Abstract
The ABC transporter, P-glycoprotein, is an integral membrane protein that mediates the ATP-driven efflux of drugs from multidrug-resistant cancer and HIV-infected cells. Anti-P-glycoprotein antibody C219 binds to both of the ATP-binding regions of P-glycoprotein and has been shown to inhibit its ATPase activity and drug binding capacity. C219 has been widely used in a clinical setting as a tumor marker, but recent observations of cross-reactivity with other proteins, including the c-erbB2 protein in breast cancer cells, impose potential limitations in detecting P-glycoprotein. We have determined the crystal structure at a resolution of 2.4 Å of the variable fragment of C219 in complex with an epitope peptide derived from the nucleotide binding domain of P-glycoprotein. The 14-residue peptide adopts an amphipathic α-helical conformation, a secondary structure not previously observed in structures of antibody–peptide complexes. Together with available biochemical data, the crystal structure of the C219-peptide complex indicates the molecular basis of the cross-reactivity of C219 with non-multidrug resistance-associated proteins. Alignment of the C219 epitope with the recent crystal structure of the ATP-binding subunit of histidine permease suggests a structural basis for the inhibition of the ATP and drug binding capacity of P-glycoprotein by C219. The results provide a rationale for the development of C219 mutants with improved specificity and affinity that could be useful in antibody-based P-glycoprotein detection and therapy in multidrug resistant cancers.
Multidrug resistance is the prevalent cause of treatment failure in cancer chemotherapy. At present, the best understood mechanism of acquired resistance to anticancer drugs is the increased expression of multidrug resistance gene 1 (MDR-1)-encoded P-glycoprotein (Pgp). Pgp is a 170-kDa integral membrane protein, originally identified by Ling and colleagues (1), that is frequently overexpressed in cancer cells subjected to prolonged chemotherapy. Pgp is a member of a large, widespread family of ATP-driven transport proteins called ATP-binding cassette (ABC) transporters.
In recent years, the members of the ABC transporter superfamily have attracted study because of their important roles in biological processes in both prokaryotes and eukaryotes, and especially because of their association with major biomedical issues such as cystic fibrosis, antigen presentation, and multidrug resistance (MDR) in cancer- and HIV-infected cells (2, 3). For understanding of the molecular basis of MDR, an atomic structure of Pgp would be of considerable importance, in particular for elucidating the basis of substrate recognition and the mechanism by which ATP drives the transport process. A structure would also contribute valuable information on other members of the ABC superfamily and the clinical problems that are associated with dysfunction of these proteins. Attempts to determine the structures of ABC transporters and their separate domains have had limited success to date. Structural information on Pgp is limited to a low resolution study by electron microscopy (4).
Based on amino acid sequence analysis, Pgp is thought to consist of two homologous halves, each containing a six-helical membrane-spanning domain involved in drug efflux and a cytosolic nucleotide-binding domain (NBD).
One approach to obtaining information on the structural aspects of the functional domains of Pgp is the use of monoclonal antibodies elicited against peptide regions of the protein. One of these antibodies, C219, was elicited against SDS-solubilized plasma membranes of multidrug-resistant Chinese hamster ovary and human cell lines (5) and binds to a conserved cytoplasmic region present in all classes of P-glycoprotein NBDs from rodents and humans. Screening of the C-terminal cytoplasmic domain with a series of hexapeptides identified a continuous region containing the core sequence of VQEALD (6). Anti-P-glycoprotein antibody C219 binds to both of the ATP-binding regions of Pgp and has been shown to inhibit its ATPase activity (7, 8). In addition, C219 has the capacity to inhibit drug binding to the membrane-spanning domains of Pgp (7).
C219 is by far the most widely used antibody for Pgp immunodetection in the clinic and the laboratory (6, 9–13). Observations of cross-reactivity between C219 and other proteins besides Pgp and recognition of classes of Pgps that are not involved in multidrug resistance, such as human MDR3 Pgp and a sister protein of Pgp, impose a potential limit on the utility of C219 in MDR diagnosis (14–19). Recent findings of cross-reactivity with p185c-erbB2, a protein that can be overexpressed in breast cancer cells, have urged caution in interpreting the results of immunodetection of Pgp in these cells. As a tool to address these problems, recombinant molecules based on C219 have been developed in our laboratory, and the crystal structure of the unliganded C219 variable fragment (Fv) has been determined (20).
