Abstract
Repeated, specific interactions between capsid protein (CP) subunits direct virus capsid assembly and exemplify regulated protein–protein interactions. The results presented here reveal a striking in vivo switch in CP assembly. Using cryoelectron microscopy, three-dimensional image reconstruction, and molecular modeling, we show that brome mosaic virus (BMV) CP can assemble in vivo two remarkably distinct capsids that selectively package BMV-derived RNAs in the absence of BMV RNA replication: a 180-subunit capsid indistinguishable from virions produced in natural infections and a previously unobserved BMV capsid type with 120 subunits arranged as 60 CP dimers. Each such dimer contains two CPs in distinct, nonequivalent environments, in contrast to the quasi-equivalent CP environments throughout the 180-subunit capsid. This 120-subunit capsid utilizes most of the CP interactions of the 180-subunit capsid plus nonequivalent CP–CP interactions. Thus, the CP of BMV, and perhaps other viruses, can encode CP–CP interactions that are not apparent from mature virions and may function in assembly or disassembly. Shared structural features suggest that the 120- and 180-subunit capsids share assembly steps and that a common pentamer of CP dimers may be an important assembly intermediate. The ability of a single CP to switch between distinct capsids by means of alternate interactions also implies reduced evolutionary barriers between different capsid structures. The in vivo switch between alternate BMV capsids is controlled by the RNA packaged: a natural BMV genomic RNA was packaged in 180-subunit capsids, whereas an engineered mRNA containing only the BMV CP gene was packaged in 120-subunit capsids. RNA features can thus direct the assembly of a ribonucleoprotein complex between alternate structural pathways.
Most isometric viruses contain capsid proteins (CPs) arranged with quasi-icosahedral symmetry (1). This strategy allows the assembly of capsids containing more than the 60 identical CPs permitted by strict icosahedral symmetry, providing the CP is polymorphic and can adapt to a specific set of alternate, quasi-equivalent environments. Progressively larger capsids with 60T subunits can be formed as the number of quasi-equivalent CP–CP interaction states, T, increases according to T = h2 + hk + k2 for integers h and k. Such CP polymorphism offers insight into regulated protein conformational changes, protein–protein interaction, and protein–nucleic acid interaction.
By expanding capsid size, quasi-equivalence allows expanding viral genomes. Virus evolution appears to have led to continuing modification of CP polymorphism because many CPs share a similar β-barrel core (2) and known capsids range from T = 1 to T = 169 (3, 4). However, little is known about the extent of assembly flexibility possible in a single CP to allow such evolutionary transitions.
One group of viruses with well-studied, quasi-icosahedral capsids is the bromovirus genus, a set of positive-strand RNA plant viruses with T = 3 capsids. Brome mosaic virus (BMV) and the closely related cowpea chlorotic mottle virus (CCMV) have been used in many studies of virion structure, assembly, and disassembly (5). BMV CP and CCMV CP are 70% identical in sequence and functionally interchangeable in vivo (ref. 6; R. Allison and P.A., unpublished work). The crystal structure of the 28-nm diameter CCMV capsid has been determined to 3.2-Å resolution (7). The only CP assemblies isolated to date from bromovirus-infected plants are 180-subunit, T = 3 particles containing viral RNA (5).
Bromovirus genomes are divided among three messenger-sense RNAs (Fig. 1A). RNA1 and RNA2 encode RNA replication factors 1a and 2a (8). RNA3 encodes the 3a cell-to-cell movement protein and CP. CP is translated from subgenomic RNA4, produced by partial transcription of the negative-sense RNA3 replication intermediate (8). This dependence of CP expression on viral RNA synthesis complicates study of several important questions, such as whether in vivo encapsidation of bromovirus RNA is coupled to viral RNA replication, as with poliovirus (9), and whether such coupling might underlie CP specificity for packaging viral RNA.
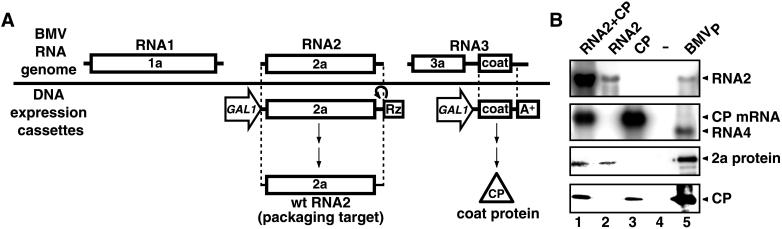
Figure 1.
