Abstract
Sterol regulatory element binding proteins (SREBPs) enhance transcription of genes encoding enzymes of cholesterol and fatty acid biosynthesis and uptake. In the current experiments, we observed a decline in the mRNA encoding one SREBP isoform, SREBP-1c, in the livers of rats that were rendered diabetic by treatment with streptozotocin. There was no change in the mRNA encoding SREBP-1a, which is derived from the same gene as SREBP-1c but uses a different promoter. The ratio of SREBP-1c:1a transcripts fell 25-fold from 5:1 in control rats to 0.2:1 in the diabetic animals. The SREBP-1c mRNA rose nearly to normal, and the 1c:1a ratio increased 17-fold when the diabetic rats were treated for 6 h with insulin. These treatments produced no change in the mRNA for SREBP-2, which is encoded by a separate gene. The SREBP-1c mRNA also fell selectively in freshly isolated rat hepatocytes and rose when the cells were treated with insulin. Considered together with recent data on hepatocytes [Foretz, M., Pacot, C., Dugal, I., et al. (1999) Mol. Cell. Biol. 19, 3760–3768], the current in vivo studies suggest that insulin may stimulate lipid synthesis in the liver by selectively inducing transcription of the SREBP-1c gene.
Keywords: sterol regulatory element binding proteins, fatty acid metabolism, gene transcription
One classic action of insulin (INS) in the liver is the stimulation of fatty acid synthesis that is mediated in part by an increase in the transcription of the genes encoding acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS), two sequential enzymes in the fatty acid biosynthetic pathway (1). This stimulation is responsible for the increase in triglyceride levels in liver and plasma that accompany hyperinsulinemic states and contribute to the development of systemic INS resistance (2). Recent evidence suggests that the transcription factor mediating these INS effects is a member of the family of proteins designated sterol regulatory element binding proteins (SREBPs) (3, 4).
SREBPs were purified and cloned from human HeLa cells as a family of three proteins that bind to sterol regulatory elements and activate transcription of genes encoding the low density lipoprotein receptor and enzymes of cholesterol biosynthesis (5, 6). One of these SREBP isoforms, designated SREBP-1c or adipocyte determination and differentiation factor-1 (ADD-1), was also cloned independently as a protein that binds to a regulatory element (E-box) in the enhancer of the FAS gene (7). Overexpression of SREBPs was shown to enhance fatty acid synthesis as well as cholesterol synthesis in cultured Chinese hamster ovary (CHO) cells and 3T3-L1 adipocytes (8–10).
The three isoforms of SREBP, each comprising ≈1,150 amino acids, share a tripartite structure (11). The NH2-terminal segment of ≈480 amino acids is a transcription factor of the basic helix–loop–helix leucine zipper family. This is followed by a membrane attachment segment of ≈80 amino acids consisting of two transmembrane helices separated by a short 31-amino acid hydrophilic loop that protrudes into the lumen of the endoplasmic reticulum and nuclear envelope. This is followed by a COOH-terminal segment of ≈590 amino acids that forms a complex with a polytopic membrane protein designated SREBP cleavage-activating protein (SCAP). During their synthesis, the SREBPs are inserted into membranes of the endoplasmic reticulum and nuclear envelope, where they form complexes with SCAP (12).
The mechanism for the regulated proteolytic release of the transcriptionally active NH2-terminal domains of SREBPs was elucidated by studies in tissue culture cells (13). In sterol-depleted cells, the SREBP–SCAP complex is cleaved by Site-1 protease, an intraluminal serine protease that cleaves a bond in the luminal loop, thereby separating the two membrane-spanning helices. The NH2-terminal helix is then cleaved by Site-2 protease, a zinc metalloenzyme that releases the soluble NH2-terminal segment so that it can enter the nucleus and activate transcription. When sterols build up in cells, SCAP is inactivated, and the Site-1 protease can no longer cleave the SREBPs. As a result, the SREBPs remain membrane-bound and transcription of the target genes declines (13).
The three SREBPs are produced by two genes. SREBP-1a and -1c are produced from a single gene through the use of separate promoters that give rise to separate first exons that are spliced into a common second exon (11). The two proteins differ in the length of the acidic transcription-activating domain at the NH2 terminus. In SREBP-1a, this segment comprises 42 amino acids, of which 12 are acidic. SREBP-1a binds transcriptional coactivators such as CBP (14), which mediate transcriptional activation. In SREBP-1c, the acidic domain contains only 24 amino acids (6 acidic). Its ability to bind coactivators has not been studied. To compare the in vitro transcriptional activity of SREBP-1a and -1c, truncated dominant-positive versions of these proteins were transfected into M19 cells, a line of cultured CHO cells that lacks its own nuclear SREBPs as a result of a defect in the Site-2 protease (10). The SREBPs were truncated immediately prior to the first transmembrane helix so that they entered the nucleus directly and without a requirement for proteolysis. In these cells, SREBP-1a was 5–12 times more potent than SREBP-1c in activating transcription of target genes, including ACC, FAS, the low density lipoprotein receptor, and multiple enzymes of cholesterol biosynthesis.
