Abstract
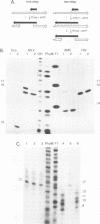
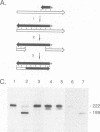
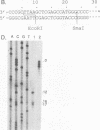
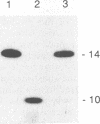
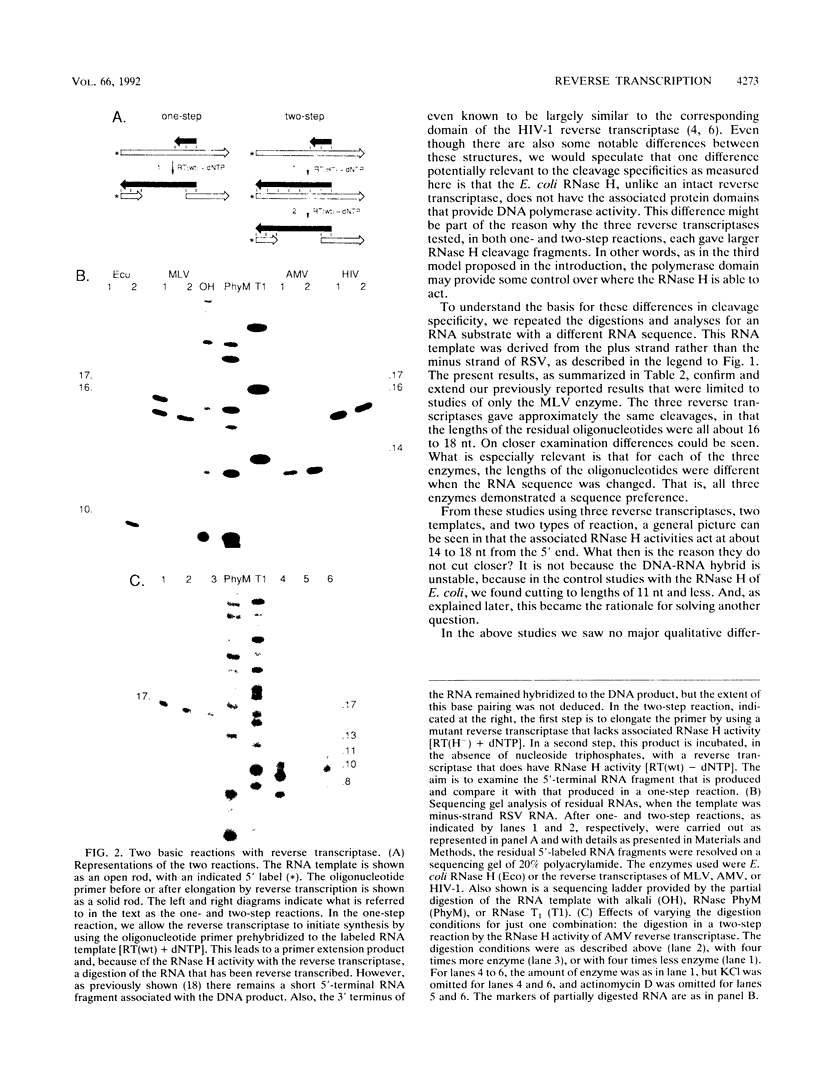
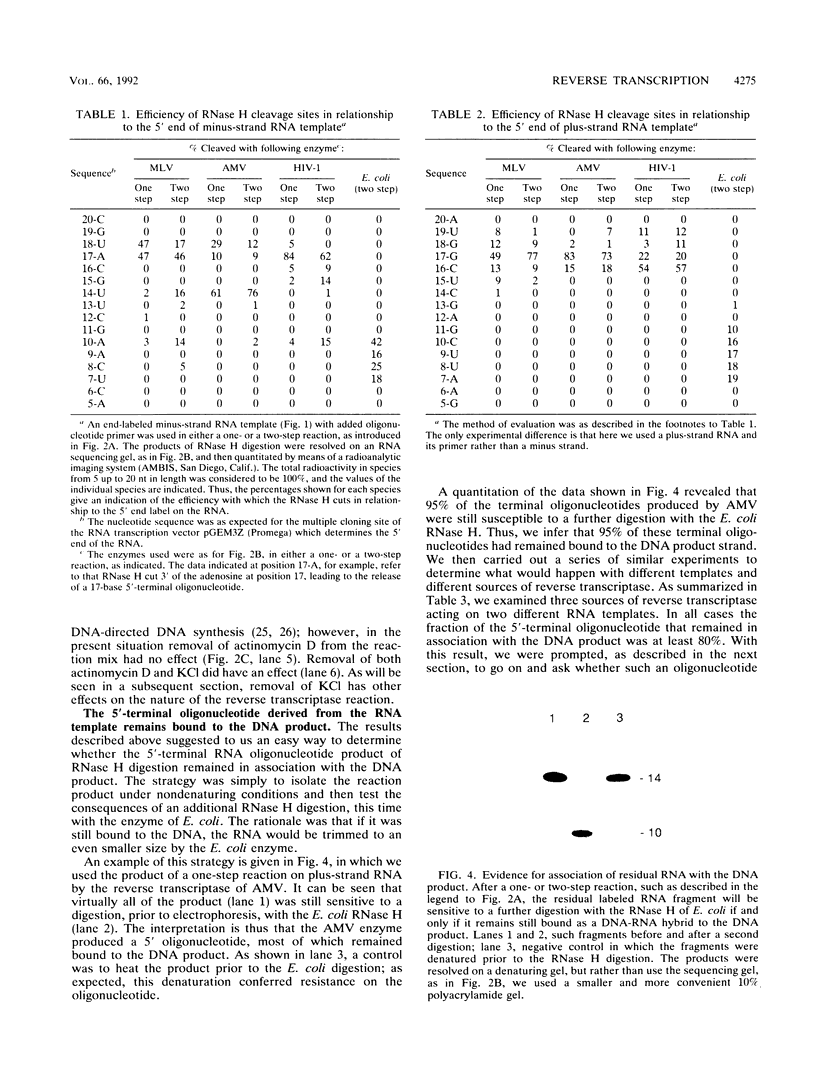
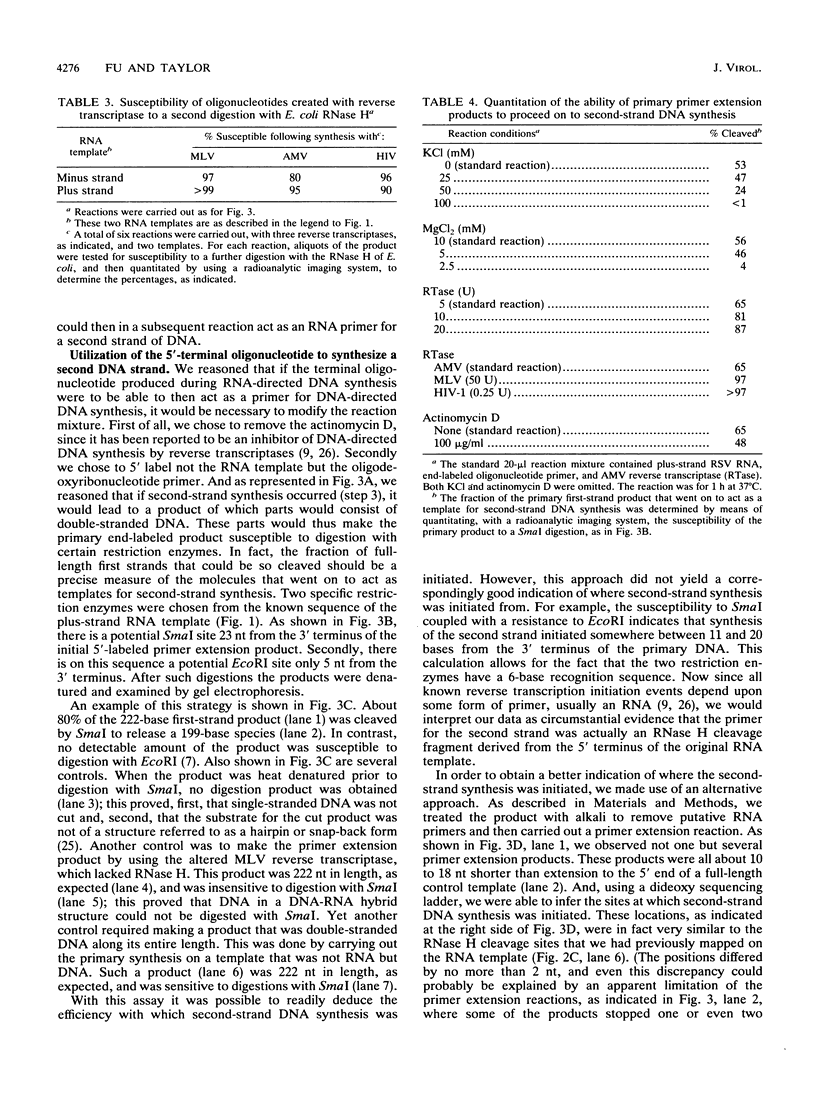
Luo and Taylor (J. Virol. 64:4321-4328, 1990) have previously shown that when, during RNA-directed DNA synthesis, a retroviral reverse transcriptase comes to a halt at the end of an RNA template, the associated RNase H produces a specific oligonucleotide that contains the 5' end of that template; in those studies the length of the oligonucleotide was predominantly 17 nucleotides. We have now investigated variables that might affect the formation and length of such a terminal oligonucleotide. We found small but significant variations in the length could be caused by the choice of reaction conditions and also the sources of reverse transcriptase and RNA template. Nevertheless, the general finding in all these situations was that RNase H acted at or about 14 to 18 nucleotides from the 5' end, thereby supporting the interpretation that in the reverse transcriptase, the cleavage site for the RNase H is held at around this distance behind the DNA polymerase activity. In other words, it appears that for the intact protein, the RNase H and reverse transcriptase activities may work in a coupled or coordinate manner. We also found that more than 80% of the residual 5' oligonucleotides remained base paired to the RNA-directed DNA product. Furthermore, under