(A) Structure of the BMV RNA genome and plasmid DNA cassettes used to express wild-type BMV RNA2 and CP. GAL1, Rz, and A+ denote the GAL1 promoter, a self-cleaving hepatitis δ virus ribozyme (18), and the yeast ADH1 polyadenylation signal, respectively (14). (B) BMV RNA and protein accumulation in yeast. Equal amounts of total RNA from yeast expressing the indicated BMV components were analyzed by Northern blotting with RNA2 (Top) and CP (Second blot) probes. RNA from plant-derived BMV virions (BMVP) was used as a control. Equal amounts of total protein from yeast expressing the indicated components were analyzed by Western blotting with anti-2a (Third blot) and anti-CP (Bottom) antisera. Controls (lane 5) were baculovirus-expressed 2a protein (19) (Third blot, lane 5) and BMVP CP (Bottom, lane 5).
To test whether BMV RNA encapsidation could be separated from viral RNA replication in vivo, we expressed BMV CP and certain BMV RNAs (Fig. 1) in Saccharomyces cerevisiae. This yeast supports infectious virion production by an animal RNA virus, flock house virus (10), and BMV RNA replication and subgenomic mRNA synthesis (11). Using this yeast system, cryoelectron microscope (cryoEM) image reconstruction, and molecular modeling, we demonstrate separation of BMV RNA replication and encapsidation in vivo, and the unprecedented RNA-controlled ability of a single CP to assemble either quasi-equivalent or nonequivalent capsids. The results have implications for virion assembly, virus evolution, and regulated protein interactions.
Materials and Methods
Yeast and Plasmids.
Yeast strain YPH500 was used throughout as described (11). BMV CP was expressed from pMKCP3, constructed as follows. Annealed oligonucleotides TCGAGATCTTTAAAATG and TCGACATTTTAAAGATC were ligated into the SalI site overlapping the CP initiation codon in pB3TP8 (12). The BglII/HindIII fragment (CP ORF) from this plasmid, EcoRI/BamHI fragment (GAL1 promoter) of pBM272 (13), and HindIII/SphI fragment (ADH1 polyadenylation signal) of pAAH5 (14) were simultaneously ligated into EcoRI/SphI-digested yCPlac33 (15). pB2NR3, which produces wild-type RNA2 under the control of the yeast GAL1 promoter, was the generous gift of J. Chen (Institute for Molecular Virology, University of Wisconsin, Madison, WI).
Virion Preparation and Analysis.
Yeast were grown to OD600 = 1.0–1.6, pelleted for 4 min at 5,000 × g, resuspended in 1/20th vol of ST [1 M sorbitol/50 mM Tris⋅HCl (pH 7.5)], repelleted, and incubated 2.5 hr at 30°C in 1/100th original culture volume of ST with 10 mM 2-mercaptoethanol and 200 units/ml lyticase (Sigma). Spheroplasts were lysed by vortexing twice for 5 min after adding 1 ml of VSB [0.1 M sodium acetate (pH 5.0)/10 mM MgCl2/5 mM CaCl2/1 mM EDTA], 0.5 ml of CHCl3, and 0.5 ml of glass beads per ml of cell suspension, and centrifuged 5 min at 21,000 × g. The supernatant was held on ice for 5 min and centrifuged as above. This clarified lysate was centrifuged for 25 min at 120,000 rpm, 10°C in a Beckman Coulter TLA-120.2 rotor. The pellet was resuspended in 0.5 ml of VSB per liter of starting culture, clarified by centrifuging 1 min at 15,000 × g, layered on a linear 5–30% (wt/vol) sucrose gradient in 0.5× VSB, and centrifuged 1.5 hr at 40,000 rpm, 10°C in a Beckman Coulter SW 41 rotor. The gradient was fractionated into 0.2-ml fractions with an ISCO gradient fractionator. Sedimentation coefficients and molecular weights were determined by using an Optima XL-A analytical ultracentrifuge (Beckman Coulter), taking the BMV partial specific volume as 0.71 cm3⋅gm−1 (16). Sedimentation coefficients were measured at 20°C at A260 = 0.8, 0.6, and 0.2 and extrapolated to zero concentration in water. Mass determinations were made at A260 = 0.4, 20°C.
RNA and Protein Analysis.
RNA was isolated as described (17) with 1 μg/ml glycogen added to virion RNA before precipitation. Northern blot analysis of RNA2 and CP mRNA was performed as described (18). For total RNA analysis, RNA was electrophoresed through 1% agarose, stained with SYBR Green II (Molecular Probes), and visualized with a FluorImager (Molecular Dynamics). Protein electrophoresis and Western blotting were as described (19) without Tween 20 and with anti-2a sera (19) diluted 1:500 or anti-CP sera (American Type Culture Collection) diluted 1:40,000.
CryoEM and Image Reconstruction.
