Abstract
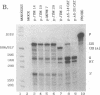
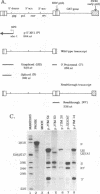
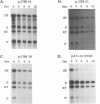
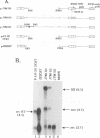
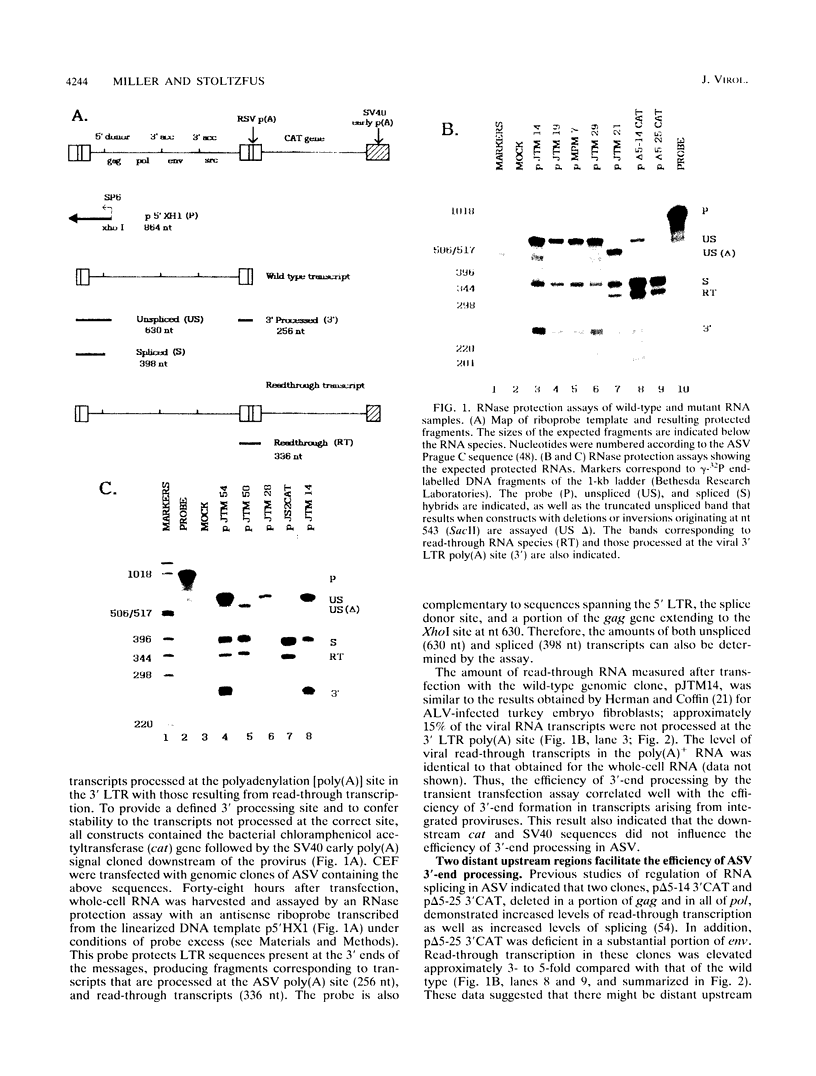
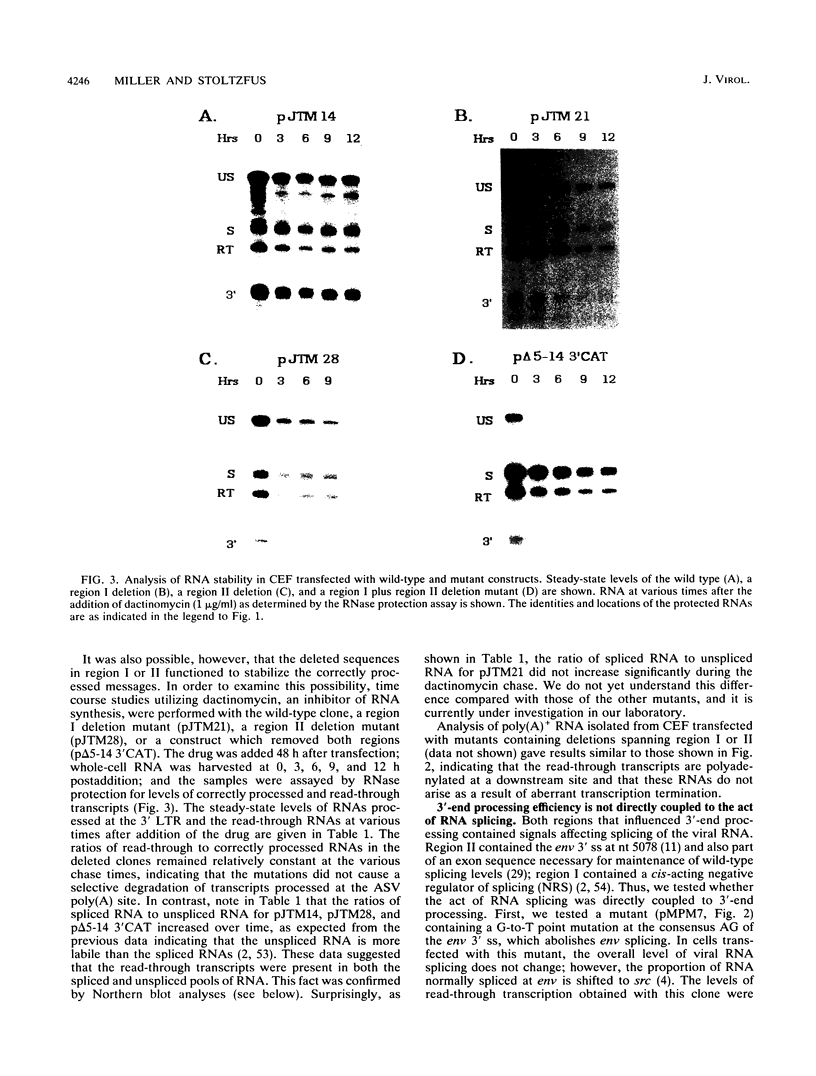
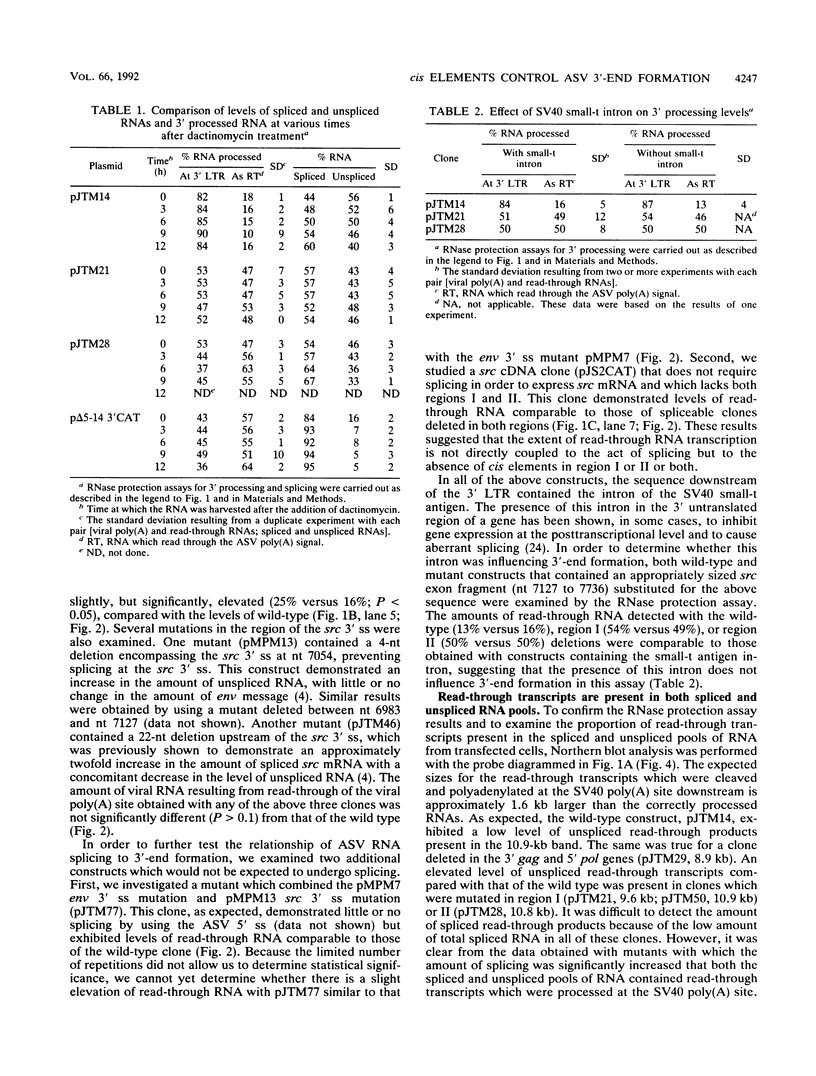
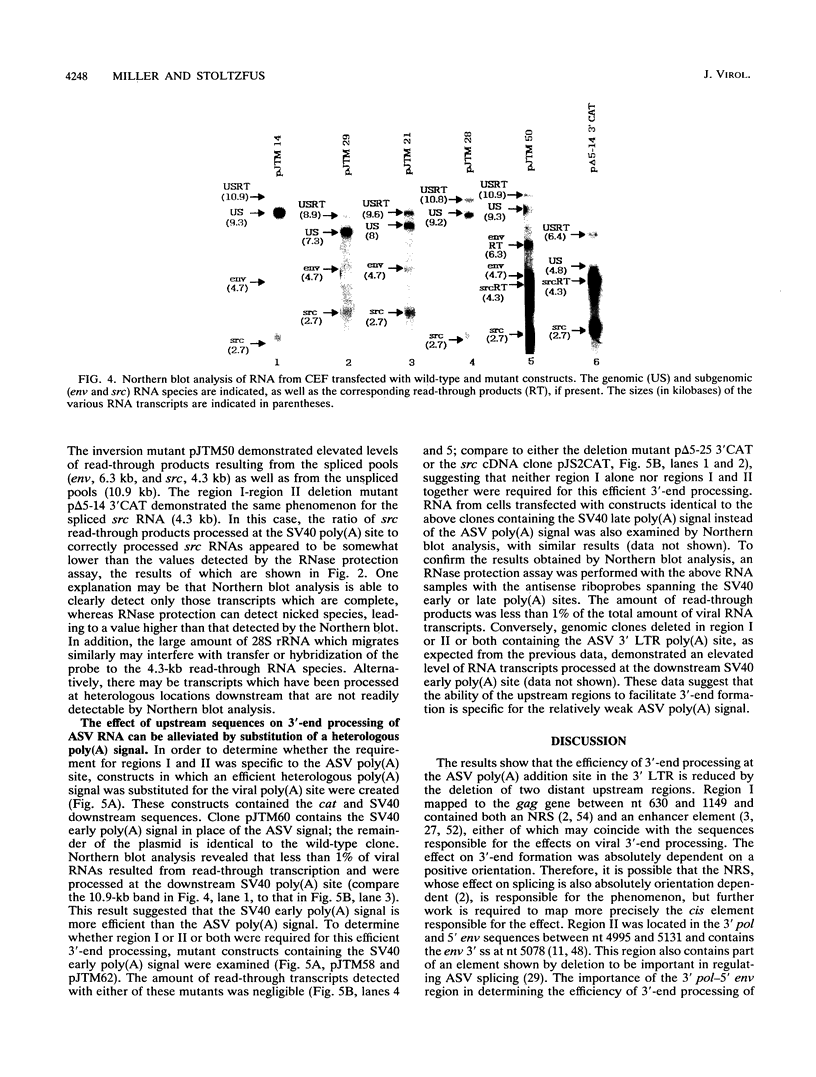
Retroviruses, pararetroviruses, and related retrotransposons generate terminally redundant RNAs by transcription of a template flanked by long terminal repeats in which initiation occurs within the 5' long terminal repeat sequences and 3'-end processing occurs within the 3' long terminal repeat sequences. Processing of avian sarcoma virus RNA is relatively inefficient; approximately 15% of the viral RNA transcripts are read-through products; i.e., they are not processed at the viral poly(A) addition site but at sites in the cellular sequence further downstream. In this report, we show that the efficiency of processing at the viral site is further reduced by deletion of two distant upstream sequences: (i) a 606-nucleotide sequence in the gag gene containing a cis-acting negative regulator of splicing and (ii) a 136-nucleotide sequence spanning the env 3' splice site. The deletion of either or both upstream regions increases the levels of read-through products of both unspliced and spliced viral RNA. In contrast, deletion of the src 3' splice site does not affect the efficiency of processing at the viral poly(A) addition site. The effects on 3'-end processing are not correlated either with distance from the promoter to the poly(A) addition site or with the overall level of viral RNA splicing. Substitution of the avian sarcoma virus poly(A) signal with the simian virus 40 early or late poly(A) signal relieves the requirement for the distant upstream sequences. We propose that cellular factors, which may correspond to splicing factors, bound to the upstream viral sequences may interact with factors bound at the avian sarcoma virus poly(A) signal to stabilize the polyadenylation-cleavage complex and allow for more efficient 3'-end processing.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo S., Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988 Nov;8(11):4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo S., Yun M., Beemon K. cis-acting regulatory elements within gag genes of avian retroviruses. Mol Cell Biol. 1987 Jan;7(1):388–397. doi: 10.1128/mcb.7.