SREBP-2 is produced by a gene that is separate from the one that produces the two SREBP-1 isoforms. The protein is ≈50% identical to the SREBP-1 isoforms, and it contains a long transcriptional acidic activation domain that resembles SREBP-1a. In M19 cells, truncated SREBP-2 activated the same genes as SREBP-1a, but the relative activities differed with respect to the two target pathways. SREBP-2 was more potent in stimulating genes of the cholesterol biosynthetic pathway, and SREBP-1a was relatively more potent in activating fatty acid synthesis (10).
Whereas SREBP-2 is expressed at approximately similar levels in all tissues studied, the relative expression of SREBP-1a and SREBP-1c differs. In most cultured cell lines, the SREBP-1a transcript is much more abundant than the SREBP-1c transcript as determined by a quantitative nuclease protection assay. In sharp contrast, in most tissues of adult animals, the SREBP-1c transcript predominates (15). This is particularly striking in liver where the SREBP 1c:1a ratio is ≈9:1. Similar results were found for mouse, hamster, and human livers (15, 16). The reason why the liver, an active lipogenic organ, should produce the less active version of SREBP-1 is not clear.
A role for SREBPs in regulating fatty acid synthesis in liver was first suggested by the finding of massive overproduction of fatty acids as well as cholesterol in livers of transgenic mice that expressed the truncated dominant-positive form of SREBP-1a in liver (17). Lipid overproduction was caused by marked increases in the mRNAs encoding ACC and FAS as well as a panel of cholesterologenic enzymes that included 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) synthase, HMG-CoA reductase, squalene synthase, and others. Lipid overproduction was facilitated by increases in the mRNAs encoding enzymes that supply the lipogenic substrates acetyl-CoA (ATP citrate lyase) and NADPH (malic enzyme, glucose 6-phosphate dehydrogenase, and 6-phosphogluconate dehydrogenase) (18). Stearoyl-CoA desaturase mRNA was also increased, and as a result the fatty acids that accumulated were mostly monounsaturated (18). These studies show that SREBP-1a is capable of activating the entire program of synthesis of unsaturated fatty acids, as well as cholesterol, in the liver. Similar results were observed in animals expressing truncated SREBP-2, although the ratio of cholesterol to fatty acid synthesis was higher in these mice than in the ones expressing SREBP-1a (19).
As predicted from the studies in cultured cells, truncated SREBP-1c was much less effective than SREBP-1a in increasing the mRNAs for all of the lipogenic enzymes in transgenic mice (20). Under conditions in which expression of the SREBP-1c and -1a transgenes was approximately matched, the mRNAs encoding ACC and FAS were increased by only 2- and 4-fold, respectively, in 1c transgenics, as compared with 9- and 16-fold in 1a transgenics, and the mRNAs for the cholesterologenic enzymes were not increased at all in the 1c transgenics.
The SREBP-1c animals did show one important regulatory abnormality, and this involved the response to fasting and refeeding (3). In normal mice, the levels of mRNAs encoding the enzymes of fatty acid synthesis declined markedly after fasting, and they increased on refeeding a high carbohydrate low-fat diet. Fasting also led to a decline in the level of nuclear SREBP-1 (presumably SREBP-1c), as determined by immunoblotting, and refeeding elevated the protein to supranormal levels. In the transgenic mice expressing truncated SREBP-1c, the mRNAs encoding ACC and FAS did not decline with fasting, and they failed to rise after refeeding. These studies raised the possibility that the decline in nuclear SREBPs is normally responsible for the fasting-induced decline in lipogenic mRNAs, and forced overexpression of nuclear SREBP-1c obviates this effect (3). Inasmuch as the decline in lipogenic mRNAs is usually attributed to the decline in INS that follows fasting, the data raised the possibility that INS withdrawal decreases nuclear SREBP-1. This in turn implies that INS may enhance fatty acid synthesis in liver by increasing nuclear SREBP-1.
