Abstract
The basal transcription machinery of Archaea corresponds to the minimal subset of factors required for RNA polymerase II transcription in eukaryotes. Using just two factors, Archaea recruit the RNA polymerase to promoters and define the direction of transcription. Notably, the principal determinant for the orientation of transcription is not the recognition of the TATA box by the TATA-box-binding protein. Instead, transcriptional polarity is governed by the interaction of the archaeal TFIIB homologue with a conserved motif immediately upstream of the TATA box. This interaction yields an archaeal preinitiation complex with the same orientation as the analogous eukaryal complex.
The basal transcription machinery of Archaea is strikingly similar to the core components of the eukaryal RNA polymerase (RNAP) II apparatus (1, 2). Archaeal promoters contain a TATA box found 25 bp upstream of the transcription start site that forms a ternary complex with the archaeal TATA-box-binding protein (aTBP) and the archaeal homologue of transcription factor TFIIB, TFB (3, 4). It is possible to reconstitute transcription from a range of archaeal promoters by using recombinant aTBP, TFB, and highly purified RNAP; no additional transcription factors are required even on highly positively supercoiled templates (5–7). The crystal structure of the ternary complex of aTBP and the C-terminal core domain of TFB (TFBc) on DNA is highly similar to the corresponding structure in eukaryotes. However, the orientation of aTBP and TFBc with respect to the transcription start site was inverted in the archaeal complex compared with the homologous eukaryal TBP/TFIIB/TATA-box structure (8, 9).
The formation of the aTBP/TFB/TATA-box ternary complex is required for recruitment of the RNAP to the transcription initiation site, downstream of the TATA box. Clearly, it is of key importance that transcription occurs in a unidirectional manner at most promoters and, thus, a mechanism must exist to ensure that RNAP is recruited in a directional manner. The simplest model to explain the directional recruitment of RNAP would be to define the polarity of the preinitiation complex at the first step; the binding of TBP to the TATA box. However, despite extensive analysis in both Eucarya and Archaea, whether and how TBP binds the TATA element in a preferred orientation and how transcriptional polarity is established are still poorly understood. Recent studies suggest that the intrinsic deformability of the TATA box may be important, indicating that the C-terminal repeat of eukaryal TBP (eTBP) preferentially contacts the more flexible end of the TATA box (10). However, the molecular basis of this preference is unclear. The highly asymmetric charge potential distribution in eTBP may also play a role (11). An alternative model suggests that the asymmetric positioning of a proline in the C-terminal repeat may contribute to orientational specificity (12). However, experiments using eTBP derivatized with a DNA scission reagent have shown that eTBP has only a modest (60:40) preference for binding the TATA box of the adenovirus major late promoter and yeast CYC1 promoters in a particular orientation (13). In eukaryal nuclei, eTBP is complexed with associated factors, TAFs. As a number of TAFs in the TFIID complex have been demonstrated to possess DNA-binding activities (e.g., see refs. 14 and 15), it is likely that TAF–DNA interactions may influence the orientation of TFIID on some promoters.
Although the above models may account for orientational specificity in TBP–TATA-box recognition in eukarya, they are unlikely to do so in archaea. aTBP is a considerably more symmetric molecule than its eukaryal homologues: the two repeats in eTBP have 28–30% identity, whereas in aTBP they are ≈40% identical. Crucially, the charge potential distribution in aTBP is highly symmetric (16) and, furthermore, the proline clash model (12) cannot apply to aTBP because the analogous positions in both repeats are occupied by proline. Finally, both biochemical and genome sequence analyses suggest that Archaea do not possess TAFs (6, 17, 18), implying that TAFs arose after the separation of archaeal and eukaryal lineages. It is likely, therefore, that the evolutionary progenitor of the eukaryal and archaeal transcription machineries relied on factor(s) other than TBP to define transcriptional polarity.
It is therefore possible that TFB and/or RNAP plays important roles in determining the orientation of transcription in Archaea. In eukarya, TFIIB can influence the orientation of binding of eTBP to the TATA box (13), and this property of TFIIB might be attributable to the recently described interaction between TFIIB and the BRE sequence element found upstream of the TATA box in some strong promoters (19). An analogous motif has been identified in the strong archaeal T6 promoter of the Sulfolobus shibatae virus SSV1 (20). In the current work, we demonstrate that the archaeal BRE is found in a range of promoters of various strengths and that the TFB–BRE interaction is the principal element which defines transcription polarity. We also demonstrate that the absolute polarity of the archaeal ternary complex is the same as the homologous eukaryal TBP–TFIIB–DNA complex.
