Abstract
Combinatorial libraries of synthetic and natural products are an important source of molecular information for the interrogation of biological targets. Methods for the intracellular production of libraries of small, stable molecules would be a valuable addition to existing library technologies by combining the discovery potential inherent in small molecules with the large library sizes that can be realized by intracellular methods. We have explored the use of split inteins (internal proteins) for the intracellular catalysis of peptide backbone cyclization as a method for generating proteins and small peptides that are stabilized against cellular catabolism. The DnaE split intein from Synechocystis sp. PCC6803 was used to cyclize the Escherichia coli enzyme dihydrofolate reductase and to produce the cyclic, eight-amino acid tyrosinase inhibitor pseudostellarin F in bacteria. Cyclic dihydrofolate reductase displayed improved in vitro thermostability, and pseudostellarin F production was readily apparent in vivo through its inhibition of melanin production catalyzed by recombinant Streptomyces antibioticus tyrosinase. The ability to generate and screen for backbone cyclic products in vivo is an important milestone toward the goal of generating intracellular cyclic peptide and protein libraries.
Large numbers of compounds and small, modular molecular architectures are desirable properties in any combinatorial library, but it is difficult to screen libraries with both of these properties. Combinatorial chemistry generates libraries of defined pharmacophores that give meaningful structure-activity relationship data, but functional and cell-based assays dictate spatial-addressing strategies that constrain libraries to modest sizes (103–105 members; refs. 1–3). Genetic encoding allows the elaboration of very large libraries (106–109 members) that can be readily interfaced with biological selection (4–8), screening (9–12), two-hybrid (4–9), and affinity methods (9–11) for target identification, but library members tend to be relatively large (>30 monomers) stand-alone molecules (aptamers or peptides; refs. 10 and 13) or variable segments fused to larger scaffolds (6–9, 11, 12). Although larger constructs help ensure stability to cellular catabolism, they reduce the utility of genetically encoded libraries by hindering pharmacophore identification and by introducing scaffolds that can produce artifacts and obscure active sequences.
A genetically encoded library of small, stable molecules would extend the utility of combinatorial methods by enabling the intracellular production of very large libraries of defined pharmacophores devoid of scaffold-dependent effects. Intracellular libraries of polyketides (14) and small peptides (15) have been described, but neither class of compound is currently amenable to the intracellular generation of large, chemically diverse libraries of stable products. We chose to pursue the intracellular production of small, constrained peptides, because they incorporate a wide array of functional groups and encode five times more information per position than nucleic acids. Head-to-tail cyclization is an attractive method to constrain peptides in vivo, because the products are compatible with any cellular environment and are significantly resistant to proteolysis (16). The ability to cyclize peptides and interrogate the resulting products in vivo is a necessary prerequisite to the generation and manipulation of intracellular libraries of cyclic peptides.
This report describes the use of inteins (internal proteins) to catalyze head-to-tail peptide and protein ligation in vivo. Inteins catalyze a multistep posttranslational protein modification through which they are excised from a precursor fusion protein while religating the flanking domains into a contiguous polypeptide joined by a single vestigial amino acid (17). In Synechocystis sp. PCC6803 (Ssp), it has been observed recently that the replicative polymerase is encoded by two distinct genes (18). The gene encoding the amino-terminal fragment of the polymerase is terminated with a sequence resembling the amino terminus of an intein (N intein or IN), and the gene encoding the carboxyl-terminal fragment of the polymerase is initiated with a sequence resembling the carboxyl terminus of an intein (C intein or IC). Coexpression of the two gene fragments in Escherichia coli results in the production of the full-length polymerase, suggesting that the two intein fragments associate in vivo to function as a heterodimeric (or split) intein.
The ability of a natural split intein to affect a protein ligation in trans suggested a mechanism for in vivo split intein-mediated circular ligation of peptide and proteins (SICLOPPS) via permutation of the order of elements in the fusion protein precursor. Trans-splicing in the context of a fusion protein precursor with the primary sequence IC–target–IN results in head-to-tail cyclization of the target sequence (Fig. 1). Herein, we describe the use of SICLOPPS for the production of a cyclic peptide and protein and demonstrate the ability to couple cyclic product generation to a functional screen.
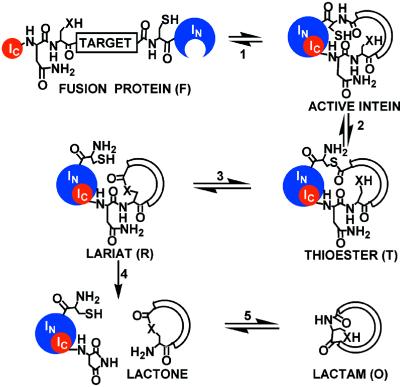
Figure 1.