Here, we present the structure of the C219 Fv fragment in complex with 14-residue NBD epitope peptide VVQEALDKAREGRT containing the core amino acid sequence of the NBD epitope. These results provide significant information on the molecular recognition by antibody C219 of Pgp’s cytosolic domains. Furthermore, the results suggest a structural basis for C219’s cross-reactivity, the inhibition of the ATP-driven drug-efflux pump, and binding of drugs to Pgp. This information provides a rationale for engineering anti-Pgp antibodies with improved specificity and affinity that could be useful in tumor imaging and therapeutic targeting strategies.
Methods
Expression of C219 Single-Chain Fv.
The anti-P-glycoprotein single-chain Fv fragment was constructed from cDNA derived from hybridoma cells secreting the monoclonal antibody C219, by cloning the variable region genes and expressing them in Escherichia coli as described (20).
Peptide Synthesis.
The 14-amino acid NBD-epitope peptide, with sequence VVQEALDKAREGRT, was synthesized by using standard solid phase technology. The peptide was purified by using a semipreparative reversed phase column on HPLC. The peptide peak was confirmed by amino acid analysis and mass spectrometry.
Crystallization, Data Collection, and Structure Determination.
Crystals of single-chain (scFv) C219 were grown in the presence of NBD-epitope peptide (ratio: 1:1) in hanging drops containing 40% (vol/vol) 2-methyl-2,4-pentanediol in 50 mM Mes buffer (pH 6.3).
A complete data set was collected from a crystal with dimensions 0.2 × 0.2 × 0.15 mm, on a MAR-Research Imaging Plate (MAR Research, Hamburg, Germany) on a Rigaku (Tokyo) Rotaflex rotating anode with a copper target and Osmic focusing optics (Osmic, Troy, Michigan). Diffraction data were processed by using denzo and scalepack (21). Data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection statistics
| Resolution, Å | 2.4 |
| Space group | P212121 |
| Cell dimensions, Å | 68.8, 82.7, 94.0 |
| Reflections, n, overall; shell 2.46–2.40 Å | 142245; 5585 |
| Unique reflections, n, overall; shell 2.46–2.40 Å | 21637; 1267 |
| Completeness, %, overall; shell 2.46–2.40 Å | 98.7; 88.8 |
| I/σI < 2, %, overall; shell 2.46–2.40 Å | 16.0; 44.0 |
| Rsymm = Σ|I − 〈I〉|/Σ〈I〉 overall; shell 2.46–2.40 Å | 0.084; 0.472 |
Two C219 scFv molecules were located in the asymmetric unit by molecular replacement in AMoRe (22, 23) using a monomer of the unliganded C219 scFv [Protein Data Bank ID code 1AP2 (20)] as a search model. Rigid body and structure refinement were performed by using the program Crystallography and NMR System [CNS (24–27)]. Structure refinement consisted of repeated rounds of simulated annealing in combination with torsion angle dynamics and positional and B-factor refinement, using a maximum likelihood target. A randomly sampled Rfree set consisting of 10% of the collected data was set aside for cross-validation and σA weighting. Refinement procedures were preceded by calculation of a bulk-solvent model and estimates of σA and weight values, followed by conjugate gradient minimization. Molecular model building along with the automated refinement procedure was performed in o (28) by using σA weighted electron density maps.
As the structures of the uncomplexed scFv and the scFv-peptide complex were refined using different refinement techniques, for good comparison of both models, the unliganded Fv fragment was re-refined by using the maximum likelihood target and torsion angle refinement method (refinement statistics for both models are shown in Table 2).
Table 2.
Refinement statistics
| C219-peptide complex, 2AP2 | Unliganded C219, 1AP2 | |
|---|---|---|
| Resolution, Å | 500–2.40 | 500–2.36 |
| R, % | 23.1 | 22.0 |
| Rfree, % | 27.4 | 27.3 |
| Atoms, n | 3956 | 3722 |
| Residues, n | 501 | 468 |
| Water molecules, n | 164 | 102 |
| rms deviation bonds, Å | 0.006 | 0.005 |
| rms deviation angles | 1.19° | 1.02° |
| Residues with unfavorable φ, Ψ | 4 | 0 |
| rmsd improper dihedrals | 0.68° | 0.71° |
| Average B-factors, Å2 | 36 | 16 |
| Crossvalidated σA error, Å | 0.50 | 0.51 |
Buried surface area values were derived from the change in solvent accessible surface area values between the uncomplexed and complexed C219 binding site and peptide according to Connolly (29), by using a probe radius of 1.7 Å.