CryoEM of 2.5 mg/ml virus-like particles (VLP) or BMV virion samples was performed essentially as described (20). The samples were examined in a Philips EM420 (at 80 kV) or a CM200 FEG (at 200 kV) transmission electron microscope, and images were recorded on film at nominal magnifications of ×49,000 or ×38,000, respectively. The diameters of vitrified 65S VLPs followed a Gaussian distribution (SD = 5%). Images of particles whose diameters varied no more than 3.5% from the mean were selected for further processing, and all images were scaled to match the mean particle diameter. Common lines procedures (21) were used to initially determine particle orientations and centers, and further refinement was performed by model-based techniques (22). Particle handedness was determined as described (23). Images of 88S VLPs were recorded with a Gatan 794 Multiscan charge-coupled device (CCD) camera with the electron beam spread just large enough to cover the CCD detector (0.4 μm on the specimen) for image recording without preirradiation (22). Reconstructions of BMV virions and the 88S and 65S VLPs were computed from 432, 1122, and 337 boxed particle images, respectively, to effective resolutions of 20, 21, and 24 Å.
Molecular Modeling.
Reconstructed electron densities of the 65S VLP and CCMV A/B CP dimer (7) were codisplayed with the program o (24). Residues disordered in the A subunit x-ray structure, up to Lys-42, were removed from the B subunit model. No adjustment of individual residues was made. Initial dimer coordinates were transformed with standard icosahedral symmetry operators and adjusted by hand to display the 120-subunit model. Because no internal magnification standard was available, the 65S VLP electron density map was scaled to optimize contacts between modeled subunits, yielding a 5% size reduction. Observed structure factor amplitudes for conjugate gradient refinement of the initial model by the program x-plor (25) were computed by Fourier transformation of the cryoEM electron density. The A and B subunits were treated as independent, rigid objects during the refinement. The overall procedure is formally comparable to a typical rigid body x-ray structure refinement, as discussed by Cheng et al. (26). Figs. 4B through 6C were generated with bobscript (27) and raster3d (28).
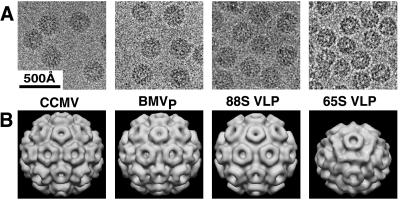
Figure 4.
CryoEM and three-dimensional image reconstruction of CCMV, BMVP, and yeast-derived VLPs. The 88S and 65S VLPs were purified by sucrose gradient sedimentation from yeast expressing RNA2 + CP and CP only, respectively. (A) Cryoelectron micrographs showing representative fields of the indicated particles. Unstained particles appear dark against the brighter (less dense) supporting layer of vitreous ice. (B) Three-dimensional image reconstructions of CCMV (7), BMVP, and the 88S and 65S VLPs from yeast.
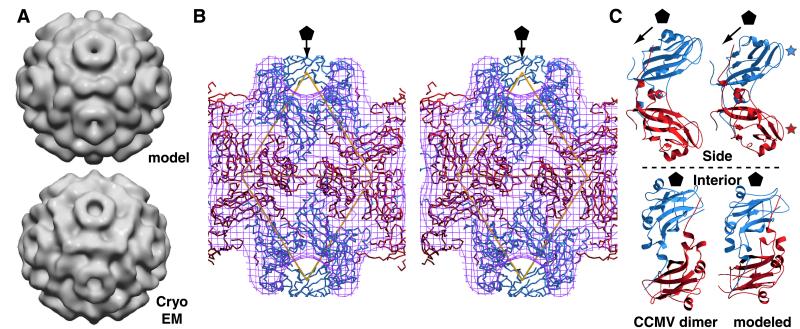
Figure 6.
(A) Surface views of modeled (Upper) and reconstructed (Lower) 65S VLP electron density. The density map of the model was computed to 25-Å resolution; the 65S VLP reconstruction is as in Fig. 4B. (B) Stereo image of modeled 65S VLP subunits fitted within the cryoEM density map. Subunits are colored as in Fig. 5. The outer envelope of the cryoEM density map is represented as a purple mesh, and the fivefold axis is indicated by a pentagon. Asymmetric units are indicated by yellow triangles. Note B/B subunit interactions around twofold and threefold axes. (C) Ribbon diagrams of A/B dimers from CCMV (Left) and the modeled 65S VLP dimer (Right). The upper images represent views tangential to the particle surfaces. Blue and red stars indicate exterior loops of A and B subunits, respectively. The lower images represent views from the particle centers.
Results
BMV RNA Encapsidation in Yeast in 88S and 65S VLPs.