A clue to the mechanism for the INS effect on SREBP-1 was elucidated in an important paper by Foretz et al. (4). These workers studied freshly isolated rat hepatocytes that were placed into culture with or without INS. By using a probe that did not distinguish between the 1a and 1c isoforms of SREBP-1, they showed that INS deprivation led to a fall in the level of mRNA for SREBP-1, and this was restored by INS administration in vitro, an effect that was enhanced at high glucose levels. Moreover, they showed that the INS-induced increase in SREBP-1 mRNA was blocked when the hepatocytes were treated with glucagon or dibutyryl cyclic AMP, which antagonizes INS action. The changes in SREBP-1 mRNA were attributed to altered transcription from one or both of the SREBP-1 promoters, as determined by nuclear run-on assays. The functional significance of these changes was established by the finding that the expression of the target genes FAS and ACC, as well as other INS-responsive genes such as S14 and L-pyruvate kinase, was reduced when the hepatocytes were infected in vitro with an adenovirus encoding a dominant-negative version of SREBP-1c, which can form heterodimers with endogenous SREBPs but cannot bind to DNA (4). These studies suggested that INS may alter transcription of lipogenic genes in liver, at least in part, by enhancing transcription of the gene encoding SREBP-1. Whether INS affects the SREBP-1a or -1c isoform, or both, and whether INS affects SREBP-2 were not addressed.
In the current studies, we have analyzed the effect of INS on the levels of the three SREBP mRNAs in livers of living rats. For this purpose, we treated rats with streptozotocin (STZ), which destroys the β-cells of the pancreas and leads to an acute INS deficiency (21). We show that this treatment leads to a profound fall in SREBP-1c mRNA, and this is restored by administration of INS. The changes in the SREBP-1c transcript were highly specific in that no changes in levels of the mRNAs encoding SREBP-1a or SREBP-2 were detected. These data support an in vivo role for INS in specifically enhancing transcription driven by the SREBP-1c promoter. The subsequent increase in levels of the SREBP-1c protein in turn plays a role in mediating the increase in lipogenic mRNAs induced by INS.
Materials and Methods
Reagents and General Methods.
We obtained restriction and modifying enzymes from New England Biolabs, Pfu DNA polymerase from Stratagene, and [α-32P]dCTP and [α-32P]CTP from Amersham. The content of cholesterol and triglyceride in the liver was measured as described (22). Plasma glucose was measured by using a Glucose (Trinder) 100 Kit (Sigma).
Animals and Treatments.
Male Sprague–Dawley rats (200–220 g; obtained from Harlan–Sprague–Dawley) were housed in colony cages, maintained on a 12-h light/12-h dark cycle, and fed Teklad 4% Mouse/Rat Diet #7001 from Harlan Teklad Premier Laboratory Diets (Madison, WI). Rats were treated with STZ (Sigma) and INS according to a protocol described by Lakshmanan et al. (21) with minor modifications, as follows. After fasting for 24 h, diabetes was induced by a single intravenous injection of 0.2–0.3 ml of 50 mM sodium citrate solution (pH 4.5) containing STZ (65 mg/kg body weight). Control rats were injected with 50 mM sodium citrate solution (pH 4.5). After injection, animals were fasted for a further 24 h, after which plasma glucose levels were checked and diabetes was confirmed (glucose level >250 mg/dl). The animals were then fed a chow diet for 12 h, after which INS was administered to the STZ+INS group. The animals received a combination of human regular INS (3 units; Eli Lilly) intraperitoneally and human NPH INS (4 units; Eli Lilly) subcutaneously, each given in 0.2 ml of Dulbecco’s PBS (catalogue no. 14190–144; GIBCO/BRL). The rats in the control and STZ groups received 0.2 ml of PBS injected both intraperitoneally and subcutaneously. After injection of INS or PBS, the animals were fasted for 6 h and then killed by halothane anesthesia.
Rat Hepatocytes.
Hepatocytes were isolated from nonfasting 250-g male Sprague–Dawley rats by the collagenase method (23). Hepatocytes were prepared as described (4, 24) with minor modifications. Animals were anesthetized with halothane, and each liver was perfused in situ via the portal vein with 100 ml of Liver Perfusion Medium (catalogue no. 17701–038; GIBCO/BRL). The medium was warmed to 37°C and infused at a rate of ≈30 ml/min. The liver was then perfused with Liver Digest Medium containing collagenase (catalogue no. 17703–034; GIBCO/BRL) for ≈5 min at a flow rate of ≈30 ml/min. The liver was removed, the hepatic capsule was stripped, and the dissociated cells were dispersed by shaking, followed by filtration at 4°C through gauze into an equal volume of ice-cold DMEM (catalogue no. 11885–084; GIBCO/BRL) containing 5% (vol/vol) fetal calf serum, 10 mM Hepes (pH 7.4), 100 units/ml of penicillin, and 100 μg/ml of streptomycin. The cells were pelleted and washed twice at 4°C with the same buffer. Aliquots of 8 × 106 cells were plated onto 100-mm rat collagen I-coated dishes (catalogue no. 354450; Becton Dickinson Labware) in Hepatocyte Attachment Medium (catalogue no. 17706–029; GIBCO/BRL) supplemented with 5% (vol/vol) FCS, 100 nM triiodothyronine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mg/ml gentamycin. After incubation at 37°C in 9% CO2 for 4 h, the cells were washed twice with PBS and incubated with Medium 199 containing Earle’s salts (catalogue no. 11150–059; GIBCO/BRL) supplemented with 100 nM dexamethasone, 1 nM INS, 100 nM triiodothyronine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cell viability, as measured by trypan blue exclusion, was always greater than 85%. After incubation for 16 h, the cells were switched to a similar medium supplemented with INS and glucose as indicated in the legends.