Results
Structure Determination.
The structure of scFv C219-peptide complex was solved by molecular replacement using the unliganded scFv C219 as a search model [PDB ID code 1AP2 (20)]. The results of the crystallographic refinement are summarized in Table 1. Geometrical parameters, analyzed by procheck (30) and what if (31) showed acceptable values for a crystal structure at 2.4 Å resolution (Table 2).
Overall Structure of the C219 NBD Peptide Complex.
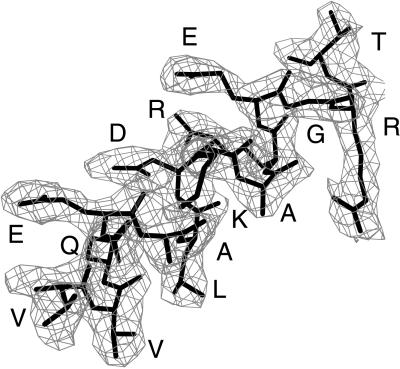
In the final model of the C219-peptide complex, the two molecules that were found in the asymmetric unit (throughout the text referred to as molecules I and II) displayed electron density for all 112 light chain and 120 heavy chain residues. Continuous and unambiguous density was also seen for the complete peptide (14 residues) in scFv C219 molecule II (Fig. 1). For molecule I, electron density was readily interpretable for residues 1–7. For residues 8–11, only backbone and Cβ atom density was observed; therefore, the side chain atoms of these residues were excluded. Electron density was poor for peptide residues 12, 13, and 14 in molecule I, and these were not included in the model.
Figure 1.
Electron density corresponding to the helical peptide epitope in molecule II. Density is calculated with coefficients |Fobs − Fcalc|ph(calc) before incorporation of the peptide into the model. The final conformation of the epitope is superimposed in thick bonds. Residues in the epitope are identified by single letter residue type. This figure was prepared with the program o (28).
At the N terminus of the light chain, three residues of the OmpA leader sequence (Phe-2L, Val-1L, and Arg 0L) could be identified in the electron density of both molecules. (For reference to residue numbers, the suffices L and H refer to antibody light and heavy chains, respectively, P refers to epitope peptide, and S refers to solvent atoms.) Near the C terminus of the heavy chain, additional electron density was observed for residues belonging to the carboxy-terminal c-myc epitope tag: Ser 121H, Gly 122H, and Ser 123H in molecule I and, additionally, Glu 124H and Gln 125H in molecule II. No density was seen for residues of the histidine tag. Although the linker peptide that connects the Fv C219 light and heavy chain was intact in the crystallization experiments (data not shown), no corresponding electron density was observable.
The C219 scFv molecules in the asymmetric unit are virtually identical. The individual light and heavy chains show rms deviations for α-carbon atoms of 0.35 and 0.39 Å, respectively, and a rms deviation of 0.52 Å was found when the complete C219 Fv fragments were superimposed.
Peptide Structure.
In accord with the peptide’s propensity in solution (20), both NBD-epitope peptides in the structure of the liganded scFv C219 exist in an amphipathic α-helical conformation. In molecule II, the peptide forms a 3.5-turn helix (Fig. 2) stabilized by additional intrapeptide hydrogen bonds between the Nɛ2 nitrogen and the carbonyl oxygen of Gln 3P and the Oδ2 of Asp 7P (Fig. 3A) and a salt bridge between the side chain Oδ1 of Asp 7P and the guanidinium nitrogen NH1 of Arg 10P. The 2.5-turn helical peptide in molecule I is virtually identical to the corresponding portion of the peptide in molecule II: Superposition of the epitope residues 1–7 of both peptides resulted in a rms deviation of 0.195 Å for Cα atoms.
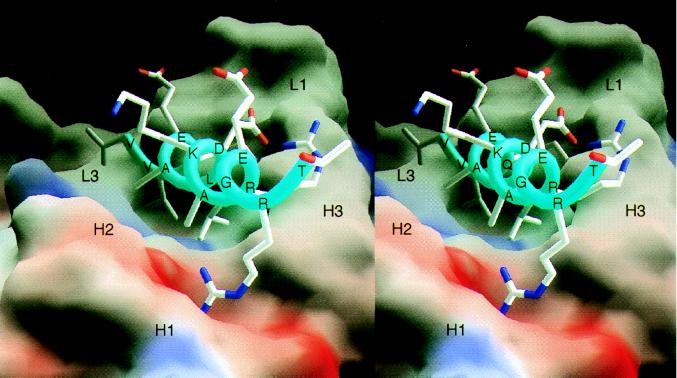
Figure 2.