BMV RNA2 and CP were expressed in yeast individually and together (Fig. 1). Coexpressing RNA2 and CP increased RNA2 accumulation 20-fold but caused little change in 2a protein levels (Fig. 1B, lanes 1–2), implying that CP stabilized RNA2 in a nontranslatable form. Coexpressing RNA2 decreased CP mRNA accumulation 4-fold (Fig. 1B, lanes 1 and 3), suggesting that RNA2 competed with CP mRNA for a common stabilizing factor.
Consistent with their possible encapsidation, RNA2 and CP mRNA selectively fractionated in a CP-associated, nuclease-resistant form. When yeast expressing BMV RNA2 and CP were lysed without phenol or other nuclease inhibitors, endogenous nuclease degraded all RNAs except RNA2, CP mRNA, and low molecular weight RNA fragments (Fig. 2A, lanes 1 and 2). Centrifuging the lysate under conditions that pellet BMV virions removed most low molecular weight RNA fragments, whereas RNA2 and CP mRNA were recovered in the pellet (Fig. 2A, lane 4) in a form resistant to S1 nuclease (data not shown). Most proteins remained in the supernatant, but the pellet was enriched for a small number of proteins, including one comigrating with BMV CP (Fig. 2B, lanes 3 and 4). Western blotting confirmed the identity of this band and revealed that CP was exclusively recovered in the high-speed pellet.
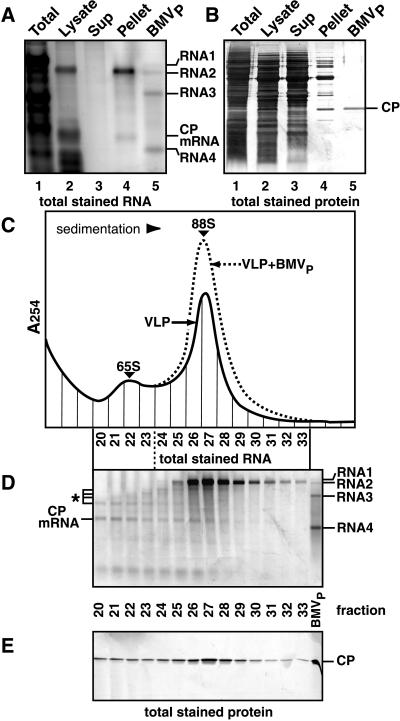
Figure 2.
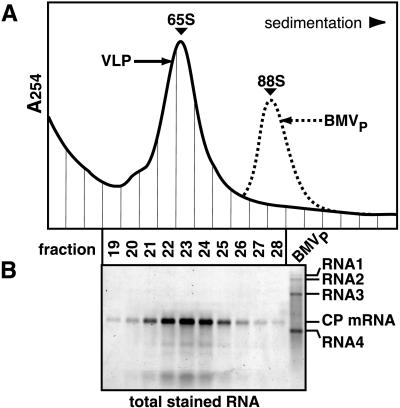
Analysis of VLPs from yeast expressing BMV RNA2 and CP. (A) RNA analysis. RNA from the indicated fractions (see text) was electrophoresed through 1% agarose, stained for total RNA, and imaged by laser fluorometry. BMVP RNA (lane 5) was included as a control. The indicated bands in lanes 2 and 4 were identified as CP mRNA and RNA2 by hybridization to CP and RNA2 probes. (B) Protein analysis. Total protein was electrophoresed and visualized by silver staining. BMVP CP (rightmost lane) was included as a control. (C) Sucrose gradient sedimentation. The resuspended pellet in lanes 4 of A and B was centrifuged through a 5–30% linear sucrose gradient, and gradient fractions were analyzed by UV absorption at 254 nm. Sedimentation profiles of yeast-derived VLPs alone (solid curve) and with BMVP added (dotted curve) are shown. (D) RNA analysis of sucrose gradient fractions, as in A. BMVP RNA (right lane) was used as a control. RNA2, RNA2 fragments (*), and CP mRNA were identified by hybridization to CP- and RNA2-specific probes. (E) Protein analysis of sucrose gradient fractions, as in B.
Sucrose-gradient sedimentation of the resuspended high-speed pellet revealed a faster sedimenting, major peak of VLPs and a slower, minor VLP peak (Fig. 2C). Plant-derived BMV virions (hereafter, BMVP) added to the resuspended pellet cosedimented with the major peak (Fig. 2C) at s20,w = 88 S (Table 1). Full-length RNA2 was the principal RNA in the major VLP peak (Fig. 2D, fractions 25–29) and was absent from slower-sedimenting fractions. In the minor VLP peak (fractions 20–23), the most abundant RNA was CP mRNA. BMV CP was the only protein detectable in either VLP (Fig. 2E). CP in the minor peak was indistinguishable from that of the major peak or BMVP, suggesting that the sedimentation difference did not result from CP modification.