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich S. L., Stoltzfus C. M. Mutations in the regions of the Rous sarcoma virus 3' splice sites: implications for regulation of alternative splicing. J Virol. 1991 May;65(5):2640–2646. doi: 10.1128/jvi.65.5.2640-2646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. ATTAAA as well as downstream sequences are required for RNA 3'-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985 Nov;5(11):3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady H. A., Wold W. S. Competition between splicing and polyadenylation reactions determines which adenovirus region E3 mRNAs are synthesized. Mol Cell Biol. 1988 Aug;8(8):3291–3297. doi: 10.1128/mcb.8.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. H., Tiley L. S., Cullen B. R. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol. 1991 Jun;65(6):3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnlein S., Hauber J., Cullen B. R. Identification of a U5-specific sequence required for efficient polyadenylation within the human immunodeficiency virus long terminal repeat. J Virol. 1989 Jan;63(1):421–424. doi: 10.1128/jvi.63.1.421-424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S., Alwine J. C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989 Oct;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. J., Stoltzfus C. M. Cloning and nucleotide sequences of cDNAs spanning the splice junctions of Rous sarcoma virus mRNAs. J Virol. 1985 Mar;53(3):969–972. doi: 10.1128/jvi.53.3.969-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou H. C., Dabrowski C., Alwine J. C. Simian virus 40 late mRNA leader sequences involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J Virol. 1991 Dec;65(12):6677–6685. doi: 10.1128/jvi.65.12.6677-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi T., Huang M., Gorman C., Jaenisch R. A generic intron increases gene expression in transgenic mice. Mol Cell Biol. 1991 Jun;11(6):3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Imperiale M. J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989 Nov;9(11):4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Kilpatrick J. E., Imperiale M. J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3' end formation. Mol Cell Biol. 1991 Mar;11(3):1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gilmartin G. M., Nevins J. R. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol. 1991 May;11(5):2432–2438. doi: 10.1128/mcb.11.