Abstract
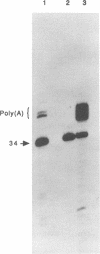
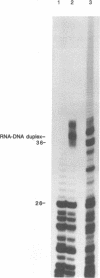
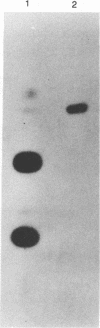
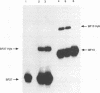
The poliovirus RNA polymerase error frequency was measured in vivo at eight sites in the poliovirus genome. The frequency at which specific G residues in poliovirion RNA changed to another base during one round of viral RNA replication was determined. Poliovirion RNA uniformly labeled with 32Pi was hybridized to a synthetic DNA oligonucleotide that was complementary to a sequence in the viral genome that contained a single internal G residue. The nonhybridized viral RNA was digested with RNase T1, and the protected RNA oligonucleotide was purified by gel electrophoresis. The base substitution frequency at the internal G residue was measured by finding the fraction of this RNA oligonucleotide that was resistant to RNase T1 digestion. A mean value of 2.0 x 10(-3) +/- 1.2 x 10(-3) was obtained at two sites. A modification of the above procedure involved the use of 5'-end-labeled RNA oligonucleotides. The mean value of the error frequency determined at eight sites in the viral genome by using this technique was 4.1 x 10(-3) +/- 0.6 x 10(-3). Sequencing two of the RNase T1-resistant RNA oligonucleotides confirmed that the internal G was changed to a C, A, or U residue in most of these oligonucleotides. Thus, our results indicated that the polymerase had a high error frequency in vivo and that there was no significant variation in the values determined at the specific sites examined in this study.
Full text
PDF





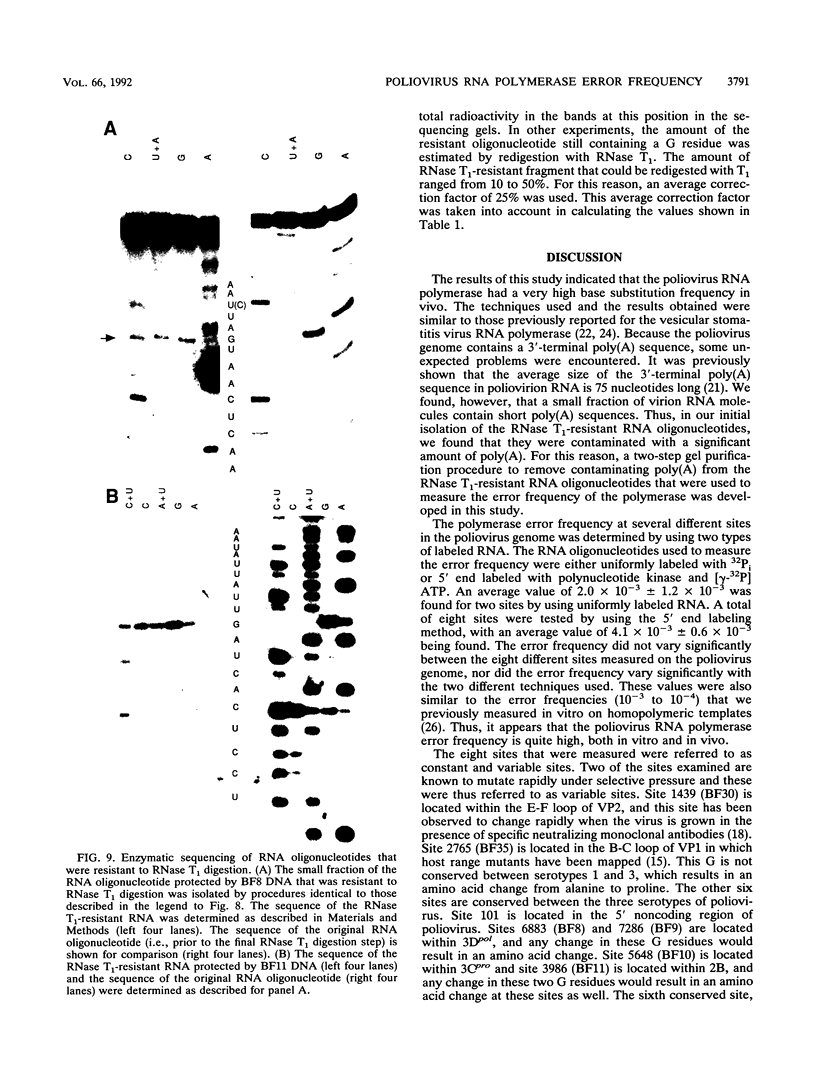


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Emini E. A., Leibowitz J., Diamond D. C., Bonin J., Wimmer E. Recombinants of Mahoney and Sabin strain poliovirus type 1: analysis of in vitro phenotypic markers and evidence that resistance to guanidine maps in the nonstructural proteins. Virology. 1984 Aug;137(1):74–85. doi: 10.1016/0042-6822(84)90010-2. [DOI] [PubMed] [Google Scholar]
- Emini E. A., Wimmer E., Jameson B. A., Bonin J., Diamond D. Neutralization antigenic sites of poliovirus and peptide induction of neutralizing antibodies. Ann Sclavo Collana Monogr. 1984;1(2):139–146. [PubMed] [Google Scholar]
- Gojobori T., Yokoyama S. Rates of evolution of the retroviral oncogene of Moloney murine sarcoma virus and of its cellular homologues. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4198–4201. doi: 10.1073/pnas.82.12.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. J., Domingo E., de la Torre J. C., Steinhauer D. A. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J Virol. 1990 Aug;64(8):3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Kew O. M., Nottay B. K., Hatch M. H., Nakano J. H., Obijeski J. F. Multiple genetic changes can occur in the oral poliovaccines upon replication in humans. J Gen Virol. 1981 Oct;56(Pt 2):337–347. doi: 10.1099/0022-1317-56-2-337. [DOI] [PubMed] [Google Scholar]
- Kuge S., Kawamura N., Nomoto A. Strong inclination toward transition mutation in nucleotide substitutions by poliovirus replicase. J Mol Biol. 1989 May 5;207(1):175–182. doi: 10.1016/0022-2836(89)90448-8. [DOI] [PubMed] [Google Scholar]