Materials and Methods
Plasmid Construction.
Plasmid pINIT was generated by annealing the self-complementary oligonucleotide INIT and ligating into BamHI-digested pBluescript. Insertion of the INIT oligonucleotides removed the BamHI sites flanking the insert. Subsequent TATA-box-containing oligonucleotides were inserted into the internal BamHI site in pINIT.
Oligonucleotides.
Name is followed by sequence; X = 5-bromodeoxyuridine.
INIT, GATCTTGAACCCTCTATCGGATCCGATAGAGGGTTCAA;
EF1-T, GATCCATGTGCGAAAGCTTTAAAAAGTAAGTTCAAAAGT;
EF1-L, GATCACTTTTGAACTTACTTTTTAAAGCTTTCGCACATAG;
TATAINV-T, GATCCTATGTGCGAAAGCTTTTTAAAGTAAGTTCAAAAGT;
TATAINV-L, GATCACTTTTGAACTTACTTTAAAAAGCTTTCGCACATAG;
FLIP2.3-T, GATCCTATGTGCGAATACTTTAAAAAGCTAGTTCAAAAGT;
FLIP2.3-L, GATCACTTTTGAACTAGCTTTTTAAAGTATTCGCACATAG;
FLIP2.6-T, GATCCTATGTGCACTTACTTTAAAAAGCTTTCTCAAAAGT;
FLIP2.6-L, GATCACTTTTGAGAAAGCTTTTTAAAGTAAGTGCACATAG;
FLIP2.10-T, GATCCTATTTGAACTTACTTTAAAAAGCTTTCGCACAAGT;
FLIP2.10-L, GATCACTTGTGCGAAAGCTTTTTAAAGTAAGTTCAAATAG;
UPINV-T, GATCCTATGTGCGAAAGCTTTAAAAAGCTTTCTCAAAAGT;
UPINV-L, GATCACTTTTGAGAAAGCTTTTTAAAGCTTTCGCACATAG;
DOINV-T, ATCCTATGTGCACTTACTTTAAAAAGTAAGTTCAAAAGT;
DOINV-L, GATCACTTTTGAACTTACTTTTTAAAGTAAGTGCACATAG;
T6WT-T, GATCTAGATAGAGTAAAGTTTAAATACTTATATAGATAGA;
T6WT-L, GATCTCTATCTATATAAGTATTTAAACTTTACTCTATCTA;
T66FL-T, GATCTAGATAGATATAAGTTTAAATACTTTACTAGATAGA;
T66FL-L, GATCTCTATCTAGTAAAGTATTTAAACTTATATCTATCTA;
16SWT-T, GATCATATAGAAGTTAGATTTATATGGGATTTCAGAACAA;
16SWT-L, GATCTTGTTCTGAAATCCCATATAAATCTAACTTCTATAT;
16S6FL-T, GATCATATAGAAAAATCCTTTATATGTCTAACCAGAACAA;
16S6FL-L, GATCTTGTTCTGGTTAGACATATAAAGGATTTTTCTATAT;
X-UP, GATCTCTATCTATATAAGTATTTAAACTXXACTCTATCTA;
X-DO, GATCTAGATAGAGTAAAGTTTAAATACXXATATAGATAGA.
Proteins and Transcription Assays.
TBP, TFB, TFBc, and RNAP were purified as described previously (6, 9). In vitro transcription reactions were essentially as previously described (6). To prevent competition between specific and nonspecific initiation sites, preliminary experiments were performed to determine at which concentration template becomes saturating. This was found to be at approximately 100 pM. Accordingly, template was added to reactions at 2 nM. After recovery of RNA, equal aliquots were taken and used in primer extension assays with either radiolabeled T7 or T3 sequencing primer.
Electrophoretic Mobility-Shift Assays (EMSAs) and DNase I Footprinting.
Binding reactions were carried out at 48°C for 20 min in 50 mM Tris⋅HCl (pH 7.5 at 25°C)/80 mM KCl/25 mM MgCl2/5% (vol/vol) glycerol/1 mM DTT/5 μg/ml poly(dG⋅dC). Products were electrophoresed on a nondenaturing 6% polyacrylamide gel. Footprinting was performed in the same buffer as described previously (20), followed by resolution of products on a denaturing 6% polyacrylamide gel.
UV-Mediated Photocrosslinking.
