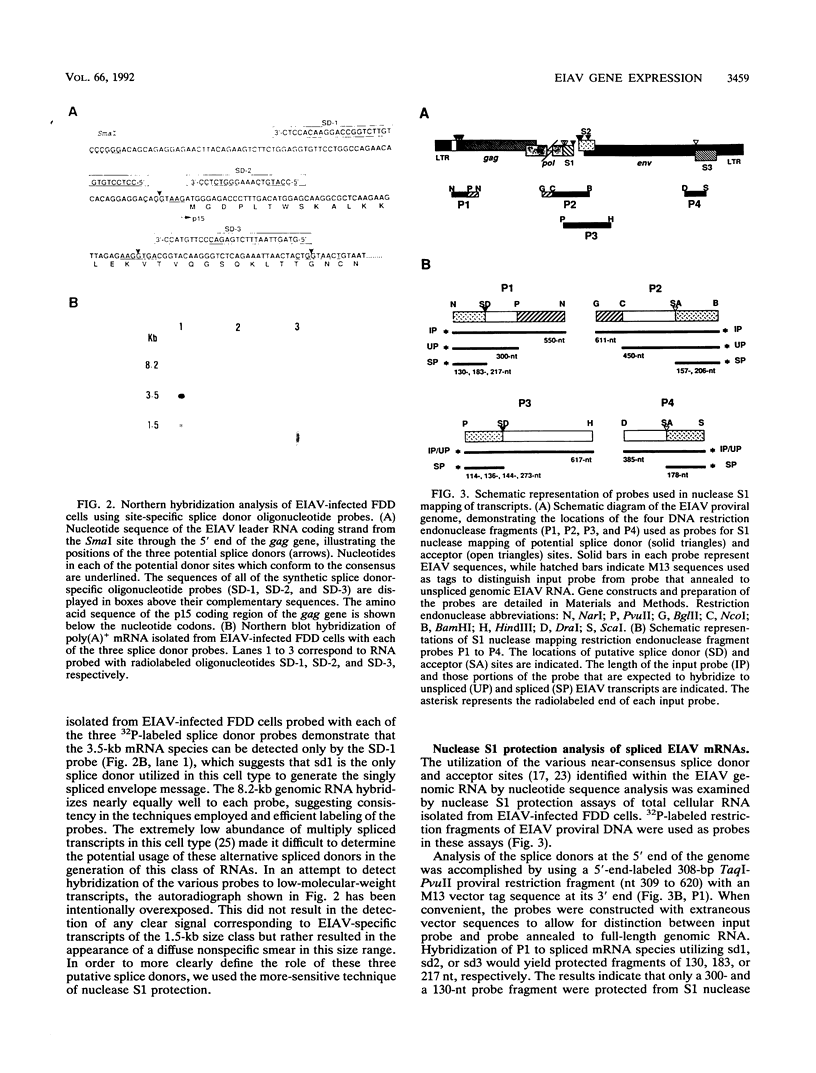
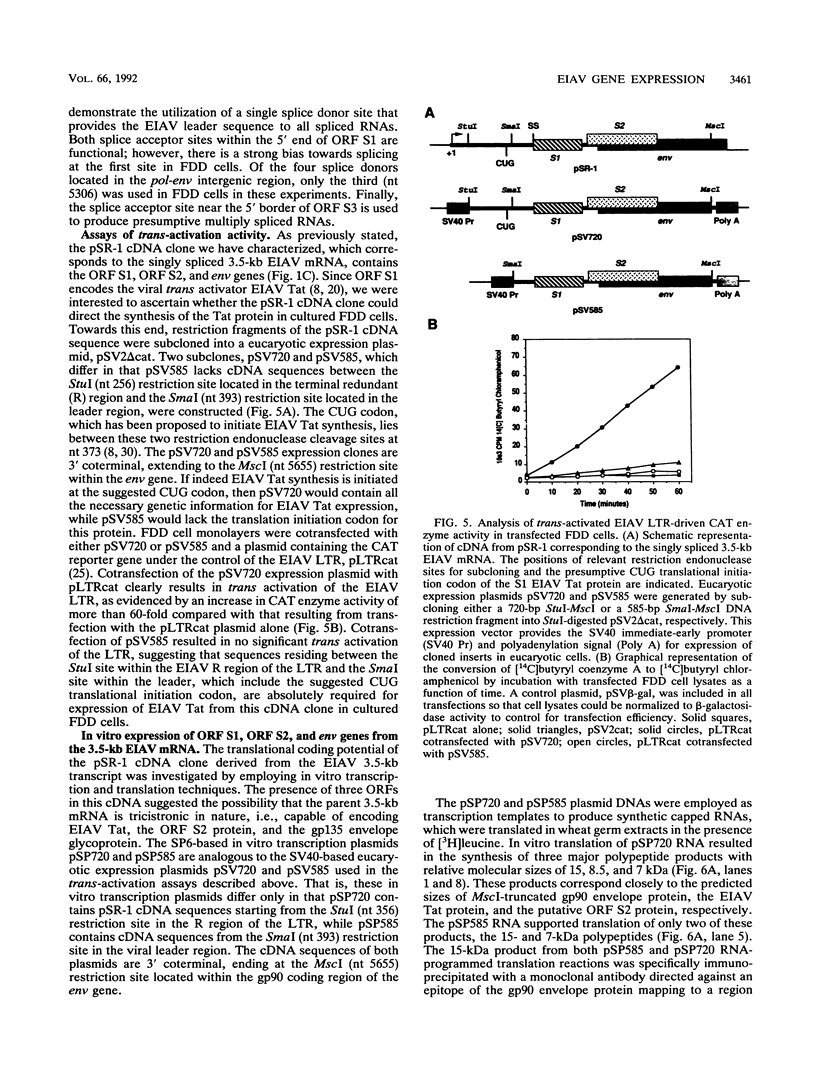
The utilization of predicted splice donor and acceptor sites in generating equine infectious anemia virus (EIAV) transcripts in fetal donkey dermal cells (FDD) was examined. A single splice donor site identified immediately upstream of the gag coding region joins the viral leader sequence to all downstream exons of spliced EIAV transcripts. The predominant 3.5-kb transcript synthesized in EIAV-infected FDD cells appears to be generated by a single splicing event which links the leader sequence to the first of two functional splice acceptor sites near the 5' end of the S1 open reading frame (ORF). The translation products encoded by the 3.5-kb transcript were examined by producing in vitro transcripts from a cDNA corresponding to this RNA followed by in vitro translation in wheat germ extracts. These transcripts directed the synthesis of three proteins: the virus trans-activator protein (EIAV Tat) encoded by ORF S1, a protein of unknown function encoded by ORF S2, and the virus envelope glycoprotein. When transfected into FDD cells, this cDNA also directed expression of EIAV Tat. Amino-terminal sequence analysis of the in vitro-synthesized S1 protein supports the suggestion that translation of EIAV Tat is initiated at a CUG codon within the virus leader region. Both in vitro-synthesized S2 protein and synthetic peptides corresponding to S2 are shown to react positively with sera obtained from EIAV-infected horses, providing the first direct evidence of expression of this protein in infected animals.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexandersen S., Carpenter S. Characterization of variable regions in the envelope and S3 open reading frame of equine infectious anemia virus. J Virol. 1991 Aug;65(8):4255–4262. doi: 10.1128/jvi.65.8.4255-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Blobel G. Immunoprecipitation of proteins from cell-free translations. Methods Enzymol. 