Table 1.
Biophysical data for BMVP and yeast-derived 65S VLPs
| Particle | s20,w, S | Mr (×106)* | Mr (×106)† | % RNA‡ |
|---|---|---|---|---|
| BMV (ref. 16) | 88 | 4.6 | NA | 21.4 |
| BMVP (this work) | 88 | 4.3 ± 0.2 | 4.5 | 20.0 ± 1.2 |
| 65S VLP | 65 | 2.7 ± 0.2 | 2.7 | 9.5 ± 1.4 |
Determined by analytical ultracentrifugation.
† Determined by scanning transmission electron microscopy. Data generously provided by M. Simon at Brookhaven National Laboratory (29).
‡ Determined from sucrose gradient-purified particles. Protein content was measured using the Hartree–Lowry or bicinchoninic acid (Pierce) assay. Extracted RNA content was measured by optical absorbance at 260 nm.
To further explore the nature of the VLP peaks in Fig. 2C, these analyses were repeated with yeast expressing CP alone. In the absence of RNA2, CP was found exclusively in a rapidly sedimenting form selectively associated with nuclease-resistant CP mRNA. Sucrose gradient centrifugation (Fig. 3A) revealed a single prominent VLP peak that cosedimented at s20,w = 65 S (Table 1), with the slower peak of Fig. 2C well behind BMVP. Within this peak, CP mRNA was the major RNA (Fig. 3B), and BMV CP was the only protein detectable (data not shown).
Figure 3.
Analysis (as in Fig. 2) of VLPs from yeast expressing BMV CP without RNA2. (A) Sucrose gradient sedimentation profiles of yeast-derived VLPs alone (solid curve) and with BMVP added (dotted curve). (B) RNA analysis of sucrose gradient fractions. CP mRNA was identified by hybridization to a CP probe.
Capsid Structure of 88S and 65S VLPs.
CryoEM and three-dimensional image reconstruction were used to visualize gradient-fractionated 88S and 65S yeast-derived VLPs and BMVP (Fig. 4). The 88S VLP and BMVP capsid reconstructions were indistinguishable from each other and from CCMV virions (7). In contrast, the 65S VLPs had an average diameter approximately 10% smaller than BMVP and a drastically different surface morphology.
The number of CP subunits per 65S VLP was determined by measuring the molecular weights (Mr) and protein:RNA ratios of 65S VLPs and BMVP controls (Table 1). Mr determinations by analytical centrifugation and scanning transmission electron microscopy (29) were in good agreement. The Mr and RNA content of the 65S VLP were 2.7 × 106 and 9.5 ± 1.4% (Table 1). These results are inconsistent with the most obvious candidate capsid smaller than T = 3, a T = 1 capsid with 60 CPs in perfect icosahedral symmetry. Although T = 1 capsids have been assembled in vitro from BMV CP (16), a T = 1 capsid with 60 CPs and one copy of the CP mRNA would have a Mr of 1.56 × 106 and contain 23% RNA. Instead, the data imply that 65S VLPs consist of one CP mRNA and 120 CPs, yielding a predicted Mr of 2.76 × 106 and RNA content of 13%.
Molecular Modeling of the 65S VLP Capsid.
The 88S, T = 3 bromovirus capsids display pentameric and hexameric CP clusters (“capsomeres”) at the fivefold and threefold symmetry axes, respectively (Fig. 4B). The 65S VLP image reconstruction revealed similar pentameric capsomeres (Fig. 4B). This implied that it might be possible to model the 65S VLP structure from the T = 3 bromovirus capsid structure, by preserving CP monomer interactions at the 5-fold axes, while exploring possible perturbations of surrounding interactions.
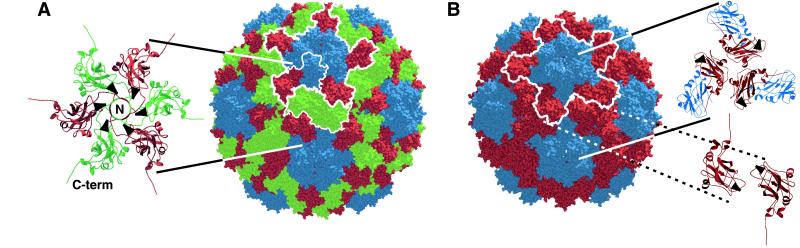
In T = 3 capsids, chemically identical CP subunits occupy three similar but distinct symmetry environments (1) denoted A, B, and C (blue, red, and green in Fig. 5A). In bromovirus capsids, each A subunit is involved with a flanking B subunit in a strong dimer interaction involving the reciprocal exchange of C-terminal arms (7) (Figs. 5A and 6C). C/C dimers (Fig. 5A) also depend on these interactions. The intimate nature of the C-terminal invasion (7), the strong tendency of bromovirus CP to form dimers in solution (16), and the effects of mutations on the dimer interaction sites (30) all implicate CP dimers as basic units of bromovirus capsid assembly.