Circular ligation mechanism. An expressed fusion protein (F) folds to form an active protein ligase (1). The enzyme catalyzes an N-to-S acyl shift (2) at the target–IN junction to produce a thioester intermediate (T), which undergoes transesterification (3) with a side-chain nucleophile (X) at the IC–target junction to form a lariat intermediate (R). Asparagine side-chain cyclization (4) liberates the cyclic product as a lactone, and an X-to-N acyl shift (5) generates the thermodynamically favored, lactam product (O) in vivo.
Materials and Methods
Vector Construction.
The gene for the Ssp DnaE IN was amplified from Ssp genomic DNA with Taq polymerase and primers introducing 5′ BglII and NsiI and 3′ PstI restriction sites. The Ssp DnaE IC gene was amplified similarly with primers introducing 5′ NcoI and 3′ NdeI and SacI restriction sites. Plasmid pDIMCP resulted from individually cloning the intein fragments into pDIMC7 [identical to pDIMC6 (ref. 19), except for conversion of a BamHI restriction site into BglII]. An alanine-to-histidine mutation in the IC gene (A35H) was affected by Quick-Change mutagenesis (Stratagene), resulting in pDIMCPAH. Excision of the intein fragments as an NcoI/PstI digest and ligation into pAR4 [derived from pAR3 (ref. 20; American Type Culture Collection no. 87026), with a unique NcoI in the multiple cloning site] generated pARCP (Fig. 2a) and pARCPAH. E. coli dihydrofolate reductase (DHFR) was amplified from pET22b-DHFR (21) with primers introducing a 5′ NdeI site followed by (CAC)6 (encoding six histidine residues) and a 3′ PstI site, digested with NdeI/PstI and ligated into NdeI/NsiI digested pARCP or pARCPAH to produce pARCP-DHFR (Fig. 2b) and pARCPAH-DHFR (Fig. 2c). A polyhistidine sequence was prepared synthetically with NdeI, NsiI, and BspMI sites and ligated into pARCPAH to produce plasmid pARCP2-6H, which encodes cyclo-[CHMHHHHHHGAGAA]. Plasmid pARCP-p was produced in three steps from pDIMCPAH. (i) Quick-Change mutagenesis introduced an AflII site into IN, generating pDIMCPMA. (ii) The pseudostellarin F gene was synthetically prepared and ligated into MfeI/AflII-digested pDIMCPMA to produce pDIMCP-p. (iii) The fusion construct was excised from pDIMCP-p as an NcoI/PstI fragment and ligated into NcoI/PstI-digested pAR4 to produce pARCP-p (Fig. 2e). To produce plasmid pARCBD-p, a KpnI site was introduced at the carboxyl terminus of the IN gene of pARCP-p by Quick-Change mutagenesis to produce pARCP-pK. The gene encoding the chitin-binding domain was amplified from plasmid pCYB1 (NEB, Beverly, MA) with primers introducing a 5′ KpnI site and a 3′ HindIII site. Both the PCR product and pARCP-pK were digested with KpnI and HindIII and ligated together to generate pARCBD-p (Fig. 2f). All enzymes were from Promega or New England Biolabs unless otherwise noted.
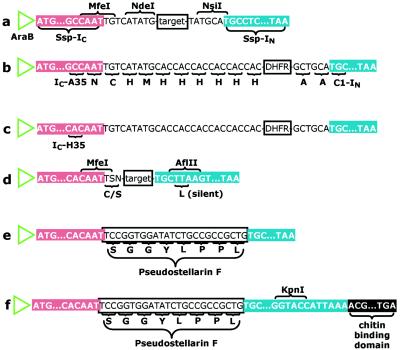
Figure 2.
SICLOPPS plasmid constructs. (a) Plasmid pARCP. A gene encoding a cyclization target is cloned in frame between genes for the two components of the Ssp DnaE split intein (IC and IN) downstream from the araB promoter. (b) Plasmid pARCP-DHFR. Six histidines are inserted between the amino-terminal methionine and isoleucine of DHFR. (c) Plasmid pARCPAH-DHFR. (d) The modified vector: a cysteine (TGY) or serine (TCN) codon can be generated by cloning into the MfeI site (N represents any nucleobase; S represents C or G; Y represents pyrimidines). (e) Plasmid pARCP-p. (f) Plasmid pARCBD-p.
DHFR Purification.
XL1-Blue cells harboring either pARCP-DHFR or pARCPAH-DHFR were grown in LB medium plus 50 μg/ml chloramphenicol at 37°C until the culture reached an OD600 of 0.7. The culture was induced with l-(+)-arabinose to a final concentration of 0.5% and grown at 28°C for 24 h. Cells were harvested by centrifugation (7,000 × g for 10 min) and frozen in liquid nitrogen. The cells were lysed, and DHFR-containing proteins were purified as described (21). The cyclic product was separated from other DHFR-containing intermediates by FPLC by using a Mono-Q column (Amersham Pharmacia) eluted with a gradient of 0–1 M NaCl in 50 mM Tris⋅HCl over 30 min. Western blotting was performed with anti-His (Qiagen, Chatsworth, CA) and goat anti-mouse-alkaline phosphatase-conjugate (Pierce) antibodies according to the manufacturers’ instructions.