Immunoblotting.
Rabbit polyclonal antibodies against rat SREBP-1 and -2 were prepared as described for antibodies directed against mouse SREBP-1 (17). For this purpose, rabbits were immunized with a His(6)-tagged fusion protein containing fragments of rat SREBP-1 (amino acids 37–187) or SREBP-2 (amino acids 32–250) that were produced in bacteria by using the Express System (Qiagen; Chatsworth, CA). For immunoblotting experiments, nuclear extracts and membrane fractions (105 g pellet) were prepared from rat livers as described for hamster livers (25). Aliquots (30 μg of protein) of the membrane and nuclear extract fractions were subjected to SDS/PAGE (8% gel), transferred to Hybond C extra membranes (Amersham), incubated with 5 μg/ml rabbit anti-rat SREBP-1 IgG or anti-rat SREBP-2 IgG. The protein content of samples was determined with the BCA Kit (Pierce). Immunoblot analysis was performed with the Enhanced Chemiluminescence (ECL) Western Blotting Detection System Kit (Amersham) with a horseradish peroxidase-conjugated donkey anti-rabbit IgG antibody (Amersham).
Blot Hybridization of RNA.
A plasmid-encoding rat phosphoenolpyruvate carboxykinase (PEPCK) was kindly provided by Daryl Granner (26). All other cDNA probes have been described (17). cDNA probes were labeled with [α-32P]dCTP (3,000 Ci/mmol) with the Megaprime DNA Labeling System (Amersham). Total RNA was prepared by using an RNA STAT-60 Kit (Tel-Test, Friendswood, TX). For Northern gel analysis, aliquots of total RNA from four rats were pooled (total 10 μg), denatured with formaldehyde and formamide, subjected to electrophoresis in a 1% formaldehyde agarose gel, and transferred to Hybond N+ membranes (Amersham). The filters were hybridized with the indicated 32P-labeled probe (106 cpm/ml) for 2 h at 65°C (17). The resulting bands were quantified by exposure of the filter to a BAS 1000 Fuji PhosphorImager (Fuji), and the values were normalized to the signal generated with a probe for cyclophilin mRNA.
RNase Protection Assay.
A cDNA fragment for rat SREBP-1a was amplified by reverse transcriptase-PCR from first-strand cDNA by using rat liver poly(A)+ RNA as a template and degenerate primers derived from conserved hamster and mouse SREBP-1a sequences, as follows: primer, 5′-ATGGACGAGCTG(G/C)CCTTCGGTGAGGCGGCT-3′; and 3′ primer, 5′-CCAGAGAGGAACCCAGGGAAGCAG-3′ (15, 16). The ends of the 5′ and 3′ primers contained HindIII and EcoRI sites, respectively. The amplified fragment corresponds to exon 1a (specific for SREBP-1a) and part of exon 2 (common to SREBP-1a and SREBP-1c). Amplified cDNA fragments were subcloned into the pGEM-3Zf(+) vector (Promega). After linearization of plasmid DNA with HindIII, antisense RNA was transcribed with [α-32P]CTP (20 mCi/ml) by using bacteriophage T7 RNA polymerase (Ambion, Austin, TX) (15).