1983;96:111–120. doi: 10.1016/s0076-6879(83)96012-3. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Human immunodeficiency virus as a prototypic complex retrovirus. J Virol. 1991 Mar;65(3):1053–1056. doi: 10.1128/jvi.65.3.1053-1056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Dorn P. L., Levy L., Stephens R. M., Rice N. R., Casey J. W. Characterization of equine infectious anemia virus long terminal repeat. J Virol. 1987 Mar;61(3):743–747. doi: 10.1128/jvi.61.3.743-747.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn P., DaSilva L., Martarano L., Derse D. Equine infectious anemia virus tat: insights into the structure, function, and evolution of lentivirus trans-activator proteins. J Virol. 1990 Apr;64(4):1616–1624. doi: 10.1128/jvi.64.4.1616-1624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias S., Ciechanover A. Post-translational addition of an arginine moiety to acidic NH2 termini of proteins is required for their recognition by ubiquitin-protein ligase. J Biol Chem. 1990 Sep 15;265(26):15511–15517. [PubMed] [Google Scholar]
- Fordis C. M., Howard B. H. Use of the CAT reporter gene for optimization of gene transfer into eukaryotic cells. Methods Enzymol. 1987;151:382–397. doi: 10.1016/s0076-6879(87)51030-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Malmquist W. A., Barnett D., Becvar C. S. Production of equine infectious anemia antigen in a persistently infected cell line. Arch Gesamte Virusforsch. 1973;42(4):361–370. doi: 10.1007/BF01250717. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiman S., Gazit A., Tori O., Sherman L., Miki T., Tronick S. R., Yaniv A. Identification of sequences encoding the equine infectious anemia virus tat gene. Virology. 1990 May;176(1):280–288. doi: 10.1016/0042-6822(90)90254-o. [DOI] [PubMed] [Google Scholar]