Endoproteinase Lys-C Digestion.
Wild-type or cyclic DHFR (50 μg) was treated with 0.5 μg of endoproteinase Lys-C in 0.1 M NH4HCO3 at 37°C. Samples were taken at 6 and 24 h, visualized on a SDS/16% PAGE gel and submitted for matrix-assisted laser desorption ionization (MALDI) time-of-flight mass spectrometry (22).
DHFR Assays.
Thermostability was assayed by preincubation of 100 nM wild-type or cyclic DHFR at either 25°C or 65°C in MTEN buffer (50 mM Mes/25 mM Tris/25 mM ethanolamine/100 mM NaCl). Aliquots were taken at various time points and equilibrated to room temperature for 5 min in the presence of 100 μM 7,8-dihydrofolate. Activity assays were initiated with NADPH as described (21).
Synthesis of Cyclo-[Ser-Gly-Gly-Tyr-Leu-Pro-Pro-Leu].
To a solution of 3.5 mg (4 μmol) of NH2-Ser-Gly-Gly-Tyr-Leu-Pro-Pro-Leu-CO2H and 1.8 mg (16 μmol) of N-hydroxysuccinimide in 20 ml of dimethylformamide was added 3.0 mg (16 μmol) of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). The reaction was stirred for 10 h at 25°C. An additional 3.0 mg of EDC was then added, and stirring was continued at 25°C for another 10 h. The solvent was removed by rotary evaporation, and the residue was dissolved in 2 ml of water for purification by RP-HPLC on a Whatman Partisil 10 ODS-3 9.4-mm × 50-cm column eluted with a linear gradient of 0–50% (vol/vol) acetonitrile in 0.1% trifluoroacetic acid/water over 30 min. The appropriate fractions were lyophilized to yield 2.8 mg (80%) of a white solid [m/z 785 (MH+)]. 1H-NMR and UV-visible spectra of the synthetically prepared material were consistent with published spectra for the isolated natural product (23).
Pseudostellarin F Purification.
E. coli strains XL1-Blue, DH5α, or BL21-DE3 harboring pARCP-p were grown and harvested as described for DHFR purification. The medium (500 ml) was extracted three times with 1-butanol (3 × 100 ml). The extracts were combined and evaporated, and the solid residue was resuspended in 2 ml of 0.1 M K2HPO4 (pH 8.0; lysis buffer). Cells were resuspended in 10 ml of lysis buffer, sonicated, and clarified by centrifugation (20,000 × g for 20 min). The lysate was extracted (3 × 5 ml of n-butanol), and extracts were combined, evaporated, and resuspended in 500 μl of lysis buffer. The recombinant product was purified from lysate and media extracts by HPLC as described above. Lyophilization of the appropriate fractions from the lysate and media extractions yielded an oily residue [m/z 785.47 (MH+), 807.43 (MNa+), and 823.44 (MK+)]. 1H-NMR and UV-visible spectra of the recombinant material were consistent with published spectra for the isolated natural product (23). Proteins fused to the chitin-binding domain were prepared through generation of the clarified lysate as described above. The lysate was passed over a chitin column (NEB) equilibrated with lysis buffer. The column was eluted isocratically, and fractions containing splicing intermediates were pooled and submitted for MALDI mass spectral analysis (22).
Tyrosinase Cloning.
The tyrosinase gene (including ORF 438) from Streptomyces antibioticus (24) was amplified with Vent polymerase from pIJ702 (ATCC no. 35287) with primers introducing 5′ NdeI and 3′ EcoRI restriction sites. The PCR product was digested with NdeI and EcoRI and ligated into similarly digested pDIMN2 (19) to generate pDIMN-Y. Transformed ligation mixtures were grown at ambient temperature for 5 days, and colonies that expressed tyrosinase were identified by pigment formation on FeCuY plates [LB agar plates containing ampicillin (200 μg/ml), FeCl3⋅6H20 (0.2 mM), CuSO4⋅5H20 (0.2 mM), l-tyrosine (0.3 mg/ml, Y), and isopropyl β-d-galactoside (1 mM)] (25).
Results
Design of Genetic Constructs.