Figure 5.
Comparison of 180- and 120-subunit capsids. (A) Space-filling model of CCMV 180-subunit capsid (7). A, B, and C subunits are blue, red, and green, respectively. One A/B dimer and one C/C dimer are each outlined with a thin white line. One of twelve pentamers of A/B dimers is outlined with a thick white line. The ribbon diagram shows B and C subunits surrounding a threefold axis. Arrowheads illustrate head-to-head CP orientation. (B) Modeled 120-subunit BMV capsid displayed as a space-filling model. A and B subunits are blue and red, respectively. One of twelve pentamers-of-dimers is outlined in white. The upper ribbon diagram illustrates the nearly tail-to-tail interaction of A/B dimers surrounding the threefold axis. The lower ribbon diagram illustrates antiparallel interaction of B subunits between threefold axes. Arrowheads are positioned on subunits as in A.
The organization of 60 high-affinity A/B dimers around the 88S virion fivefold axes and the related pentameric capsomeres of 65S VLPs (Fig. 4B) suggested that the 120 CP monomers in each 65S VLP might consist of 60 A/B dimers arranged with T = 1 symmetry. An initial 60-dimer model of the 120-subunit capsid was derived from the CCMV T = 3 structure (7) (Fig. 5A) by removing the C/C dimers, rotating the remaining pentamers of A/B dimers 30° counterclockwise, and translating them radially down the fivefold axes of symmetry until the B subunits contacted. This created a closed shell that reproduced the major features of the 65S VLP reconstruction (see below).
The calculated structure factor amplitudes from the hand-fitted model and the observed amplitudes of the cryoEM reconstruction differed on average by 54%. Treating each CP subunit as a rigid body, we then computationally refined the modeled positions of the A and B subunits (see Materials and Methods). After 40 cycles of refinement, the calculated amplitudes differed from the observed values by only 36%, a value comparable to that obtained when a known x-ray structure is modeled into its corresponding cryoEM density (26). As shown by shaded-surface representations of the modeled and reconstructed density maps (Fig. 6A), the model displays both the expected pentameric capsomeres and the dodecahedral arrangement of ridges connecting adjacent threefold axes. The fit of the modeled subunits to the reconstructed cryoEM electron density is further illustrated in the stereo image of Fig. 6B. This shows, e.g., how a shoulder between five- and threefold axes (Fig. 6A) is formed by the A/B dimer interaction (Fig. 6B). Small differences between the model and the reconstruction consist primarily of variations in the surface topography of the ridge connecting threefold axes. Each modeled ridge bears two humps derived from the two B subunits, whereas the corresponding ridge in the reconstruction bears a broader central hump (Fig. 6A). As the size of these differences is at the resolution limit of the reconstruction, they could reflect residual noise in the reconstruction or imperfections from modeling the CP monomers as rigid units.
Major Features of the 120-Subunit 65S VLP Capsid.
In the refined 120-subunit capsid model, the A/B dimer differs only slightly from the starting CCMV dimer (Fig. 6C). Compared with the CCMV dimer, the modeled A and B subunits are rotated approximately 20° relative to each other, moving the exterior loops of the A subunit (Fig. 6C, blue star) slightly closer to the reader and the exterior B subunit loops (Fig. 6C, red star) slightly away. Because the C-terminal extensions linking CP dimers show flexibility in virion swelling (7), such rotation should be possible while maintaining dimer integrity.
Despite only slight perturbation of the A/B interaction, the B subunit environment in the 120-subunit capsid is dramatically different from that in 180-subunit bromovirus capsids. In such T = 3 capsids, pentamers of A/B dimers are separated by C/C dimers. Alternating B and C subunits assume a radial, head-to-head pattern around threefold axes (Fig. 5A), forming the hexameric capsomeres (Fig. 4B). The absence of C subunits in 120-subunit capsids requires that pentamers of A/B dimers contact directly through A/B and B/B interactions. Specifically, three B subunits directly contact in a radial, tail-to-tail interaction about each threefold axis (Fig. 5B). B subunits also make additional antiparallel interactions at the twofold axes (Fig. 5B), resulting in the dodecahedral ridges that connect threefold axes (Fig. 6A).
These A/B and B/B interactions described above have no counterparts in the T = 3 bromovirus capsid. Such interactions are an inevitable consequence of the inability of 120 subunits to form a closed shell by using only quasi-equivalent interactions (1).