Molecular surface representation of the scFv C219 (molecule II) binding site with the bound α-helical P-glycoprotein epitope peptide. The molecular surface is colored for electrostatic potential (red for negative charge, blue for positive charge). Peptide residues and the approximate locations of C219 heavy (H) and light chain (L) hypervariable loops are indicated. Fig. 2 was produced with the program grasp (32).
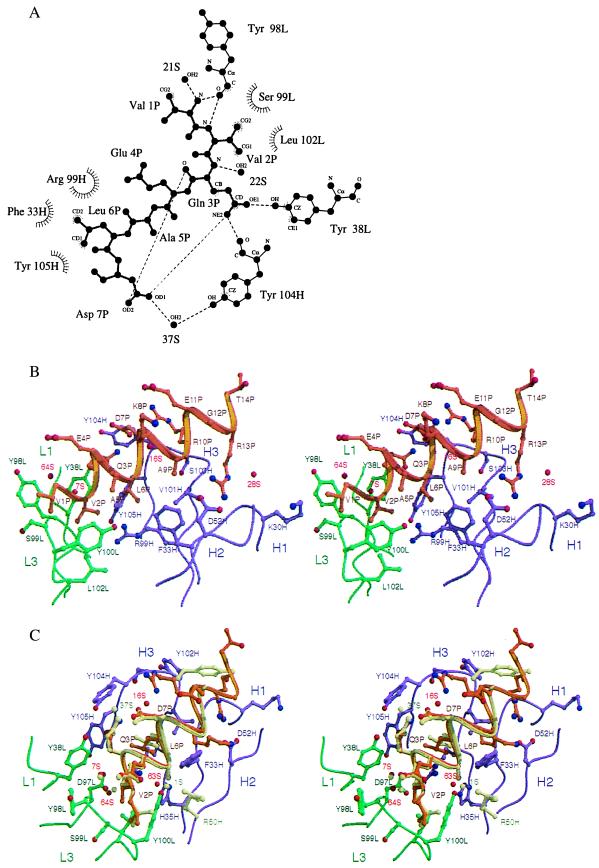
Figure 3.
Interactions between the α-helical peptide and the C219 binding site. (A) Two-dimensional ligplot (33) representation of the interactions between residues of the minimal NBD-epitope peptide (P), C219 heavy (H) and light chain (L) residues, and solvent molecules (S), as seen in molecule I. The residues that form van der Waals contacts with the peptide are depicted as labeled arcs with radial spokes pointing toward the peptide atoms with which they interact. C219 residues that form hydrogen bonds are shown in a ball-and-stick representation, and the hydrogen bonds are presented as dashed lines. Of all of the intrapeptide hydrogen bonds present in the structure, only the bonds between Gln 3P and Asp 7P are shown. (B) Stereoplot of the Fv-peptide interactions seen in molecule II. (C) Comparison of the bound NBD-epitope peptide in molecule I and II. In B and C, light (L) and heavy chain (H) residues and backbone positions of the scFv C219 are shown in green and magenta. Peptide backbone and side chains are shown in khaki for molecule I and in gold for molecule II. Positions of water molecules are indicated as red spheres. Different positions of binding site residues and water molecules in molecule I are also colored khaki. B and C were generated by using molscript (34) and raster3d (35).
The C219 NBD-Peptide Interface.