The genes encoding Ssp IC and IN were amplified from Ssp genomic DNA by standard molecular biology methods (26) and serially ligated into pDIMC7 (19). The resulting cyclization precursor fragment was excised from pDIMC7 and cloned adjacent to the araB promoter of pAR3 (20) to generate the pARCP vector series (Fig. 2). These vectors activate the expression of cyclization precursors in the presence of arabinose. The E. coli DHFR gene was cloned between the NdeI and NsiI sites of pARCP to create an in-frame fusion with each of the split intein genes (Fig. 2 b and c). The PCR primer used to amplify DHFR also introduced a sequence encoding a six-histidine tag at the 5′ end of the DHFR gene to allow immunodetection of the region to be cyclized. Two DHFR constructs were assembled to investigate the role of the penultimate residue of IC in acid/base catalysis of asparagine side-chain cyclization. Plasmid pARCP-DHFR (Fig. 2b) encodes the wild-type IC gene, which has an alanine residue neighboring the terminal asparagine. Plasmid pARCPAH-DHFR (Fig. 2c) incorporates an alanine-to-histidine mutation at the penultimate position in the IC gene. To produce pseudostellarin F (cyclo-[SGGYLPPL]), the vector was modified by silent mutation to create an AflII site at the 5′ end of the IN gene (Fig. 2d). An MfeI site occurs naturally at the 3′ end of the wild-type IC gene. Ligation of a synthetically prepared, double-stranded insert encoding pseudostellarin F into the modified vector produced plasmid pARCP-p (Fig. 2e). A KpnI site was introduced at the 3′ end of the IN gene to fuse the gene for the chitin-binding domain to the pseudostellarin-producing construct (Fig. 2f).
Production and Characterization of Cyclic DHFR.
DHFR cyclization was readily apparent by SDS/PAGE on arabinose induction of pARCP-DHFR (Fig. 3a). Bands with apparent molecular masses corresponding to the linear (L, 23 kDa) and cyclic (O, 21 kDa) DHFR products, the fusion protein (F, 37 kDa), IN (14 kDa), and IC (4 kDa) were clearly visible, as were bands tentatively assigned as the thioester (T, 36 kDa) and lariat intermediates (R, 26 kDa) (Fig. 3a, lane 2). Mutation of the penultimate residue of IC (A35) from alanine to histidine (Fig. 2c) improved the yield of cyclic DHFR (Fig. 3a, lane 3). Methotrexate-agarose affinity chromatography of the crude lysate (Fig. 3a, lane 4) confirmed that the majority of the induced bands contained correctly folded DHFR. Although IN is not covalently attached to DHFR, it was retained on the methotrexate column presumably because of noncovalent complex formation with the IC–DHFR lariat intermediate (R). The methotrexate-agarose eluant was fractionated by FPLC, allowing purification of 5 mg of the cyclic product per liter of culture (Fig. 3a, lane 5). Western blotting (not shown) with an anti-His antibody indicated the presence of the polyhistidine linker sequence (Fig. 2d) in the FPLC-purified protein. The protein migrated more rapidly in SDS/PAGE analyses than did recombinant DHFR (Fig. 3a, lane 6), despite the extra 11-amino acid linker sequence (Fig. 2b), implying an additional topological constraint. Furthermore, no reaction was detected when the FPLC-purified protein was reacted with phenylisothiocyanate (27), suggesting that the amino terminus was unavailable.
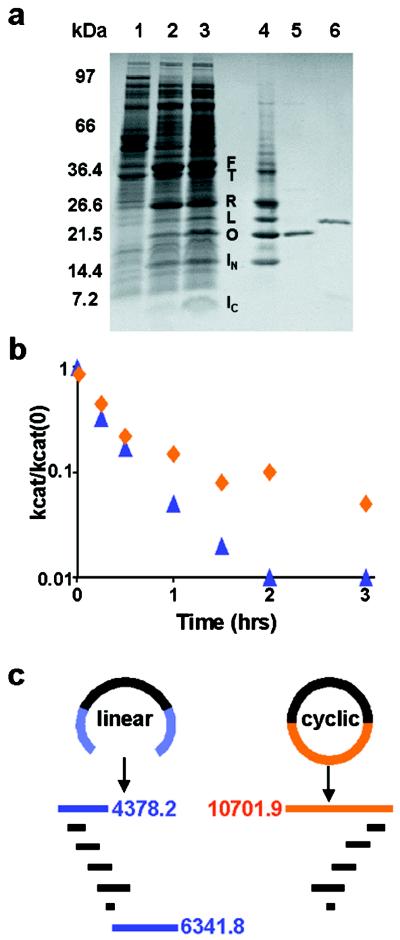
Figure 3.
(a) SDS/PAGE analysis of DHFR cyclization on a 10–20% gradient Tris/glycine ready-gel (Bio-Rad). F, IC–DHFR–IN fusion protein; T, IC–DHFR–IN fusion thioester intermediate; R, IC–DHFR lariat intermediate; L, linear DHFR; O, cyclic DHFR. (Lane 1) Uninduced XL1-Blue/pARCP-DHFR. (Lane 2) Arabinose-induced XL1-Blue/pARCP-DHFR. (Lane 3) Arabinose-induced XL1-Blue/pARCPAH-DHFR. (Lane 4) Lane 3 crude lysate after methotrexate agarose. (Lane 5) Lane 4 material after FPLC. (Lane 6) Wild-type DHFR. (b) Effect of preincubation at 65°C on wild-type (blue triangles) and cyclic (red diamonds) DHFR activity. kcat is determined after preincubation of the enzyme at 65°C; kcat(0) is determined after preincubation at 25°C. (c) Expected endoproteinase Lys-C digestion pattern for linear and cyclic DHFR.