Discussion
Alternate BMV CP–CP Interactions.
The results presented here for BMV reveal a polymorphism in the assembly of a single CP, allowing in vivo assembly of 180 CPs in quasi-equivalent environments or 120 CPs in two nonequivalent environments. Both capsids selectively encapsidate viral RNA and contain 12 similar pentamers of CP dimers (Fig. 5). In the 180-subunit capsid, these pentamers are joined through additional CP dimers by using quasi-equivalent interactions throughout. In the 120-subunit capsid, the pentamers directly interact through distinct, nonequivalent CP–CP interactions around the two- and threefold axes. These results suggest that CPs of other viruses might also encode functional interactions not apparent from the mature virion. Such interactions might play important roles in assembly or other infection steps.
Strong structural similarities between the two capsid types and their common ability to selectively encapsidate viral RNA suggest that their assembly may share steps or intermediates. Assembly of 180-subunit bromovirus capsids has been suggested to initiate with a hexamer-of-dimers (7), but this structure is absent from the 120-subunit capsid. Instead, both capsids may initiate assembly with the shared pentamer-of-dimers (Fig. 5). Viral RNA might nucleate the formation of pentamers-of-dimers, which otherwise have only relatively weak hydrophobic interactions (7).
The ability of a single CP to form two distinct capsids using alternate interactions could also provide an intermediate for virus evolution. Thus, evolutionary barriers between capsids with significantly different CP–CP interactions may be lower than suspected.
RNA Control of Capsid Assembly.
The in vivo switch directing the type of BMV capsid formed was controlled by the RNA packaged: a natural BMV genomic RNA was packaged in 180-subunit capsids (Fig. 2), whereas an engineered mRNA containing only the BMV CP gene was packaged in 120-subunit capsids (Fig. 3). Thus, viral RNAs can not only initiate interaction with CP and contribute to virion stability but also guide the assembly pathway and determine the structure of the final product. This suggests that other RNAs might similarly regulate assembly of other viral and cellular ribonucleoprotein complexes.
Poliovirus, another positive-strand RNA virus with a 180-subunit capsid, encapsidates only RNAs newly synthesized by the viral RNA replication complex (9). In contrast, RNA replication was dispensable for selective encapsidation of BMV RNAs in vivo (Figs. 2 and 3). Thus, replication-independent mechanisms must exist in vivo to select BMV RNAs for encapsidation. This is consistent with selective binding of bromovirus CP to its cognate RNAs in vitro (5, 31). While present data do not preclude interactions with RNA replication contributing additional packaging selectivity, no preference for encapsidating replication-derived BMV RNA3 or RNA4 over DNA-transcribed BMV RNAs has been observed in yeast (M.K. and P.A., unpublished work).
Relation to Other Capsids.
Though predicted (1), T number polymorphism has been observed for only a few quasi-icosahedral viruses. Hepatitis B virus forms T = 3 and T = 4 capsids (32), and bacteriophage P22 forms T = 4 and T = 7 capsids (33). Auravirus had been suggested to form T = 3 and T = 4 capsids (34), but cryoEM reconstructions show all Auravirus capsids to have T = 4 symmetry (35). Under certain conditions in vitro, bromovirus CP can assemble T = 1 and larger T = 4 or T = 7 particles in vitro (16). None of these polymorphisms require the breakdown of quasi-equivalency, as must occur to form the BMV 120-subunit capsid.
The only other known capsids with 120 subunits are from double-stranded (ds) RNA viruses, such as yeast L-A virus capsid (36) and the cores of mammalian rotavirus (37) and bluetongue virus (38). These viruses use a notably different design than the BMV 65S VLP. The dsRNA virus capsids contain 60 CP dimers with paired A and B subunits aligned in a nearly parallel orientation. In contrast, in the BMV 120-subunit capsid, A and B subunits dimerize in an antiparallel configuration (Figs. 5B and 6C). Also, in dsRNA viruses, A and B subunits adopt significantly different conformations to accommodate their distinct environments (36, 38), whereas the A and B subunits of the BMV 65S VLP were modeled with nearly identical conformations (Figs. 5B and 6C). Though limited conformational differences, such as those that distinguish CCMV A, B, and C subunits, may occur in the A and B subunits of the 65S VLP, BMV CP appears to accommodate a range of self-interactions with less rearrangement than the dsRNA virus CPs.