The binding of the 14-residue NBD peptide to the C219 binding site buries 700 Å2 (compared with 900–1,100 Å2 in other Fab-peptide complexes) of highly complementary, predominantly hydrophobic surface area from both binding partners (342 Å2, from the peptide; 354 Å2 from the binding site in molecule II). The C219 binding site is a shallow groove that is flanked on one side by an aromatic wall composed of tyrosine residues (loop H1 and H3) and by an open basic patch (loops H1 and H2) on the other side (Fig. 2). (Complementarity determining regions from the heavy chain are indicated by the prefix H, and from the light chain by the prefix L.) The floor of the groove is composed of residues Leu 102L, Tyr 100L, Arg 99H, Phe 33H, and Val 101H. The binding side is lined by residues Ser 99L, Tyr 98L, Tyr 38L, Tyr 105H, Tyr 104H, and Ser 103H. The C219 binding site mainly interacts with the hydrophobic face of the NBD-peptide formed by the side chains Val 1P, Val 2P, Leu 6P, and Ala 9P (Fig. 3 B and C). The side chain of Val 2P at the N-terminal side of the peptide is deeply embedded in a hydrophobic slot, making van der Waals contacts with the side chains of hypervariable loop L3 residues Ser 99L, Tyr 100L, and Leu 102L (Fig. 3B). The side chain of Leu 6P is similarly isolated within the C219 binding groove and makes van der Waals contacts with the side chains of Phe 33H, Arg 99H, and Tyr 105H. The side chains of Val 1P and Ala 9P face the hydrophobic patches on either side of the binding groove (Fig. 2). The aromatic wall of the binding side composed of tyrosine residues Tyr 38L, Tyr 98L, Tyr 104H, and Tyr 105H interacts with the N terminus of the peptide through hydrophobic stacking interactions and by hydrogen bonding (Fig. 3A). The backbone oxygen of Tyr 98L is involved in a capping interaction with the N-terminal nitrogen of the peptide and forms a hydrogen bond with the amide nitrogen of Val 2P; the Oɛ1 oxygen of Gln 3P forms a hydrogen bond with the hydroxyl oxygen of Tyr 38L and with the carbonyl oxygen of Tyr 104H. In molecule I, the latter interaction involves the Nɛ2 nitrogen of Gln P3 (Fig. 3A). Two water molecules are found to contribute to the shape complementarity between the N terminus of the peptide and the aromatic wall. Both interact with the backbone of the peptide, forming a hydrogen bond with the amide nitrogen of Gln 3P and a capping interaction with the terminal nitrogen of Val 1P (Fig. 3 B and C).
In the two noncrystallographically related molecules, different interactions are seen in the hydrogen bonding pattern of Asp 7P (Fig. 3C). In both molecules, a hydrogen bond interaction exists between the Oδ1 group of Asp 7P and Tyr 104H, mediated by a water molecule. In molecule I, this water molecule (37S) bonds with the hydroxyl oxygen of Tyr 104H; in molecule II, the water molecule (16S) interacts with the carbonyl oxygen of the same tyrosine residue (Fig. 3C). In molecule II, the Oδ1 oxygen of Asp 7P is furthermore involved in an intrapeptide charge interaction with guanidinium nitrogen NH1 of Arg 10. In addition, the aliphatic moiety of the Arg 10P forms a hydrophobic stacking interaction with the backbone of loop H3 residue Ser 103H. Only sparse density was seen for the Arg 10P side chain in molecule I. At the C-terminal end of the peptide in molecule II, the helix is tightly anchored to the C219 binding site through a salt bridge between the guanidinium group of Arg 13P and the side chain of Asp 52H. The interaction is strengthened further by hydrogen-bonding interaction with the backbone Lys 30H and by van der Waals interactions with Val 101H. The two different conformations of Tyr 102H (loop H3) in molecule I and II shown in Fig. 3C are attributable to different crystal packing interactions.
The minimal epitope of C219 has been mapped to the amino acid sequence VQEALD in the C-terminal half of Pgp and VQAALD in the N-terminal half (6). These epitope mapping results correspond well with the electron density seen in the crystal structure for the peptide in C219 molecule I. It is very likely that the binding motif seen in molecule I is a more typical binding mode for the epitope because it is not biased by the involvement of its peptide residues in crystal contacts, as is the case in molecule II.
Crystal Contacts.
The C terminus of the peptide in molecule II is involved in 13 crystal contacts, which is 10% of all of the crystal packing interactions. Heavy chain residues of three symmetry-related copies of molecule I play a dominant role in the contacts with the peptide. The packing interactions mediated by the peptide residues from molecule II include nine van der Waals interactions with Lys P8, Glu P11, Gly P12, Arg P13, and Thr P14; two hydrogen bonds with the carbonyl oxygen of Gly P12 and Oγ1 oxygen of Thr P14; and two charge interactions with the Nζ nitrogen of Lys P8 and with the Oɛ2 oxygen of Glu P11.
Comparison Between Unliganded and Complexed C219 Structures.