certain conditions, these short RNAs could act as efficient primers for an associated DNA-directed DNA synthesis in the reverse direction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brewer L. C., Wells R. D. Mechanistic independence of avian myeloblastosis virus DNA polymerase and ribonuclease H. J Virol. 1974 Dec;14(6):1494–1502. doi: 10.1128/jvi.14.6.1494-1502.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E., Boyer H. W. Solubility and dialysis limits of DNA oligonucleotides. Biochim Biophys Acta. 1972 Mar 14;262(2):116–124. doi: 10.1016/0005-2787(72)90224-9. [DOI] [PubMed] [Google Scholar]
- Condreay L. D., Wu T. T., Aldrich C. E., Delaney M. A., Summers J., Seeger C., Mason W. S. Replication of DHBV genomes with mutations at the sites of initiation of minus- and plus-strand DNA synthesis. Virology. 1992 May;188(1):208–216. doi: 10.1016/0042-6822(92)90751-a. [DOI] [PubMed] [Google Scholar]
- Crouch R. J. Ribonuclease H: from discovery to 3D structure. New Biol. 1990 Sep;2(9):771–777. [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Furfine E. S., Reardon J. E. Reverse transcriptase.RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991 Jan 5;266(1):406–412. [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Fu T. B., Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992 May;66(5):3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., Richardson C. C. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1990 Jun 25;265(18):10565–10573. [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- Krug M. S., Berger S. L. Ribonuclease H activities associated with viral reverse transcriptases are endonucleases. Proc Natl Acad Sci U S A. 1989 May;86(10):3539–3543. doi: 10.1073/pnas.86.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien J. M., Aldrich C. E., Mason W. S. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol. 1986 Jan;57(1):229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Hirsch R. C., Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 1991 Nov;10(11):3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Sharmeen L., Taylor J. Specificities involved in the initiation of retroviral plus-strand DNA. J Virol. 1990 Feb;64(2):592–597. doi: 10.1128/jvi.64.2.592-597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason W. S., Taylor J. M., Hull R. Retroid virus genome replication. Adv Virus Res. 1987;32:35–96. doi: 10.1016/s0065-3527(08)60474-1. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Cywinski A., Smith J. K. Discontinuities in the DNA synthesized by an avian retrovirus. J Virol. 1983 Dec;48(3):654–659. doi: 10.1128/jvi.48.3.654-659.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Faras A. J., Varmus H. E., Levinson W. E., Bishop J. M. Ribonucleic acid directed deoxyribonucleic acid synthesis by the purified deoxyribonucleic acid polymerase of Rous sarcoma virus. Characterization of the enzymatic product. Biochemistry. 1972 Jun 6;11(12):2343–2351. doi: 10.1021/bi00762a021. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990 Nov 6;29(44):10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]