Cyclic DHFR had steady-state kinetic parameters and substrate, cofactor, and methotrexate dissociation constants that were indistinguishable from those of the wild-type enzyme at 25°C. Activity assays conducted after 65°C preincubation of wild-type and cyclic DHFR indicated that cyclization improved the thermostability of the enzyme (Fig. 3b). Endoproteinase Lys-C digestion was used to demonstrate unambiguously that the FPLC-purified protein was cyclic DHFR. Digestion of the wild-type enzyme produces amino-terminal (4.4 kDa) and carboxyl-terminal (6.3 kDa) fragments; in a cyclic protein, these two fragments would be joined, resulting in a 10.7-kDa digestion product (Fig. 3c). The FPLC-purified material was resistant to proteolysis compared with the wild-type enzyme, and mass spectral analysis of the digestion mixtures identified a 10.7-kDa peak in the SICLOPPS product, which was absent in the wild-type enzyme (data not shown).
Production and Characterization of Pseudostellarin F.
Pseudostellarin F production was readily detected in vivo through inhibition of recombinant S. antibioticus tyrosinase (Fig. 4). Coexpression of pseudostellarin F in tyrosinase expressing cells dramatically reduced pigment formation (Fig. 4d). Expression of an unrelated cyclic peptide from pARCP2-6H failed to inhibit tyrosinase (Fig. 4 a and b), and inhibition absolutely required arabinose induction (Fig. 4, compare c and d). SDS/PAGE analysis of arabinose-induced pARCP-p in several bacterial strains (BL21-DE3, DH5α, and XL1-Blue) allowed the visualization of bands corresponding to the fusion protein (F), thioester intermediate (T), and IN. An intense, low-molecular-mass band was also visible, but the resolution was insufficient to separate the lariat intermediate (R) and IC (data not shown). Although pseudostellarin F was too small to be visualized by SDS/PAGE, mass spectral analysis indicated its presence in both the crude cell lysate and medium. Approximately 30 μg of the recombinant cyclic peptide was isolated from the cell lysate per gram of wet cell mass. Pseudostellarin F was also isolated from the medium by 1-butanol extraction followed by HPLC, with a yield that varied between 2 mg/liter (XL1-Blue) and 20 mg/liter (BL21-DE3), depending on the expression strain. The NMR spectrum of the recombinant material was consistent with that reported for the natural product (23), and the retention time of the bacterially expressed cyclic peptide was identical to a synthetically prepared standard. The recombinant material failed to react with ninhydrin (28), indicating a backbone cyclic peptide (lactam) rather than a lactone product. Neither HPLC nor mass spectral analysis provided any evidence for production of the linear parent peptide.
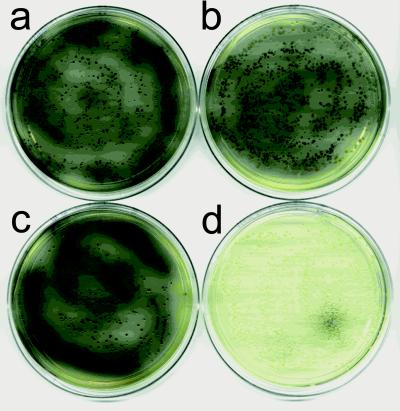
Figure 4.
In vivo tyrosinase inhibition by pseudostellarin F. XL1-Blue cells cotransformed with pDIM-NY and either pARCP2-6H (a and b) or pARCP-p (c and d). Cells were plated on FeCuY plates (25) with chloramphenicol (50 μg/ml), either without (a and c) or with (b and d) l-(+)-arabinose (0.5%).
A chitin-binding domain was fused to the carboxyl terminus of IN to affinity-purify intermediates of the intein-mediated ligation reaction and characterize them by MALDI mass spectrometry (Table 1). All of the intermediates of the splicing reaction, including IC, were retained when the crude cell lysate from arabinose-induced pARCBD-p in XL1-Blue was passed over a chitin affinity column. Pseudostellarin F was recovered from the unretained material by 1-butanol extraction. The observed molecular masses for the fusion protein (F), the thioester intermediate (T), and IN were in excellent agreement with the values predicted from the gene sequence. The mass of IC was consistent with the asparagine cyclized form as predicted from the proposed mechanism of product release. The molecular mass of the lariat intermediate (R) was more consistent with the linear IC–pseudostellarin F fusion product than with the branched lactone product expected from the transesterification reaction.
Table 1.
Mass spectral characterization of pseudostellarin F cyclization intermediates
| Reaction component | Mass, Da
|
||
|---|---|---|---|
| Linear | Cyclic | Observed | |
| F, T | 24,380.5 | NA | 24,380.4 |
| IN | 19,642.0 | NA | 19,642.3 |
| R | 4,756.5 | 4,738.5 | 4,756.2 |
| Ic | 3,969.2 | 3,951.2 | 3,953.0 |
| Pseudostellarin F | 802.4 | 784.4 | 784.4 |
F, fusion protein; T, thioester; R, lariat intermediate; NA, not aplicable.