Aliquots of total RNA (10 μg) from each sample were incubated with the above SREBP-1 cRNA probe plus a cRNA probe for the mRNA of β-actin and the reagents contained in the HybSpeed RPA Kit (Ambion). In preparing the probes, we adjusted the specific activity of the [α-32P]CTP to give comparable signals for β-actin and total SREBP-1. The specific activities ranged between 1.7–2.6 × 109 cpm/μg for SREBP-1 and 5.3–8.1 × 108 cpm/μg for β-actin. After digestion with RNase A/T1, protected fragments were separated on 8 M urea/4.8% polyacrylamide gels, and the gels were dried and subjected to autoradiography by using reflection film and intensifying screens (DuPont). The protected fragments corresponding to SREBP-1a (257 bp) and SREBP-1c (160 bp) were visualized on gels analyzed quantitatively with a Fuji PhosphorImager as described above. The level of β-actin mRNA in each sample was used to normalize signals obtained for the SREBP mRNAs. For comparison of rat SREBP-1a and -1c mRNA levels, the results were corrected for the difference in number of 32P-labeled CTP atoms in each protected fragment (72 and 39, respectively).
Results
To study the effects of INS on hepatic SREBPs, we modified a published protocol in which rats are starved for 24 h, treated with STZ or a control solution, and then starved for an additional 24 h to enhance the toxic effect of STZ on pancreatic beta cells (21). The animals were then allowed access to food for 12 h after which they were treated either with INS or a control saline solution. To eliminate the effects of INS on food intake, we withdrew food from all of the rats and killed the animals 6 h later. Table 1 shows that STZ treatment caused a significant decrease in body and liver weight and a marked increase in plasma glucose. The content of cholesterol and triglyceride in the liver increased. The animals treated with STZ plus INS showed a slightly higher body weight, a higher liver weight, and a reduced content of plasma glucose, hepatic cholesterol, and hepatic triglycerides when compared with the animals treated with STZ alone.
Table 1.
Metabolic parameters in control rats, streptozotocin-treated rats (STZ), and streptozotocin-treated diabetic rats supplemented with insulin (STZ+INS)
| Parameter | Treatment
|
||
|---|---|---|---|
| Control | STZ | STZ+INS | |
| Body weight, g | 268 ± 7.6 | 210 ± 3.4* | 224 ± 12.1† |
| Liver weight, g | 9.3 ± 0.09 | 5.8 ± 0.37* | 8.1 ± 0.31† |
| Liver weight/body weight, % | 3.5 ± 0.14 | 2.8 ± 0.17* | 3.6 ± 0.24† |
| Plasma glucose, mg/dl | 168 ± 13 | 425 ± 51* | 218 ± 34† |
| Liver cholesterol content, mg/g | 2.2 ± 0.08 | 2.7 ± 0.13* | 1.8 ± 0.07† |
| Liver triglyceride content, mg/g | 2.7 ± 0.55 | 4.3 ± 0.30‡ | 2.1 ± 0.52† |
Each value represents the mean ± SEM of four male rats.
*P < 0.01 between control and STZ groups.
†P < 0.01 between STZ and STZ+INS groups.
‡P < 0.05 between control and STZ groups.
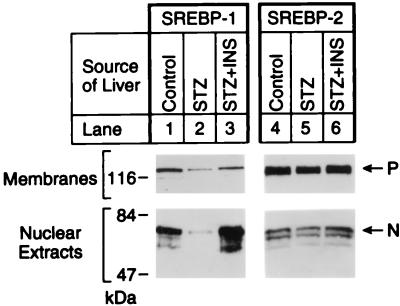
Nuclear extracts from the livers of control rats showed abundant amounts of the nuclear form of SREBP-1 and -2 as determined by SDS/PAGE and immunoblotting (Fig. 1). The antibody used for these studies does not distinguish between SREBP-1a and -1c, but previous studies have shown that the vast majority of SREBP-1 in livers of mice, hamsters, and humans is the 1c isoform (15, 16). STZ caused a marked decrease in nuclear SREBP-1, but no change in SREBP-2. INS reversed the effect of STZ on SREBP-1, but had no effect on SREBP-2. The membrane-bound precursor of SREBP-1 decreased slightly after STZ treatment and increased slightly after INS. Again, we observed no effect on the precursor for SREBP-2.
Figure 1.
Immunoblot analysis of SREBP-1 and -2 in membranes and nuclear extracts from livers of control rats and rats treated with STZ without or with INS. Livers from the three groups of rats in Table 1 (four male rats per group) were pooled, and aliquots of the membrane pellet (30 μg of protein) and nuclear extract (30 μg) were subjected to SDS/PAGE (8% gel). Immunoblot analysis was performed with 5 μg/ml rabbit anti-rat SREBP-1 IgG (lanes 1, 2, and 3) or anti-rat SREBP-2 IgG (lanes 4, 5, and 6) as the primary antibody and 0.25 μg/ml horseradish peroxidase-coupled donkey anti-rabbit IgG as the secondary antibody. Filters were exposed to Reflection NEF496 film for 15 s (lanes 1, 2, and 3) or 30 s (lanes 4, 5, and 6) at room temperature. P and N denote the precursor and cleaved nuclear forms of SREBP, respectively.