- Li W. H., Wu C. I., Luo C. C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985 Mar;2(2):150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Minor P. D., Schild G. C., Bootman J., Evans D. M., Ferguson M., Reeve P., Spitz M., Stanway G., Cann A. J., Hauptmann R. Location and primary structure of a major antigenic site for poliovirus neutralization. Nature. 1983 Feb 24;301(5902):674–679. doi: 10.1038/301674a0. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Bradley J., Yang X. F., Wimmer E., Moss E. G., Racaniello V. R. Poliovirus host range is determined by a short amino acid sequence in neutralization antigenic site I. Science. 1988 Jul 8;241(4862):213–215. doi: 10.1126/science.2838906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottay B. K., Kew O. M., Hatch M. H., Heyward J. T., Obijeski J. F. Molecular variation of type 1 vaccine-related and wild polioviruses during replication in humans. Virology. 1981 Jan 30;108(2):405–423. doi: 10.1016/0042-6822(81)90448-7. [DOI] [PubMed] [Google Scholar]
- Page G. S., Mosser A. G., Hogle J. M., Filman D. J., Rueckert R. R., Chow M. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J Virol. 1988 May;62(5):1781–1794. doi: 10.1128/jvi.62.5.1781-1794.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin J. D., Moscona A., Pan W. T., Leider J. M., Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986 Aug;59(2):377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy J. M., Capone J. P., RajBhandary U. L., Sharp P. A. An inducible mammalian amber suppressor: propagation of a poliovirus mutant. Cell. 1987 Jul 31;50(3):379–389. doi: 10.1016/0092-8674(87)90492-2. [DOI] [PubMed] [Google Scholar]
- Spector D. H., Baltimore D. Polyadenylic acid on poliovirus RNA. II. poly(A) on intracellular RNAs. J Virol. 1975 Jun;15(6):1418–1431. doi: 10.1128/jvi.15.6.1418-1431.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Direct method for quantitation of extreme polymerase error frequencies at selected single base sites in viral RNA. J Virol. 1986 Jan;57(1):219–228. doi: 10.1128/jvi.57.1.219-228.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D. A., de la Torre J. C., Holland J. J. High nucleotide substitution error frequencies in clonal pools of vesicular stomatitis virus. J Virol. 1989 May;63(5):2063–2071. doi: 10.1128/jvi.63.5.2063-2071.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D. A., de la Torre J. C., Meier E., Holland J. J. Extreme heterogeneity in populations of vesicular stomatitis virus. J Virol. 1989 May;63(5):2072–2080. doi: 10.1128/jvi.63.5.2072-2080.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Komaroff L., McDowell M., Baltimore D., Lodish H. F. Translation of reovirus mRNA, poliovirus RNA and bacteriophage Qbeta RNA in cell-free extracts of mammalian cells. Methods Enzymol. 1974;30:709–723. doi: 10.1016/0076-6879(74)30068-7. [DOI] [PubMed] [Google Scholar]
- Ward C. D., Stokes M. A., Flanegan J. B. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol. 1988 Feb;62(2):558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. C., Dunn B. M., Tobin G. J., Flanegan J. B. Anti-VPg antibody precipitation of product RNA synthesized in vitro by the poliovirus polymerase and host factor is mediated by VPg on the poliovirion RNA template. J Virol. 1986 Jun;58(3):715–723. doi: 10.1128/jvi.58.3.715-723.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre J. C., Wimmer E., Holland J. J. Very high frequency of reversion to guanidine resistance in clonal pools of guanidine-dependent type 1 poliovirus. J Virol. 1990 Feb;64(2):664–671. doi: 10.1128/jvi.64.2.664-671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]