Discussion
The trans-splicing ability of the naturally occurring DnaE split intein from Ssp (18) has been harnessed to create a general method for in vivo peptide and protein backbone cyclization. Expression from a construct in which the gene for any target sequence is inserted in-frame between genes encoding the two components of the Ssp split intein (Fig. 2) results in the production of a fusion protein that spontaneously cyclizes the target in vivo (Fig. 1). The cyclization vector is compatible with both large and small target sequences and has been optimized for versatility and yield. It was designed with restriction sites both between (Fig. 2a) and within (Fig. 2d) the intein genes to enable the cloning of a wide variety of cyclization targets. An arabinose-inducible expression vector (20) was chosen specifically for the production of fusion proteins to provide high-level expression of cyclization precursors (Fig. 3a), enable the use of various bacterial expression strains, and ensure compatibility with published bacterial two-hybrid systems (4, 5, 29–32). The modular nature of the expression construct should enable the engineering of analogous vectors for use in yeast and mammalian cells. Two expression constructs were generated to test whether the imidazole side chain of histidine catalyzes the asparagine cyclization reaction that liberates the cyclic product from the lariat intermediate (Fig. 1). Cyclic DHFR was produced with both the alanine-containing wild-type (Fig. 2b) and alanine-to-histidine mutant (Fig. 2c) constructs. Because the mutant delivered approximately double the yield of cyclic product (Fig. 3a), the alanine-to-histidine mutation was incorporated into all subsequent constructs. A cysteine, serine, or threonine residue is mechanistically essential to serve as a nucleophile in the split intein catalyzed transesterification reaction and remains behind in the cyclic product (Fig. 1). The MfeI site that occurs naturally at the 3′ end of the wild-type IC gene is perfectly situated such that ligation of appropriate compatible cohesive sticky ends can generate either cysteine or serine (Fig. 2d). The vector places no other constraints on target length or composition.
DHFR was chosen as a protein cyclization target, because it has been successfully circularly permuted (33–35) and even cyclized via a disulfide bond (36). Circular permutation and cyclization studies with DHFR have shown that a linker of greater than three amino acids between the amino and carboxyl termini of the wild-type enzyme is required to retain activity (34–36). DHFR was cloned in-frame into the NdeI and NsiI sites located between the two intein genes so that the cysteine residue introduced by the splicing reaction and the additional sequence encoded by the restriction sites (Fig. 2 b and c) could provide the necessary flexibility to ensure the catalytic integrity of the cyclized product. The purified product migrated more rapidly than the wild-type enzyme in SDS/PAGE analyses (Fig. 3a), was resistant to amino-terminal modification and proteolysis, gave a unique proteolytic fragment (Fig. 3c), and displayed enhanced thermostability (Fig. 3b). All of the results are consistent with the production of cyclic DHFR. The yield of cyclic protein was comparable to that obtained with in vitro intein-mediated protein cyclization methods (37, 38). The resistance of cyclic DHFR to proteolysis is the first indication that cyclization may have the intended effect of conferring resistance to cellular catabolism. The enhanced stability of the protein is also reflected in enzyme-activity measurements. Long (>10-amino acid) linkers between the amino and carboxyl termini of DHFR had no effect on activity at optimal temperatures and conferred stability to the cyclic enzyme at elevated temperatures that were sufficient to denature the wild-type enzyme (Fig. 3b). This result is consistent with previous studies that showed that the thermostability of DHFR was enhanced when the amino and carboxyl termini were constrained through the formation of a disulfide bond (36).
The tyrosinase inhibitor pseudostellarin F (cyclo-[SGGYLPPL]; ref. 23) was an attractive cyclic peptide target, because it is composed entirely of standard L amino acids and lacks posttranslational modification. The serine residue in pseudostellarin F provided an important test of the general utility of the Ssp split intein for protein splicing. The wild-type Ssp IC uses cysteine as the nucleophile for the transesterification portion of the protein ligation mechanism, and studies with DHFR showed that cysteine was capable of fulfilling a similar role in circular ligation (Fig. 1). The pseudostellarin F sequence was cloned into the restriction sites located within the two intein genes (Fig. 2e), and its expression was verified by NMR and mass spectroscopy. Milligram-scale production of pseudostellarin F in bacteria indicates that serine is a viable mechanistic substitute for cysteine in Ssp split intein-mediated protein ligation. The majority of the product was isolated from the medium, which is reasonable for an uncharged, low-molecular-mass cyclic peptide. There was some strain-dependent variation in cyclic peptide production, but the yield of pseudostellarin F from all bacterial expression strains represents a dramatic improvement over the yields reported for in vitro intein-mediated cyclic peptide production (37) and actually rivals the yield from natural sources (23).