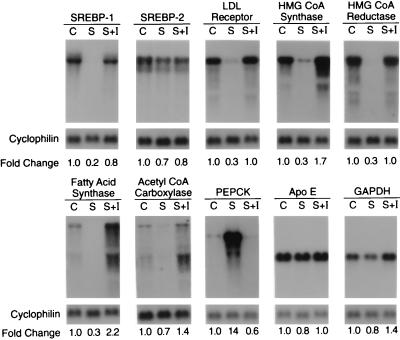
In the STZ-treated rats, the decrease in nuclear SREBP-1 without an accompanying increase in the precursor form raised the possibility that the rate of synthesis of the SREBP-1 precursor was reduced, most likely owing to a decrease in SREBP-1 mRNA. To check this possibility, we measured the amounts of mRNA for SREBP-1 and -2 by Northern blotting of RNA isolated from liver extracts (Fig. 2). The results revealed an 80% reduction in SREBP-1 mRNA in the STZ-treated rats that was restored nearly completely by INS treatment. There was no significant change in SREBP-2 mRNA. STZ significantly reduced the amounts of mRNAs derived from SREBP target genes, including the low density lipoprotein receptor, HMG-CoA synthase, HMG-CoA reductase, and fatty acid synthase. There was a smaller reduction in the mRNA for ACC. The amount of each of these mRNAs was increased to control values or above when the STZ-treated animals received INS. In contrast, the mRNA for phosphoenolpyruvate carboxykinase, a gluconeogenic enzyme, was increased by STZ treatment, and this was reversed by INS. Two other mRNAs, encoding apolipoprotein E and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were not significantly changed by STZ or INS.
Figure 2.
Amounts of various mRNAs in livers of control rats and rats treated with STZ without or with INS as measured by blot hybridization. The rats used in this experiment are described in Table 1. For each experimental group, total RNA isolated from the livers of four male rats were pooled, and 10 μg aliquots were subjected to electrophoresis and blot hybridization with the indicated 32P-labeled cDNA probe. The amount of radioactivity in each band was quantified as described in Materials and Methods. The -fold change in each mRNA of the STZ-treated diabetic group (S) and STZ-treated diabetic group supplemented with INS (S+I) relative to that of the control group (C) was calculated after correction for loading differences by measurement of cyclophilin mRNA. These values are shown below each blot.
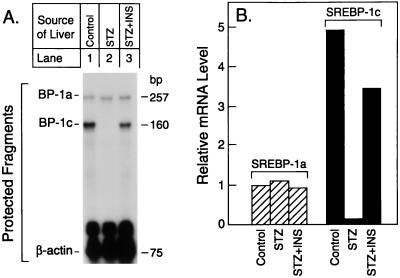
To determine whether the changes in total SREBP-1 mRNA reflected changes in the amount of SREBP-1a or SREBP-1c, we used a nuclease protection assay that distinguishes between these two transcripts (Fig. 3A). In livers of control animals, the amount of nuclease-protected fragment corresponding to SREBP-1c was 5-fold higher than the amount corresponding to SREBP-1a as judged by densitometric scans (Fig. 3B). STZ treatment caused a near-total decline in the amount of SREBP-1c mRNA without affecting the amount of SREBP-1a mRNA. This change was reversed by INS.
Figure 3.
(A) Relative amounts of mRNAs for SREBP-1a and -1c in the livers of rats treated with STZ without or with INS as measured by RNase protection. Total RNA was isolated from the livers of the three groups of rats described in Table 1. Aliquots of pooled RNA (10 μg) were hybridized in solution for 10 min at 68°C to a mixture of 32P-labeled cRNA probes for SREBP-1 and β-actin as described in Materials and Methods. After RNase digestion, the protected fragments were separated by gel electrophoresis and exposed to film for 16 h at −80°C. (B) Quantification of hepatic mRNAs for SREBP-1a and -1c. The data in (A) were quantified as described in Materials and Methods and normalized relative to the β-actin signal. The data for SREBP-1c mRNA are plotted as the -fold change relative to the SREBP-1a mRNA level in the control group.