Binding reactions were set up as described above with 5 fmol (≈20,000 cpm) of probe and 1 pmol of TBP and/or TFB as indicated. Reactions were incubated for 10 min at 48°C before irradiation at 310 nm for 18 min with a UV transilluminator (Photodyne, New Berlin, WI). Samples were boiled in SDS/PAGE loading buffer before electrophoresis on an SDS/12% polyacrylamide gel.
Results
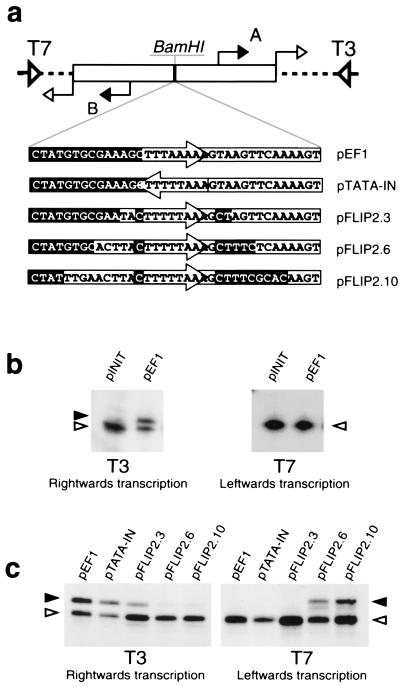
To identify the sequences and factors responsible for determining the orientation of transcription from archaeal promoters, we used the TATA-box region from the promoter for the Pyrococcus woesei ef1α gene, previously used in structural studies (9). We first tested whether the initiation site of transcription has a role in determining transcriptional polarity. Thus, we generated pINIT, the plasmid construct shown in Fig. 1a, in which two copies of an 18-nucleotide region encompassing the characterized initiation site of the Sulfolobus shibatae virus (SSV1) T6 gene were present in inverted repeat configuration flanking a BamHI restriction site. This BamHI site was then used to create pEF1 by inserting 36 nucleotide residues corresponding to the ef1α gene TATA box and flanking regions, such that the midpoint of the TATA box was equidistant from the upstream and downstream initiation sites. RNAs arising from in vitro transcription of these plasmids in either orientation can be detected by primer extension, using either T3 or T7 sequencing primers (see Fig. 1a). In vitro transcription assays, using recombinant aTBP and TFB and highly purified RNAP, were performed with the parental pINIT construct and with the construct (pEF1) containing wild-type ef1α sequences inserted in the BamHI site. Analysis with the T3 primer revealed that transcription of the pEF1 construct yields an RNA species specifically initiated at the downstream start site, A. This is indicated by a solid arrowhead in Fig. 1b. The species indicated by open arrowheads represent nonspecific initiation by RNAP at the junction of the cloned sequences and plasmid polylinker. To prevent potential interference between the specific and nonspecific initiation sites, reactions were carried out under conditions of excess template; see Materials and Methods for details. Importantly, no transcripts initiating at the specific upstream start site, B, were observed upon analysis with the T7 primer (Fig. 1b). Therefore, sequences encompassing the TATA box and its flanks, and not the initiation site, appear to define the unique polarity of transcription.
Figure 1.
Sequences flanking the TATA box are important for defining the orientation of transcription. (a) Diagram of plasmid constructs used in transcription assays. pINIT was created by ligating oligonucleotides corresponding to an inverted repeat of the T6 transcription initiation region into pBluescript. Subsequently, other oligonucleotides corresponding to derivatives of the Pyrococcus woesei ef1α promoter were inserted into the BamHI site of pINIT. The sequence of these inserts is shown. Sequences upstream of the TATA box in the wild-type promoter are shown in reverse shading; sequences naturally occurring downstream of the TATA box are boxed; and the TATA box itself is contained within an arrow. The T3 and T7 sequencing primer annealing sites are shown by open wide triangles and the T6 start sites are indicated by filled arrows and labeled A and B. The open arrows indicate the unexpected additional start sites arising at the junction of the cloned sequences and the parental polylinker that are recognized by the RNA polymerase alone and are totally independent of TBP and TFB. (b) The ef1α TATA box and flanking sequences direct unidirectional transcription. pINIT and pEF1 were used in in vitro transcription assays and transcripts detected by using primer extension with either T3 or T7 sequencing primer, as indicated. The transcript initiating at the downstream T6 start site is indicated with a solid arrowhead. The second, factor-independent, start site is indicated by an open arrowhead. The identities of the start sites were confirmed by electrophoresis adjacent to dideoxynucleotide DNA sequence ladders prepared with the radiolabeled T3 and T7 primers. (c) Sequences flanking the TATA box govern orientation of transcription. The various plasmid constructs shown in a were used in in vitro transcription assays. Annotation is as above.