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Cheley S., Kuismanen E., Finn L. A., Yamawaki-Kataoka Y. Nonmuscle and muscle tropomyosin isoforms are expressed from a single gene by alternative RNA splicing and polyadenylation. Mol Cell Biol. 1986 Nov;6(11):3582–3595. doi: 10.1128/mcb.6.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman S. A., Coffin J. M. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986 Nov;60(2):497–505. doi: 10.1128/jvi.60.2.497-505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman S. A., Coffin J. M. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science. 1987 May 15;236(4803):845–848. doi: 10.1126/science.3033828. [DOI] [PubMed] [Google Scholar]
- Huang M. T., Gorman C. M. Intervening sequences increase efficiency of RNA 3' processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990 Feb 25;18(4):937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. T., Gorman C. M. The simian virus 40 small-t intron, present in many common expression vectors, leads to aberrant splicing. Mol Cell Biol. 1990 Apr;10(4):1805–1810. doi: 10.1128/mcb.10.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Temin H. M. The efficiency of RNA 3'-end formation is determined by the distance between the cap site and the poly(A) site in spleen necrosis virus. Genes Dev. 1990 Dec;4(12B):2299–2307. doi: 10.1101/gad.4.12b.2299. [DOI] [PubMed] [Google Scholar]
- Kanter M. R., Smith R. E., Hayward W. S. Rapid induction of B-cell lymphomas: insertional activation of c-myb by avian leukosis virus. J Virol. 1988 Apr;62(4):1423–1432. doi: 10.1128/jvi.62.4.1423-1432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnitz L., Faber S., Chalkley R. Specific nuclear proteins interact with the Rous sarcoma virus internal enhancer and share a common element with the enhancer located in the long terminal repeat of the virus. Nucleic Acids Res. 1987 Dec 10;15(23):9841–9859. doi: 10.1093/nar/15.23.9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Kotler M., Skalka A. M. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988 Aug;62(8):2686–2695. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990 Feb;10(2):696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg J. M., Pepling M. E., Mount S. M. Polyadenylylation in copia requires unusually distant upstream sequences. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3038–3042. doi: 10.1073/pnas.88.8.