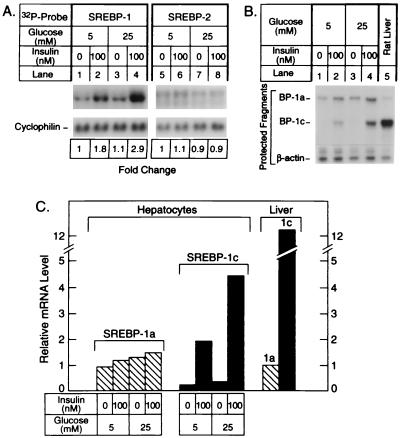
To determine whether INS has a direct effect on SREBP expression in hepatocytes, we isolated hepatocytes from livers of control rats and incubated them for 16 h in medium containing INS, dexamethasone, and triiodothyronine, a mixture that preserves the differentiated state of freshly isolated hepatocytes (4). We then incubated the cells for a further 6 h in the same medium with or without INS, and with either low glucose (5 mM) or high glucose (25 mM). The hepatocytes were then harvested, and we measured the amounts of total SREBP-1 and -2 mRNA (Fig. 4A) and the amounts of the specific SREBP-1a and -1c transcripts (Figs. 4 B and C). INS produced an increase in the total amount of SREBP-1 mRNA, and the effect was greatest (2.9-fold) at the high glucose concentration. There was no significant change in the amount of SREBP-2 mRNA. In livers of ad libitum-fed Sprague–Dawley rats, the ratio of SREBP-1c:1a transcripts was 12:1 (Fig. 4C). This ratio fell by 60-fold to 0.2:1 when hepatocytes were incubated in the absence of INS. When the hepatocytes were treated with INS plus high glucose, the 1c:1a ratio rose by 15-fold to 3:1 as a result of a marked increase in the SREBP-1c transcript and only a slight increase in the SREBP-1a transcript.
Figure 4.
(A) Amounts of mRNAs for total SREBP-1 (lanes 1–4) and SREBP-2 (lanes 5–8) in rat hepatocytes incubated with low or high glucose with or without INS. Hepatocytes were isolated from rat livers and incubated for 16 h with Medium 199 with Earle’s salts (5 mM glucose) containing 100 nM dexamethasone, 1 nM INS, 100 nM triiodothyronine, and antibiotics as described in Materials and Methods. The cell monolayers were then incubated for 6 h in the same medium containing the indicated concentration of INS and glucose. The cells were homogenized, and aliquots of total RNA (10 μg) were subjected to blot hybridization as described in the legend to Fig. 2. (B) Relative amounts of mRNAs for SREBP-1a and -1c as measured by RNase protection. Aliquots of total RNA were isolated from the hepatocytes and subjected to the RNase protection assay as described in the legend to Fig. 3 (lanes 1–4). As a control, a 10-μg aliquot of total RNA pooled from the livers of four ad libitum-fed male Sprague–Dawley rats was used (lane 5). (C) Quantification of mRNAs for SREBP-1a and -1c. The data from B were quantified as described in Materials and Methods and normalized relative to the β-actin signal. The data for SREBP-1a and -1c mRNA are plotted as the -fold change relative to the SREBP-1a mRNA level in rat liver.
Discussion
The current results in the livers of intact rats with STZ-induced diabetes, together with those of Foretz et al. (4) in isolated rat hepatocytes, indicate that INS selectively induces SREBP-1c gene transcription in liver. The increase in SREBP-1c mRNA leads to an increase in the nuclear form of SREBP-1c, and this in turn leads to an increase in the transcription of SREBP-1c target genes, including ACC and FAS. These findings appear to explain, at least in part, the long-appreciated ability of INS to stimulate the synthesis of fatty acids in liver. All of the studies reported in this paper were performed at least twice with similar results.
Most, if not all, of the known actions of INS can be attributed to the receptor-mediated phosphorylation of IRS-1 and IRS-2, which in turn triggers kinase cascades involving the phosphatidylinositol 3-kinase/Akt pathway and the MAP kinase pathway (27, 28). Which, if any, of these pathways mediates the increase in transcription of SREBP-1c is unknown. Whatever the pathway, the action is highly selective. INS does not activate the other promoter of the SREBP-1 gene, namely, the one that gives rise to the SREBP-1a transcript, and it also fails to activate the closely related SREBP-2 gene.