Sequences Flanking the TATA Box Define Transcription Polarity.
Next, derivatives of the ef1α promoter were generated in which either the TATA box was inverted or regions flanking the TATA box were swapped (depicted in Fig. 1a). Transcription of these plasmids gave the surprising result that, although reducing overall transcription to some degree, neither inverting the TATA box nor swapping the first 3 bases upstream and downstream of the TATA box altered transcriptional polarity (Fig. 1c). However, an absolute shift in the polarity of transcription was observed in constructs in which the first 6 or the first 10 bases upstream and downstream of the TATA box were swapped. Because the first base pairs on either side of the TATA box in the natural promoter are rotationally symmetric, these results indicate that sequences located between 2 and 6 base pairs from the TATA element are important for defining transcriptional orientation. It is apparent that pFLIP2.10 gives rise to higher levels of transcription than does pFLIP2.6, suggesting that bases between 7 and 10 nucleotides upstream and/or downstream of the TATA element may contribute to promoter strength; however, we have not analyzed these sequences in further detail. Attempts were made to delimit further the regions within 2–6 base pairs of the TATA box that are important for imposing polarity by generating constructs in which sequences 3–6 nucleotides upstream and downstream of the TATA box were swapped. However, this construct was highly inefficient at mediating factor-dependent transcription in either direction (data not shown), indicating that the entire region 2–6 base pairs upstream and/or downstream from the TATA box is important for transcription orientation.
Interaction of TFB with Sequences Upstream of the TATA Box Has a Key Role in Defining Transcription Orientation.
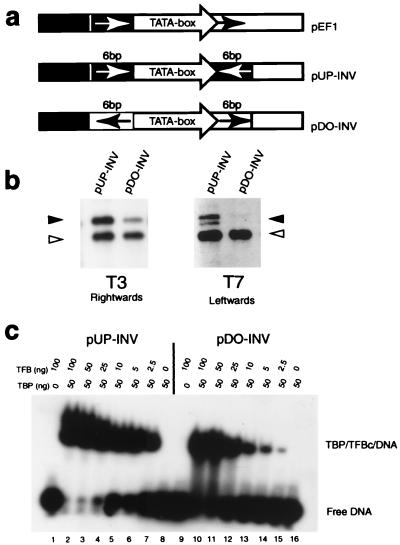
To determine whether the sequences defining transcriptional polarity are located upstream or downstream of the TATA box, or both, two further constructs were generated (Fig. 2a). These plasmids contained the wild-type ef1α TATA sequences surrounded by inverted repeats of either the six bases found upstream (pUP-INV) or downstream (pDO-INV) of the TATA box in the natural ef1α promoter. Significantly, in vitro transcription of pUP-INV gave strong bidirectional transcription, initiating at both upstream and downstream T6 start sites (detected by T7 and T3 primers, respectively; Fig. 2b). By contrast, pDO-INV gave much weaker transcription, with this low level of initiation occurring only at the downstream start site (detected by the T3 primer; Fig. 2b). These data therefore strongly suggest that there is an important cis-acting element in the sequences just upstream of the TATA element. When this motif is absent, transcription is considerably reduced, but interestingly appears to be unidirectional. Thus, while the motif upstream of the TATA box plays a dominant role in establishing the polarity of transcription, there may be additional, as-yet-unidentified, sequences that make a lesser contribution.
Figure 2.
The six base pairs upstream of the TATA box define polarity of transcription. (a) Diagram of plasmid constructs based on pINIT with derivatives of the ef1α promoter in which the six nucleotides found upstream (pUP-INV) or downstream (pDO-INV) of the TATA box in the natural ef1α promoter are present as inverted repeats flanking the TATA box. (b) In vitro transcription assays using pUP-INV and pDO-INV. Transcription products were detected by using the T3 or T7 sequencing primers. (c) EMSAs using TFB and TBP on double-stranded oligonucleotides corresponding to the pUP-INV or pDO-INV TATA box and flanking sequences.
The crystal structure of aTBP with the core domain of TFB (TFBc) and the TATA box shows that this 6-bp region upstream of the TATA box cannot be contacted by aTBP (9), implying an involvement of either TFB or RNAP in this interaction. TFB appears to be the most likely candidate since, like eukaryal TFIIB, it interacts with this DNA region in the archaeal ternary complex structure (9). If TFB contacts this region in a sequence-dependent manner, it would be predicted that the ternary complex would form with greater affinity when these sequences are present. Accordingly, EMSAs were employed to study ternary complex formation on pUP-INV and pDO-INV promoter fragments (Fig. 2c). Notably, these assays show that, while the TBP/TFBc ternary complex forms readily on pUP-INV sequences, approximately 10-fold higher concentrations of TFBc are required for analogous levels of complex formation on pDO-INV. These data strongly suggest that there is an element located upstream of the TATA box in the wild-type ef1α promoter that is recognized in a sequence-dependent manner by TFBc.