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Hart R. P., Wong W. W., Nevins J. R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986 Nov;5(11):2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Niwa M., Rose S. D., Berget S. M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990 Sep;4(9):1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Sandgren E. P., Avarbock M. R., Allen D. D., Brinster R. L. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Gimmi E. R., Perry R. P. The developmentally regulated shift from membrane to secreted mu mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol Cell Biol. 1991 Apr;11(4):2324–2327. doi: 10.1128/mcb.11.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Raines M. A., Maihle N. J., Moscovici C., Crittenden L., Kung H. J. Mechanism of c-erbB transduction: newly released transducing viruses retain poly(A) tracts of erbB transcripts and encode C-terminally intact erbB proteins. J Virol. 1988 Jul;62(7):2437–2443. doi: 10.1128/jvi.62.7.2437-2443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R. H. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-independent, and additive. Nucleic Acids Res. 1991 Dec 11;19(23):6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R., Ganem D. Sequences 5' to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990 May;4(5):764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., Alwine J. C. Identification of a sequence element on the 3' side of AAUAAA which is necessary for simian virus 40 late mRNA 3'-end processing. Mol Cell Biol. 1985 Oct;5(10):2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H., Brodmann P., Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991 Jan;5(1):141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- Sanfaçon H., Hohn T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature. 1990 Jul 5;346(6279):81–84. doi: 10.1038/346081a0. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L. M., Danna K. J. Efficient infection of monkey cells with DNA of simian virus 40. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7575–7578. doi: 10.1073/pnas.78.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M., Chang L. J., Cripe T. P., Turek L. P. Efficient transformation by Prague A Rous sarcoma virus plasmid DNA requires the presence of cis-acting regions within the gag gene. J Virol. 1987 Nov;61(11):3401–3409. doi: 10.1128/jvi.61.11.3401-3409.