- Noiman S., Yaniv A., Sherman L., Tronick S. R., Gazit A. Pattern of transcription of the genome of equine infectious anemia virus. J Virol. 1990 Apr;64(4):1839–1843. doi: 10.1128/jvi.64.4.1839-1843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiman S., Yaniv A., Tsach T., Miki T., Tronick S. R., Gazit A. The Tat protein of equine infectious anemia virus is encoded by at least three types of transcripts. Virology. 1991 Oct;184(2):521–530. doi: 10.1016/0042-6822(91)90422-8. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Orrego A., Issel C. J., Montelaro R. C., Adams W. V., Jr Virulence and in vitro growth of a cell-adapted strain of equine infectious anemia virus after serial passage in ponies. Am J Vet Res. 1982 Sep;43(9):1556–1560. [PubMed] [Google Scholar]
- Rasty S., Dhruva B. R., Schiltz R. L., Shih D. S., Issel C. J., Montelaro R. C. Proviral DNA integration and transcriptional patterns of equine infectious anemia virus during persistent and cytopathic infections. J Virol. 1990 Jan;64(1):86–95. doi: 10.1128/jvi.64.1.86-95.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow K., Olsen K., Stiegler G., Payne S. L., Montelaro R. C., Issel C. J. Lentivirus genomic organization: the complete nucleotide sequence of the env gene region of equine infectious anemia virus. Virology. 1986 Dec;155(2):309–321. doi: 10.1016/0042-6822(86)90195-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Felber B. K., Fenyö E. M., Pavlakis G. N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol. 1990 Nov;64(11):5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Felber B. K., Pavlakis G. N. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is Rev-dependent and regulated by splicing. Virology. 1991 Aug;183(2):677–686. doi: 10.1016/0042-6822(91)90996-o. [DOI] [PubMed] [Google Scholar]
- Stephens R. M., Derse D., Rice N. R. Cloning and characterization of cDNAs encoding equine infectious anemia virus tat and putative Rev proteins. J Virol. 1990 Aug;64(8):3716–3725. doi: 10.1128/jvi.64.8.3716-3725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnav Y. N., Wong-Staal F. The biochemistry of AIDS. Annu Rev Biochem. 1991;60:577–630. doi: 10.1146/annurev.bi.60.070191.003045. [DOI] [PubMed] [Google Scholar]