Polarity Is Defined at the Level of Ternary Complex Formation.
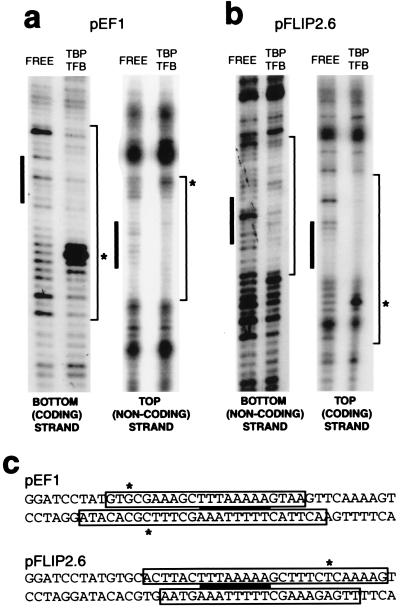
To determine whether the aTBP/TFBc complex alone has a defined polarity, DNase I footprinting assays were performed, analyzing aTBP/TFBc complexes on the pEF1 and pFLIP2.6 constructs. The ternary complex on pEF1 shows protection from DNase I cleavage from 13 nucleotides to the left (upstream) to six nucleotides to the right (downstream) of the TATA box (Fig. 3a; depicted in Fig. 3c). Furthermore, TBP/TFBc binding to pEF1 induces marked DNase I hypersensitivity (asterisks) within the region upstream but not the region downstream of the TATA box. Notably, the footprint on pFLIP2.6, which directs transcription in the opposite orientation to pEF1, demonstrates an analogous protection and hypersensitivity profile to pEF1, but rotated by 180° around the midpoint of the TATA box (Fig. 3 b and c). These data imply that transcriptional polarity is already defined at the level of ternary complex formation and, in conjunction with our EMSA data, suggest that this is primarily by means of the interaction of TFB with an element upstream of the TATA box. This TFB-responsive element (BRE) has been described previously as an important motif in the promoter of the SSV1 T6 gene (20).
Figure 3.
Polarity is defined at the level of ternary complex formation. (a) DNase I footprinting of TBP/TFBc complex on pEF1. Reaction mixtures contained free probe or 50 ng of TBP and 50 ng of TFBc where indicated. The TATA box is shown by a filled rectangle, and protected sequences are indicated at the right of each gel. Asterisks indicate protein-induced hypersensitivity to DNase I cleavage. (b) DNase I footprinting as in a on pFLIP2.6. (c) Summary of DNase I footprinting data. The sequences of the inserts in pEF1 and pFLIP2.6 are shown with the regions protected from DNase I cleavage boxed. The TATA box is indicated by a solid black bar. Sites of protein-induced hypersensitivities are indicated by asterisks.
The BRE Is Found in Many Archaeal Promoters.
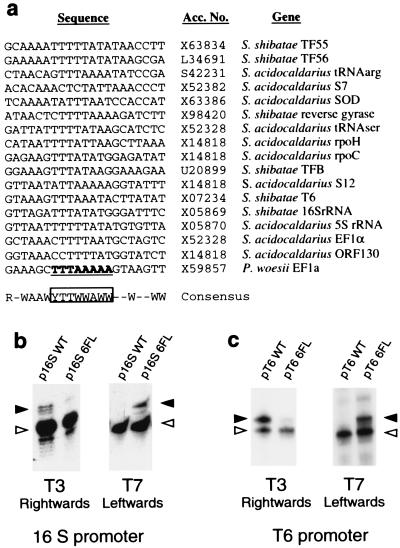
An alignment of the sequence of TATA boxes and flanking sequences of Sulfolobus promoters with characterized start sites is presented in Fig. 4a. Strikingly, there is conservation of sequence in the region that our experiments indicate is important for defining polarity; specifically, the six base pairs upstream of the TATA box have the consensus RNWAAW, (R = purine, W = A or T, N = any base). No sequence conservation is detected further upstream of the TATA element, and only weak conservation is found downstream of the TATA box. To determine whether this conserved motif is important for defining transcriptional polarity in other promoters, we extended our analyses by studying the SSV1 T6 and S. shibatae 16S rRNA promoters. In strong agreement with our data on the ef1α promoter, swapping the 6 base pairs flanking the TATA boxes of these promoters also confers an absolute switch in the orientation of transcription (Fig. 4 b and c).