Fusion of a chitin-binding domain to the carboxyl terminus of IN had little effect on pseudostellarin F yield (data not shown) but enabled the affinity purification of all of the intermediates of the circular ligation reaction. The molecular masses of intermediates fused to affinity tagged IN were generally consistent with the proposed SICLOPPS mechanism (Table 1), with the notable exception of the IC lariat (R), which seems to be hydrolyzed. This species cannot be on the product formation pathway, and it is not clear at this point whether hydrolysis is a kinetically competitive, nonproductive branch point in the splicing mechanism or whether it is an artifact of sample preparation or mass spectrometry. In addition to corroborating the circular ligation mechanism, affinity chromatography also provided information on the strength of the IN–IC complex. IN was retained on methotrexate agarose, although it was not covalently attached to DHFR, and IC was retained on a chitin column without being covalently fused to the chitin-binding domain. These results suggest that Ssp IN and IC associate very tightly. Because target cyclization is driven by this favorable protein–protein interaction, strained lactam and even lactone products may be accessible by the SICLOPPS method.
Pseudostellarin F production in bacteria was readily screened in vivo through inhibition of melanin production catalyzed by recombinant S. antibioticus tyrosinase (Fig. 4). E. coli grown in the presence of tyrosine, iron, and copper have been shown to produce melanin on the expression of S. antibioticus tyrosinase from a heat-inducible promoter (25). Thermal induction was a problem for our studies, because initial reports suggested that the Ssp split intein had poor activity at elevated temperatures (18). Tyrosinase expression was therefore placed under the control of a lac promoter (19), and a similar phenotype was observed on induction with isopropyl β-d-galactoside (data not shown). The influence of pseudostellarin F, arabinose, and the SICLOPPS vector on tyrosinase-dependent melanin production was investigated by introducing plasmids encoding pseudostellarin F (pARCP-p) or a control peptide into bacteria producing the recombinant tyrosinase. The control plasmid had no affect on melanin production in either the presence or absence of arabinose (Fig. 4 a and b). Plasmid pARCP-p also had no affect on melanin production in the absence of arabinose (Fig. 4c) but completely inhibited melanin production in the presence of the transcriptional activator (Fig. 4d). The controls clearly show that neither arabinose nor the two intein fragments alone prevent pigment formation; the pseudostellarin F sequence either in the context of the cyclic product or displayed in the myriad intermediate fusions must be uniquely responsible for tyrosinase inhibition.
SICLOPPS has been developed as a general method for backbone cyclization of peptides and proteins in vivo. The method has been used to cyclize E. coli DHFR and to produce the cyclic, eight-amino acid tyrosinase inhibitor pseudostellarin F. The cyclization of both products was confirmed by mass spectral analysis, and each was isolated with yields of milligrams per liter of bacterial culture. Cyclization could be detected for DHFR by increased thermostability in vitro and for pseudostellarin F by inhibition of recombinant S. antibioticus tyrosinase in vivo. The production/screening of pseudostellarin F in bacteria serves as a concrete example of the integration of intracellular cyclization and functional screening and provides a foundation for the elaboration and deconvolution of combinatorial libraries of cyclic products in vivo.
Acknowledgments
We thank Dr. Donald Bryant (Penn State University) for Ssp genomic DNA, Dr. Andrew Nixon (Dyax, Inc.) for plasmid pAR4, Dr. Marc Ostermeier (Penn State University) for plasmids pDIM-C7 and pDIM-N2, Dr. A. Daniel Jones (Penn State University) for mass spectral analyses, and Dr. Anne Stanley (Hershey Medical Center) for synthesis of NH2-Ser-Gly-Gly-Tyr-Leu-Pro-Pro-Leu-CO2H. C.P.S. was supported in part by National Institutes of Health Grant GM19891-01.