Acknowledgments
We thank J. Speir for CCMV data; J.-Y. Sgro, B. Sheehan, and A. Kumar for assistance with computer files and images; and R. Ashmore for graphics programs. Analytical ultracentrifugation was performed at the Biophysical Instrumentation Facility, Biochemistry Department, University of Wisconsin-Madison with assistance from D. McCaslin. Scanning transmission electron microscope data were generously provided by M. Simon of Brookhaven National Laboratory. This research was supported by grants from the National Institutes of Health (to P.A., J.J., and T.S.B.) and a National Science Foundation Shared Instrumentation grant (to T.S.B.). P.A. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- CP
capsid protein
- BMV
brome mosaic virus
- CCMV
cowpea chlorotic mottle virus
- cryoEM
cryoelectron microscopy
- VLP
virus-like particle
- BMVP
plant-derived BMV
References
- 1.Caspar D L D, Klug A. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 2.Rossmann M G, Johnson J E. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 3.Larson S B, Day J, Greenwood A, McPherson A. J Mol Biol. 1998;277:37–59. doi: 10.1006/jmbi.1997.1570. [DOI] [PubMed] [Google Scholar]
- 4.Yan X, Olson N H, Etten J L V, Baker T S. In: Proceedings: Microscopy and Microanalysis. Bailey G W, Alexander K B, Jerome W G, Bond M G, McCarthy J J, editors. Atlanta: Springer; 1998. pp. 948–949. [Google Scholar]
- 5.Fox J M, Johnson J E, Young M J. Semin Virol. 1994;5:51–60. [Google Scholar]
- 6.Osman F, Grantham G L, Rao A L. Virology. 1997;238:452–459. doi: 10.1006/viro.1997.8849. [DOI] [PubMed] [Google Scholar]
- 7.Speir J A, Munshi S, Wang G, Baker T S, Johnson J E. Structure (London) 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan M, Ahlquist P. Semin Virol. 1997;8:221–230. [Google Scholar]
- 9.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price B D, Rueckert R R, Ahlquist P. Proc Natl Acad Sci USA. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda M, Ahlquist P. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 12.Janda M, French R, Ahlquist P. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 13.Johnston M, Davis R W. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romanos M A, Scorer C A, Clare J J. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 15.Geitz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 16.Lane L. In: Handbook of Plant Virus Infections and Comparative Diagnosis. Kurstak E, editor. Amsterdam: Elsevier/North–Holland; 1981. pp. 334–376. [Google Scholar]
- 17.Elder R T, Loh E Y, Davis R W. Proc Natl Acad Sci USA. 1983;80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa M, Janda M, Krol M A, Ahlquist P. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Restrepo-Hartwig M A, Ahlquist P. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson N H, Baker T S. Ultramicroscopy. 1989;30:281–297. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller S D, Butcher S J, Cheng R H, Baker T S. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 22.Baker T S, Cheng R H. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 23.Belnap D M, Olson N H, Baker T S. J Struct Biol. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]
- 24.Jones T A, Bergdoll M, Kjeldgaard M. In: Crystallographic and Modeling Methods in Molecular Design. Ealick S, Bugg C, editors. New York: Springer; 1990. pp. 189–199. [Google Scholar]
- 25.Brunger A T. x-plorVersion 3.1 Manual. New Haven, CT: Yale Univ.; 1993. [Google Scholar]
- 26.Cheng R, Reddy V, Olson N, Fisher A, Baker T, Johnson J. Structure (London) 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esnouf R M. J Mol Graphics. 1997;15:132. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 28.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 29.Thomas D, Schultz P, Steven A C, Wall J S. Biol Cell. 1994;80:181–192. doi: 10.1111/j.1768-322x.1994.tb00929.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhao X, Fox J M, Olson N H, Baker T S, Young M J. Virology. 1995;207:486–494. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- 31.Duggal R, Hall T C. J Virol. 1993;67:6406–6412. doi: 10.1128/jvi.67.11.6406-6412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 33.Thuman-Commike P A, Greene B, Malinski J A, King J, Chiu W. Biophys J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rumenapf T, Brown D T, Strauss E G, Konig M, Rameriz-Mitchel R, Strauss J H. J Virol. 1995;69:1741–1746. doi: 10.1128/jvi.69.3.1741-1746.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Olson N H, McKinney R J, Kuhn R J, Baker T S. In: Proceedings: Microscopy and Microanalysis. Bailey G W, Alexander K B, Jerome W G, Bond M G, McCarthy J J, editors. Atlanta: Springer; 1998. pp. 946–947. [Google Scholar]
- 36.Caston J R, Trus B L, Booy F P, Wickner R B, Wall J S, Steven A C. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawton J A, Zeng C Q, Mukherjee S K, Cohen J, Estes M K, Prasad B V. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimes J M, Burroughs J N, Gouet P, Diprose J M, Malby R, Zientara S, Mertens P P C, Stuart D I. Nature (London) 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]