Figure 4.
The BRE is highly conserved and defines transcription polarity on other archaeal promoters. (a) Alignment of characterized Sulfolobus promoter sequences. R = purine; Y = pyrimidine; lack of conservation is indicated by a horizontal bar. (b) In vitro transcription assays using plasmid constructs with oligonucleotides corresponding to the wild-type S. shibatae 16S rRNA gene TATA box and flanks (16S WT) or with the 6 base pairs flanking the TATA box swapped (16S 6FL). The RNA specifically initiating at the T6 start site in a factor-dependent manner is shown by a filled arrowhead. The open arrowheads indicate factor-independent initiation by the RNAP; see Fig. 1 for details. (c) In vitro transcription assays using plasmid constructs with oligonucleotides corresponding to the wild-type S. shibatae T6 gene TATA box and flanks (T6 WT) or with the 6 base pairs flanking the TATA box swapped (T6 6FL). The RNA specifically initiating at the T6 start site in a factor-dependent manner is shown by a filled arrowhead.
The Archaeal and Eukaryal Ternary Complexes Have the Same Polarity.
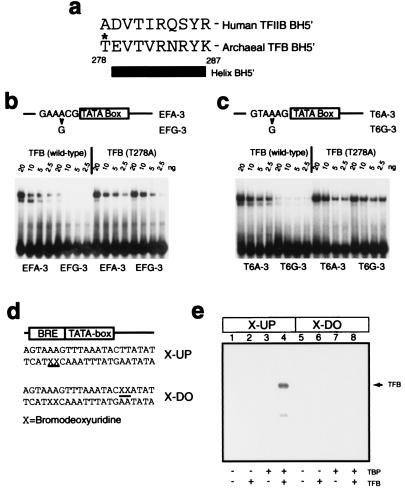
While our data indicate that the BRE–TFB interaction is of key importance in defining transcription polarity, they do not address the absolute polarity of the archaeal preinitiation complex. To resolve this issue, we used two further approaches. First, the Reinberg and Ebright laboratories demonstrated that mutation of certain residues within helix BH5′ of TFIIB resulted in loss of specificity in BRE recognition (19). We performed similar experiments in which we created derivatives of the ef1α and T6 promoters in which the highly conserved A⋅T base pair 3 bases upstream of the TATA box was changed to G⋅C. For both ef1α and T6 promoters, this substitution caused the promoter fragment to be recognized in EMSAs with approximately 1/16th the efficiency of the wild-type promoter (Fig. 5 b and c). Next, a variety of TFB mutants were generated with alanine substitutions in, and immediately preceding, helix BH5′. Notably, one of these mutant TFBs, T278A, could not discriminate between wild-type and mutant promoter fragments, strongly suggesting that this residue is important in mediating the specificity of the TFB–BRE interaction. Interestingly, the recent determination of the crystal structure of TBP and TFBc bound to a DNA molecule containing TATA box and BRE (accompanying paper by Littlefield et al., ref. 21) reveals that T278 contacts the phosphate at position −3 relative to the TATA box. Furthermore, T278 makes van der Waals contacts with the neighboring V280, restricting its rotameric configuration. V280 contacts the 5-methyl group of the thymidine at position −3. It is probable, therefore, that mutagenesis of T278 to alanine removes the backbone contact at −3 and may also alter the manner in which V280 recognizes the base at this position, resulting in TFB (T278A) being unable to discriminate between an A⋅T and a G⋅C base pair.
Figure 5.