Abbreviations
- DHFR
dihydrofolate reductase
- Ssp
Synechocystis sp. PCC6803
- IN
amino-terminal intein fragment
- IC
carboxyl-terminal intein fragment
- SICLOPPS
split intein-mediated circular ligation of peptides and proteins
- MALDI
matrix-assisted laser desorption ionization
References
- 1.Gallop M A, Barrett R W, Dower W J, Fodor S P A, Gordon E M. J Med Chem. 1994;37:1233–1251. doi: 10.1021/jm00035a001. [DOI] [PubMed] [Google Scholar]
- 2.Gordon E M, Barrett R W, Dower W J, Fodor S P A, Gallop M A. J Med Chem. 1994;37:1385–1401. doi: 10.1021/jm00036a001. [DOI] [PubMed] [Google Scholar]
- 3.Czarnik A W. Curr Opin Chem Biol. 1997;1:60–66. doi: 10.1016/s1367-5931(97)80109-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Murphy A, Hu J C, Kodadek T. Curr Biol. 1999;9:417–420. doi: 10.1016/s0960-9822(99)80188-2. [DOI] [PubMed] [Google Scholar]
- 5.Pelletier J N, Arndt K M, Pluckthun A, Michnick S W. Nat Biotechnol. 1999;17:683–690. doi: 10.1038/10897. [DOI] [PubMed] [Google Scholar]
- 6.Caponigro G, Abedi M R, Hurlburt A P, Maxfield A, Judd W, Kamb A. Proc Natl Acad Sci USA. 1998;95:7508–7513. doi: 10.1073/pnas.95.13.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer C R, Colman-Lerner A, Brent R. Proc Natl Acad Sci USA. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman T C, Smith D L, Sorger P K, Drees B L, O’Rourke S M, Hughes T R, Roberts C J, Friend S H, Field S, Murray A W. Science. 1999;285:591–595. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 9.Colas P, Cohen B, Jessen T, Grishnina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 10.Osborne S E, Matsummura I, Ellington A D. Curr Opin Chem Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 11.Stricker N L, Schatz P, Li M. Methods Enzymol. 1999;303:451–468. doi: 10.1016/s0076-6879(99)03027-x. [DOI] [PubMed] [Google Scholar]
- 12.Yoo Y, Rote K, Reichsteiner M. J Biol Chem. 1989;264:17078–17083. [PubMed] [Google Scholar]
- 13.Davidson A R, Sauer R T. Proc Natl Acad Sci USA. 1994;91:2146–2150. doi: 10.1073/pnas.91.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Betlach M, Ashley G. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenson T, Herrera J V, Kloss P, Guarneros G, Mankin A S. J Bacteriol. 1999;181:1617–1622. doi: 10.1128/jb.181.5.1617-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh T, Li S, Friedman T M, Wiaderkiewicz R, Korngold R, Huang Z W. Biochem Biophys Res Commun. 1996;224:438–443. doi: 10.1006/bbrc.1996.1045. [DOI] [PubMed] [Google Scholar]
- 17.Perler F B, Olsen G J, Adam E. Nucleic Acids Res. 1997;25:1087–1093. doi: 10.1093/nar/25.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Hu Z, Liu X Q. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostermeier M, Nixon A E, Shim J H, Benkovic S J. Proc Natl Acad Sci USA. 1999;96:3562–3567. doi: 10.1073/pnas.96.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Perez J, Gutierrez J. Gene. 1995;158:141–142. doi: 10.1016/0378-1119(95)00127-r. [DOI] [PubMed] [Google Scholar]
- 21.Miller G P, Benkovic S J. Biochemistry. 1998;37:6327–6335. doi: 10.1021/bi972922t. [DOI] [PubMed] [Google Scholar]
- 22.Moore W T. Methods Enzymol. 1997;289:520–542. doi: 10.1016/s0076-6879(97)89062-3. [DOI] [PubMed] [Google Scholar]
- 23.Morita H, Kayashita T, Kobata H, Gonda A, Takeya K, Itokawa H. Tetrahedron. 1994;50:9975–9982. [Google Scholar]
- 24.Bernan V, Filpula D, Herber W, Bibb M, Katz E. Gene. 1985;37:101–110. doi: 10.1016/0378-1119(85)90262-8. [DOI] [PubMed] [Google Scholar]
- 25.Della-Cioppa G, Garger S J, Sverlow G G, Turpen T H, Grill L K. Biotechnology. 1990;8:634–638. doi: 10.1038/nbt0790-634. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Edman P. Acta Chem Scand. 1950;4:283–293. [Google Scholar]
- 28.Gordon A J, Ford R A. The Chemist’s Companion: A Handbook of Practical Data, Techniques, and References. New York: Wiley Interscience; 1972. [Google Scholar]
- 29.Karimova G, Pidoux J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dmitrova M, Younes-Cauet G, Oertel-Buchheit P, Porte D, Schnarr M, Granger-Schnarr M. Mol Gen Genet. 1998;257:205–212. doi: 10.1007/s004380050640. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Piston D W, Johnson C H. Proc Natl Acad Sci USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi F, Charlton C A, Blau H M. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchwalder A, Szadkowski H, Kirschner K. Biochemistry. 1992;31:1621–1630. doi: 10.1021/bi00121a006. [DOI] [PubMed] [Google Scholar]
- 34.Uversky V N, Kutyshenko V P, Protasova N Y, Rogov V V, Vassilenko K S, Gudkov A T. Protein Sci. 1996;5:1844–1851. doi: 10.1002/pro.5560050910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwakura M, Nakamura T. Protein Eng. 1998;11:707–713. doi: 10.1093/protein/11.8.707. [DOI] [PubMed] [Google Scholar]
- 36.Iwakura M, Honda S. J Biochem (Tokyo) 1996;119:414–420. doi: 10.1093/oxfordjournals.jbchem.a021257. [DOI] [PubMed] [Google Scholar]
- 37.Evans T C, Benner J, Xu M Q. J Biol Chem. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 38.Camerero J A, Muir T W. J Am Chem Soc. 1999;121:5597–5598. [Google Scholar]