According to current information, a simple increase in SREBP-1c mRNA would not be sufficient to lead to an increase in transcription of the target genes. The SREBP-1c mRNA encodes a membrane-bound precursor that must be cleaved proteolytically in order for the active NH2-terminal segment to enter the nucleus. Thus far, there is no evidence that the liver produces a truncated form of SREBP-1c that bypasses the need for such proteolysis. Indeed, the size of the INS-induced SREBP-1c mRNA transcript is consistent with the coding of a full-length SREBP-1c protein; therefore, it appears that INS increases the efficiency of proteolytic processing of SREBP-1c as well as increasing its mRNA. Remarkably, however, the amount of nuclear SREBP-2 does not rise after INS treatment (Fig. 1). This finding is consistent with previous evidence that the proteolytic cleavage of SREBP-1c and -2 may be regulated independently in the liver. When the livers of hamsters and mice were depleted of cholesterol by treatment with a bile acid-binding resin and an HMG-CoA reductase inhibitor, the amount of nuclear SREBP-2 increased, and there was a paradoxical decrease in nuclear SREBP-1 (now known to be SREBP-1c) (16, 25). These in vivo results contrast with the findings in cultured cells in which cholesterol depletion leads to a parallel increase in the nuclear content of SREBP-2 and SREBP-1 (largely SREBP-1a) (11). Whether the liver has a separate protease that is specific for SREBP-1c, or whether a common Site-1 protease is controlled independently with respect to its SREBP substrates, remains to be determined.
Another unresolved question relates to the level of activity of the nuclear SREBP-1c protein in liver. SREBP-1c has a very short acidic activation domain, and it is a very weak activator of transcription when overexpressed in nonhepatic cells such as CHO cells and human kidney 293 cells (10, 20). It is possible that INS or high glucose levels lead to an alteration of SREBP-1c that increases its transcriptional activity. Such an alteration could involve phosphorylation of SREBP-1c, heterodimerization with another basic helix–loop–helix leucine zipper transcription factor, or formation of a complex with another INS-regulated transcription factor.
Acknowledgments
We thank Richard Gibson for invaluable help with animal care; Scott Clark for excellent technical assistance; and our colleague Dr. Morihiro Matsuda for help with primary hepatocytes. This work was supported by National Institutes of Health Grant HL20948, and grants from the Moss Heart Foundation and Perot Family Foundation. S.I. is the recipient of a Research Fellowship from the National Institutes of Health and Nutrition in Japan.
Abbreviations
- ACC
acetyl-CoA carboxylase
- FAS
fatty acid synthase
- SREBP
sterol regulatory element binding protein
- STZ
streptozotocin
- INS
insulin
- HMG
3-hydroxy-3-methylglutaryl
References
- 1.Hillgartner F B, Salati L M, Goodridge A G. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 2.McGarry J D. Science. 1992;258:766–770. doi: 10.1126/science.1439783. [DOI] [PubMed] [Google Scholar]
- 3.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Lièpvre X L, Berthelier-Lubrano C, Spiegelman B, Kim J B, Ferré P, et al. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Briggs M R, Hua X, Yokoyama C, Goldstein J L, Brown M S. J Biol Chem. 1993;268:14497–14504. [PubMed] [Google Scholar]
- 6.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 7.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J B, Spiegelman B M. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 9.Bennett M K, Lopez J M, Sanchez H B, Osborne T F. J Biol Chem. 1995;270:25578–25583. doi: 10.1074/jbc.270.43.25578. [DOI] [PubMed] [Google Scholar]
- 10.Pai J, Guryev O, Brown M S, Goldstein J L. J Biol Chem. 1998;273:26138–26148. doi: 10.1074/jbc.273.40.26138. [DOI] [PubMed] [Google Scholar]
- 11.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 12.Sakai J, Nohturfft A, Goldstein J L, Brown M S. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 13.Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naar A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tjian R. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M S. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimomura I, Bashmakov Y, Shimano H, Horton J D, Goldstein J L, Brown M S. Proc Natl Acad Sci USA. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimomura I, Shimano H, Korn B S, Bashmakov Y, Horton J D. J Biol Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 19.Horton J D, Shimomura I, Brown M S, Hammer R E, Goldstein J L, Shimano H. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakshmanan M R, Nepokroeff C M, Porter J W. Proc Natl Acad Sci USA. 1972;69:3516–3519. doi: 10.1073/pnas.69.12.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishibashi S, Brown M S, Goldstein J L, Gerard R D, Hammer R E, Herz J. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry M N, Friend D S. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton J D, Shimano H, Hamilton R L, Brown M S, Goldstein J L. J Clin Invest. 1999;103:1067–1076. doi: 10.1172/JCI6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng Z, Otani H, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beale E G, Chrapkiewicz N B, Scoble H A, Metz R J, Quick D P, Noble R L, Donelson J E, Biemann K, Granner D K. J Biol Chem. 1985;260:10748–10760. [PubMed] [Google Scholar]
- 27.Taha C, Klip A. J Membr Biol. 1999;169:1–12. doi: 10.1007/pl00005896. [DOI] [PubMed] [Google Scholar]
- 28.Virkamäki A, Ueki K, Kahn C R. J Clin Invest. 1999;103:931–943. doi: 10.1172/JCI6609. [DOI] [PMC free article] [PubMed] [Google Scholar]