The orientation of the archaeal TBP/TFB/DNA complex is the same as that of the eukaryal ternary complex. (a) Amino acid sequence of helix BH5′ and preceding residue in human TFIIB and archaeal TFB. The T278 residue is indicated with an asterisk. (b) EMSAs employing double-stranded oligonucleotides corresponding to the wild-type ef1α promoter (EFA-3) or ef1α promoter with an A⋅T to G⋅C substitution 3 base pairs upstream from the TATA box (EFG-3). Reaction mixtures were incubated with 20 ng of TBP and the indicated amount of TFB or TFB (T278A). (c) EMSAs employing double-stranded oligonucleotides corresponding to the wild-type T6 promoter (T6A-3) or T6 promoter with an A⋅T to G⋅C substitution 3 base pairs upstream from the TATA box (T6G-3). Reaction mixtures were incubated with 20 ng of TBP and the indicated amount of TFB or TFB (T278A). (d) Partial sequence of probes used in UV crosslinking experiments. The positions of 5-bromodeoxyuridine (BrdUrd) substitutions for thymidine are indicated by X and underlined. (e) The presence of BrdUrd substitutions upstream of the TATA box allows UV-mediated crosslinking of TFB to DNA. Five femtomoles (≈20,000 cpm) of double-stranded oligonucleotide containing radiolabeled X-UP (lanes 1–4) or X-DO (lanes 5–8) were incubated with 20 ng of the indicated proteins and irradiated as described in Materials and Methods prior to electrophoresis on an SDS/12% polyacrylamide gel. The position of TFB crosslinked to the X-UP-containing oligonucleotide probe is indicated by an arrow; the faint signal below TFB corresponds to a proteolytic fragment of TFB, as confirmed by Western blotting (data not shown).
In a second approach, we exploited the observation that TFB makes contacts across the DNA major groove on one side of the TATA box and the minor groove on the other side (8, 9, 19, 22). Although the previous crystal structures of TBP and TFB/TFIIB do not reveal base-specific contacts on either side of the TATA box, these structures were derived by using oligonucleotides that lack BRE sequences (8, 9). Thus, in light of the data above, we reasoned that these structures were likely to be representative of nonspecific TFB/TFIIB–DNA interactions. Indeed, the recent elucidation of the crystal structure of TBP/TFB/DNA on BRE-containing oligonucleotides reveals base-specific interactions between TFB and the BRE (see accompanying paper, ref. 21). Accordingly, we synthesized oligonucleotides corresponding to the T6 promoter with 5-bromodeoxyuridine (BrdUrd) at positions 2 and 3 bases upstream (X-UP) or positions 2 and 3 bases downstream (X-DO) of the TATA box. BrdUrd has been shown previously to mediate crosslinking of proteins in the major groove (for example, see ref. 14). These oligonucleotides were radiolabeled and annealed to nonsubstituted complementary oligonucleotides before incubation with TBP and/or TFB. These probes both formed ternary complex with comparable affinities (data not shown). After irradiation with UV light and resolution by SDS/PAGE, TFB could be crosslinked to the X-UP probe in the presence of TBP (Fig. 5e, lane 4). However, under the conditions used, no crosslinking of any protein species was observed with the X-DO oligonucleotides (Fig. 5e, lanes 5–8).
The crosslinking data and the mutation studies therefore indicate that the major groove of DNA upstream of the TATA box is recognized by TFB, and that residues within the C-terminal repeat of TFB are important for this sequence-specific recognition. Taken together with the accompanying paper (21), these data demonstrate that the orientation of the archaeal ternary complex is the same as that of its eukaryal counterpart.
Discussion
All crystal structures of promoter complexes containing the C-terminal core of eukaryal TBP (eTBPc) reveal eTBPc bound with the C-terminal “stirrup” contacting the upstream end of the TATA box (8, 11, 23–26). However, recent experiments using eTBP derivatives containing a DNA-cleavage reagent suggest that eTBP binds with only a modest (60:40) orientation preference (13). This is not surprising because eTBPc contains an imperfect sequence repeat that creates an essentially dyad symmetric DNA contact surface within a nearly twofold rotationally symmetric three-dimensional structure. The modest difference in binding energy for the two conformations might reflect a slight asymmetry in the details of the contact surface (12). Alternatively, the significantly asymmetric charge potential distribution of eTBP could match the need for a stronger deforming force on the downstream half of the TATA box (11). Notably, archaeal TBP is more symmetric than its eukaryal counterpart, in both its TATA box contact surface and its charge potential distribution (16). This is in good agreement with the proposal that TBP evolved by gene duplication from a homodimer, which, by definition, would be fully symmetrical. Such a situation may correspond closely to that in present-day Archaea, where TBP is still highly symmetrical and, consequently, there is a heavy reliance on TFB to define transcriptional polarity.
