Abstract
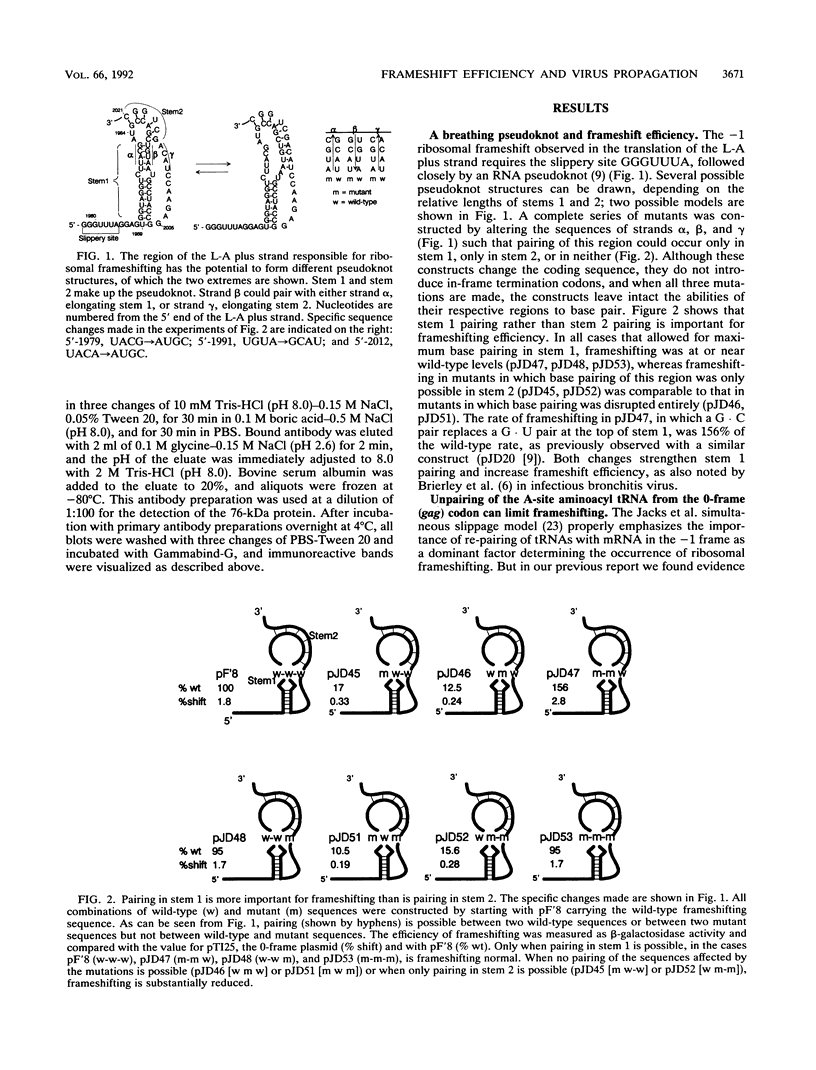
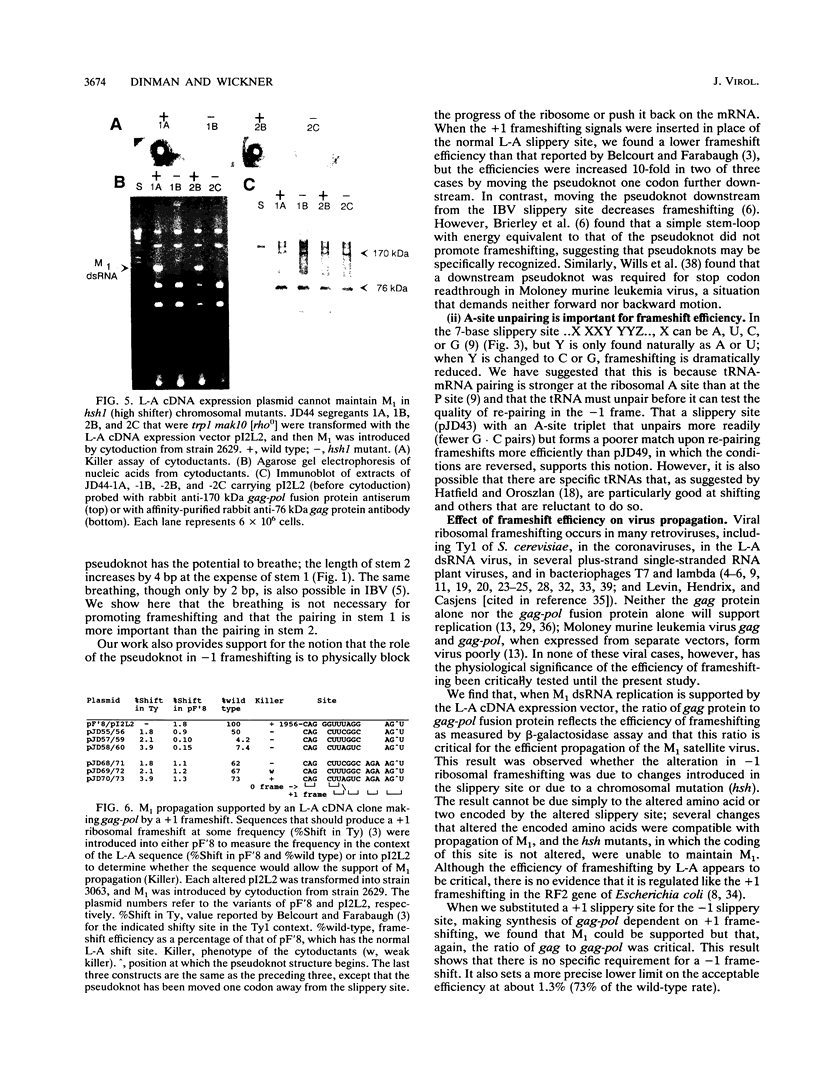
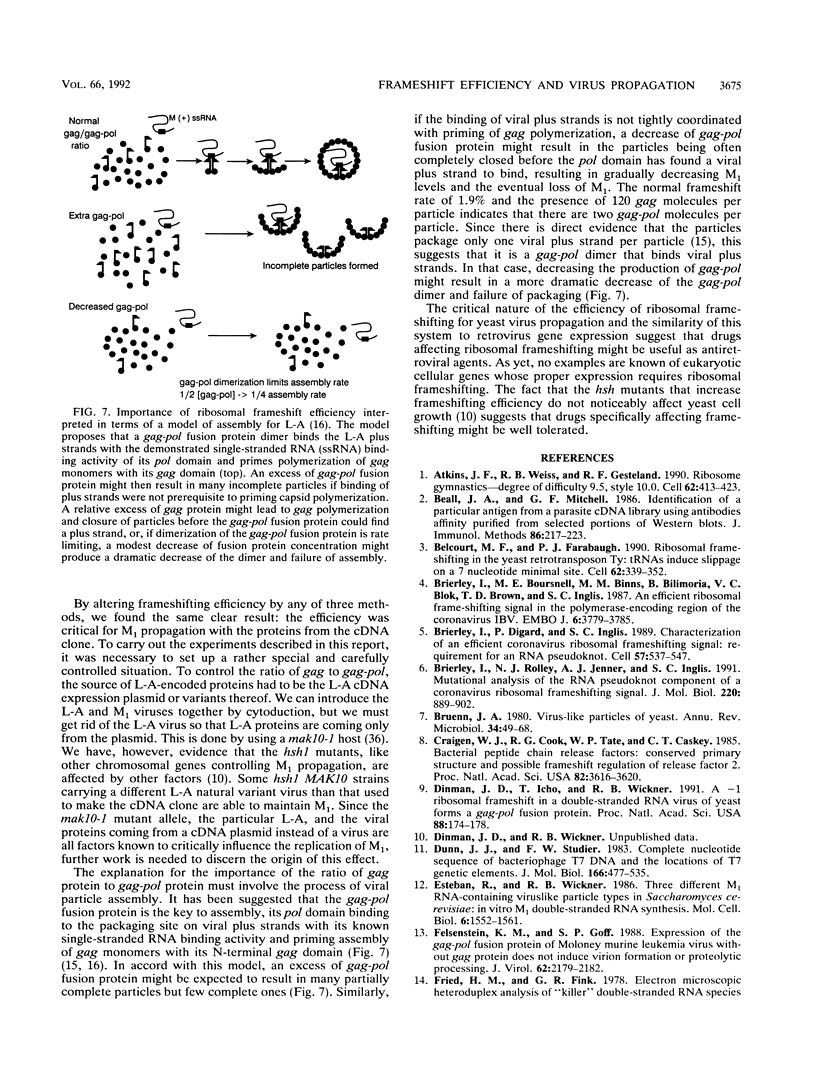
About 1.9% of ribosomes translating the gag open reading frame of the yeast L-A double-stranded RNA virus positive strand undergo a -1 frameshift and continue translating in the pol open reading frame to make a 170-kDa gag-pol fusion protein. The importance of frameshifting efficiency for viral propagation was tested in a system where the M1 (killer toxin-encoding) satellite RNA is supported by a full-length L-A cDNA clone. Either increasing or decreasing the frameshift efficiency more than twofold by alterations in the slippery site disrupted viral propagation. A threefold increase caused by a chromosomal mutation, hsh1 (high shifter), had the same effect. Substituting a +1 ribosomal frameshift site from Ty1 with the correct efficiency also allowed support of M1 propagation. The normal -1 frameshift efficiency is similar to the observed molar ratio in viral particles of the 170-kDa gag-pol protein to the 70-kDa gag gene product, the major coat protein. The results are interpreted in terms of a packaging model for L-A.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall J. A., Mitchell G. F. Identification of a particular antigen from a parasite cDNA library using antibodies affinity purified from selected portions of Western blots. J Immunol Methods. 1986 Feb 12;86(2):217–223. doi: 10.1016/0022-1759(86)90456-4. [DOI] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987 Dec 1;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991 Aug 20;220(4):889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J. A. Virus-like particles of yeast. Annu Rev Microbiol. 1980;34:49–68. doi: 10.1146/annurev.mi.34.100180.000405. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Esteban R., Wickner R. B. Three different M1 RNA-containing viruslike particle types in Saccharomyces cerevisiae: in vitro M1 double-stranded RNA synthesis. Mol Cell Biol. 1986 May;6(5):1552–1561. doi: 10.1128/mcb.6.5.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K. M., Goff S. P. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988 Jun;62(6):2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Fink G. R. Electron microscopic heteroduplex analysis of "killer" double-stranded RNA species from yeast. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4224–4228. doi: 10.1073/pnas.75.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T., Esteban R., Esteban L. M., Wickner R. B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990 Aug 24;62(4):819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Fujimura T., Wickner R. B. Gene overlap results in a viral protein having an RNA binding domain and a major coat protein domain. Cell. 1988 Nov 18;55(4):663–671. doi: 10.1016/0092-8674(88)90225-5. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A., Henderson L. E., Copeland T. D., Sowder R. C., Hixson C. V., Oroszlan S. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7041–7045. doi: 10.1073/pnas.84.20.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T., Wickner R. B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989 Apr 25;264(12):6716–6723. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- Mendenhall M. D., Leeds P., Fen H., Mathison L., Zwick M., Sleiziz C., Culbertson M. R. Frameshift suppressor mutations affecting the major glycine transfer RNAs of Saccharomyces cerevisiae. J Mol Biol. 1987 Mar 5;194(1):41–58. doi: 10.1016/0022-2836(87)90714-5. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Waterhouse P. M., Gerlach W. L. Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res. 1988 Jul 11;16(13):6097–6111. doi: 10.1093/nar/16.13.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Morrow C. D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991 Sep;65(9):5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley S. P., Sommer S. S., Wickner R. B. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984 Apr;4(4):761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991 Oct;7(7):657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Snijder E. J., den Boon J. A., Bredenbeek P. J., Horzinek M. C., Rijnbrand R., Spaan W. J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990 Aug 11;18(15):4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veidt I., Lot H., Leiser M., Scheidecker D., Guilley H., Richards K., Jonard G. Nucleotide sequence of beet western yellows virus RNA. Nucleic Acids Res. 1988 Nov 11;16(21):9917–9932. doi: 10.1093/nar/16.21.9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Ribosomal frameshifting from -2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Icho T., Fujimura T., Widner W. R. Expression of yeast L-A double-stranded RNA virus proteins produces derepressed replication: a ski- phenocopy. J Virol. 1991 Jan;65(1):155–161. doi: 10.1128/jvi.65.1.155-161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Leibowitz M. J. Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics. 1976 Mar 25;82(3):429–442. doi: 10.1093/genetics/82.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N. M., Gesteland R. F., Atkins J. F. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Lommel S. A. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology. 1989 Aug;171(2):543–554. doi: 10.1016/0042-6822(89)90624-7. [DOI] [PubMed] [Google Scholar]