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M., Dimock K., Horikami S., Ficht T. A. Stabilities of avian sarcoma virus RNAs: comparison of subgenomic and genomic species with cellular mRNAs. J Gen Virol. 1983 Oct;64(Pt 10):2191–2202. doi: 10.1099/0022-1317-64-10-2191. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Fogarty S. J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989 Apr;63(4):1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M. Synthesis and processing of avian sarcoma retrovirus RNA. Adv Virus Res. 1988;35:1–38. doi: 10.1016/s0065-3527(08)60707-1. [DOI] [PubMed] [Google Scholar]
- Strohman R. C., Moss P. S., Micou-Eastwood J., Spector D., Przybyla A., Paterson B. Messenger RNA for myosin polypeptides: isolation from single myogenic cell cultures. Cell. 1977 Feb;10(2):265–273. doi: 10.1016/0092-8674(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Mechanism of transduction by retroviruses. Science. 1992 Feb 14;255(5046):841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- Valsamakis A., Zeichner S., Carswell S., Alwine J. C. The human immunodeficiency virus type 1 polyadenylylation signal: a 3' long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pederson T. A 62,000 molecular weight spliceosome protein crosslinks to the intron polypyrimidine tract. Nucleic Acids Res. 1990 Oct 25;18(20):5995–6001. doi: 10.1093/nar/18.20.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H. Deletion in the 3' pol sequence correlates with aberration of RNA expression in certain replication-defective avian sarcoma viruses. J Virol. 1985 May;54(2):446–459. doi: 10.1128/jvi.54.2.446-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichs an der Glon C., Monks J., Proudfoot N. J. Occlusion of the HIV poly(A) site. Genes Dev. 1991 Feb;5(2):244–253. doi: 10.1101/gad.5.2.244. [DOI] [PubMed] [Google Scholar]
- Weiss E. A., Gilmartin G. M., Nevins J. R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991 Jan;10(1):215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990 Jul;15(7):277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Zhang F., Denome R. M., Cole C. N. Fine-structure analysis of the processing and polyadenylation region of the herpes simplex virus type 1 thymidine kinase gene by using linker scanning, internal deletion, and insertion mutations. Mol Cell Biol. 1986 Dec;6(12):4611–4623. doi: 10.1128/mcb.6.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]