The important role of TFB in determining directionality of transcription further demonstrates the key importance of TFB/TFIIB in preinitiation complex assembly. TFIIB contacts the core polymerase (27), possesses DNA-binding activity (19), can associate with the polymerase before recruitment (28), and leaves the polymerase shortly after initiation (29). This range of properties of TFB/TFIIB, taken together with the observations that transcription can be initiated in the absence of TBP (30, 31), suggests that the eukaryal and archaeal transcription machineries may have evolved from a progenitor in which TFB/TFIIB was the principal transcription factor responsible for promoter recognition and polymerase recruitment. In contrast, TBP may have played an accessory role in transcription in the evolutionary predecessor of the Archaea and Eucarya, the juxtaposition of a TFB and a TBP recognition sequence being stimulatory for polymerase binding. The subsequent evolution of protein–protein interactions between TBP and TFB may then have led to the state in modern Archaea and Eucarya, where ternary complex stability is a consequence of the sum of binding energies contributed by multiple interactions within the architecture of the ternary complex. This dependence on multiple intermolecular interactions between TBP, TFB, and DNA may serve as an important mechanism for ensuring the fidelity of transcription initiation site selection.
Finally, given the fundamental similarities between transcription in Archaea and Eucarya, it is tempting to speculate that TFIIB helps define transcriptional polarity in the eukaryal RNAP II system. In support of this idea, it has been reported recently that TFIIB possesses an intrinsic sequence-specific DNA-binding activity (19) and can induce TBP to bind DNA in a preferential orientation (13). Further investigations into the mode of promoter recognition by TFB and its eukaryotic relatives are thus likely to provide significant insights into the molecular basis for promoter orientation selectivity and into evolutionary relationships between the archaeal and eukaryal transcription systems.
Acknowledgments
We are grateful to various members of the S.P.J. and P.B.S. laboratories for stimulating discussions. In particular, we thank Jessica Downs, Yakov Korkhin, and Otis Littlefield. In addition, we thank M. Waring, M. Webb, and J. O. Thomas for assistance with the crosslinking experiments. The work in the S.P.J. laboratory was supported by the Leverhulme Trust and the Wellcome Trust. Work in the P.B.S. laboratory was supported in part by a grant from the National Institutes of Health (GM15225). P.L.K. was a National Institutes of Health predoctoral trainee.
Abbreviations
- RNAP
RNA polymerase
- TBP
TATA-box-binding protein
- aTBP and eTBP
archaeal and eukaryal TBPs
- TFBc
C-terminal core domain of TFB
- TAF
TBP-associated factor
- BRE
TFB-responsive element
- EMSA
electrophoretic mobility-shift assay
References
- 1.Bell S D, Jackson S P. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 2.Reeve J N, Sandman K, Daniels C J. Cell. 1997;89:999–1002. doi: 10.1016/s0092-8674(00)80286-x. [DOI] [PubMed] [Google Scholar]
- 3.Rowlands T, Baumann P, Jackson S P. Science. 1994;264:1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- 4.Hausner W, Wettach J, Hethke C, Thomm M. J Biol Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 5.Hethke C, Geerling A C M, Hausner W, de Vos W M, Thomm M. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qureshi S A, Bell S D, Jackson S P. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell S D, Jaxel C, Nadal M, Kosa P F, Jackson S P. Proc Natl Acad Sci USA. 1998;95:15218–15222. doi: 10.1073/pnas.95.26.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 9.Kosa P F, Ghosh G, DeDecker B S, Sigler P B. Proc Natl Acad Sci USA. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grove A, Galeone A, Yu E, Mayol L, Geiduschek E P. J Mol Biol. 1998;282:731–739. doi: 10.1006/jmbi.1998.2058. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y C, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 12.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 13.Cox J M, Hayward M M, Sanchez J F, Gegnas L D, van der Zee S, Dennis J H, Sigler P B, Schepartz A. Proc Natl Acad Sci USA. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 15.Burke T W, Kadonaga J T. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeDecker B S, O’Brien R, Fleming P J, Geiger J H, Jackson S P, Sigler P B. J Mol Biol. 1996;264:1072–1084. doi: 10.1006/jmbi.1996.0697. [DOI] [PubMed] [Google Scholar]
- 17.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 18.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–379. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 19.LaGrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qureshi S A, Jackson S P. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- 21.Littlefield O, Korkhin Y, Sigler P B. Proc Natl Acad Sci USA. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Hahn S. Nature (London) 1995;376:609–612. doi: 10.1038/376609a0. [DOI] [PubMed] [Google Scholar]
- 23.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 24.Nikolov D B, Chen H, Halay E D, Hoffmann A, Roeder R G, Burley S K. Proc Natl Acad Sci USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 26.Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 27.Ha I, Roberts S, Maldonado E, Sun X, Kim L-U, Green M, Reinberg D. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 28.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 29.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 30.Usheva A, Shenk T. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 31.Wieczorek E, Brand M, Jacq X, Tora L. Nature (London) 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]