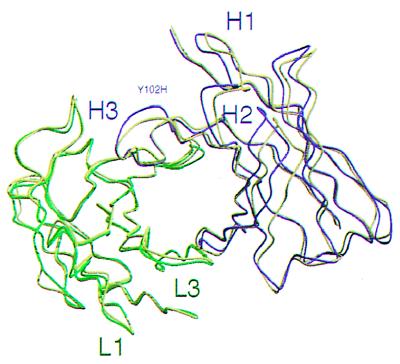
Superposition of the variable light chain framework regions (Fig. 4) reveals a 4° rotation between the variable heavy chains of the unliganded C219 Fv (PDB ID code 1AP2) and the C219 Fv peptide complex (PDB ID code 2AP2) about an axis parallel to the VL-VH pseudo-two-fold axis. This rotation leads to backbone rms deviations of 3.7 Å when the complete Fv fragments are superimposed. The largest coordinate displacements are found in loops H1 and H2. Backbone atoms of hypervariable loop H3 maintain their position on peptide binding except for Tyr 102H.
Figure 4.
Relative positions of the variable light and heavy chains in the dimers of the unliganded scFv C219 and the scFv C219-peptide complex, after superposition of the variable light chain framework regions. Light and heavy chain backbone positions of the C219-peptide complex are shown in green and magenta. The backbone of the unliganded scFv C219 is represented in yellow.
In the uncomplexed scFv C219 an initial stage of a so-called “false floor” is seen in the binding site in the shape of a salt bridge between Arg 99H of hypervariable loop H3 and Asp 97L of loop L3 (20). Water molecule 38S is involved in hydrogen bonding interactions orienting Arg 99H toward Tyr 100L (L3) and His 35H (H1). Another water molecule (12S) mediates a hydrogen bond between Tyr 100L and Arg 99H. On peptide binding, all three residues move into direct hydrogen bonding distance with each other, thereby coordinating a water molecule (1S in molecule I and 63S in molecule II) in a plane (Fig. 3C). This water molecule is located in the same position as 38S in the uncomplexed scFv C219. The water molecule mediating this false floor formation is very well defined in both complexed C219 molecules and is associated with the lowest B-factors found in the structure (1S, 13.8 Å2; 63S, 5.21 Å2). In molecule I, a fourth residue (Arg 50H) is involved in the formation of this false floor (Fig. 3C). The formation of the false floor, directed by Arg 99H, locks hypervariable loops L3, H1, H2, and H3 into a highly stable and highly complementary conformation holding the peptide in a tight grip.
Discussion
The structure of the scFv C219-peptide complex is characterized by a very unusual recognition of an α-helical peptide by an antibody. In previous structures of antibody Fab-peptide complexes, the ligand takes up either an extended or β-turn secondary structure (36). Antibodies are known that bind to protein epitopes that are α-helical [lysozyme (37, 38) and Taq DNA polymerase (39)] or mostly α-helical [myohemerythrin (40)]. In the structures of Fabs HyHEL10 and F9.13.7 in complex with hen egg white lysozyme and Guinea fowl lysozyme, a helix is involved as part of a discontinuous epitope and contributes exposed side chains, as the helix is largely buried within the lysozyme structure. In contrast to the antilysozyme antibodies, C219 interacts largely with the nonpolar face of the amphipathic helix. Crystal structures of epitope peptides from myohemerythrin and Taq DNA polymerase in complex with an antibody binding site display little or no α-helical character (41) and mostly adopt tight turn conformations. Other complexes of proteins and helical peptides are characterized by the formation of a compact globular assembly of α-helices, as seen in the calmodulin–target complexes (42).
A VL-VH domain rotation has been identified previously on binding of ligands to antibody fragments, illustrating the intrinsic flexibility of antibodies in adapting to the shape of an antigen. In most structures of Fab-peptide complexes, this conformational change is accompanied by a loss of VL-VH interface contacts that is mainly ascribed to the movement of the H3 loop out of the binding site (40, 43, 44). In the case of the binding of an epitope peptide by antimeningococcal antibody MN12H2 (45), the relative disposition of the variable domains resulted in a closer association of the binding site without major backbone rearrangements of the H3 loop, as is seen in the C219 Fv peptide complex. As in the MN12H2-peptide complex, the VL-VH domain shift and the resulting closure of the binding site is facilitated by the formation of a false floor at the bottom of the binding site.
Cross-Reactivity.
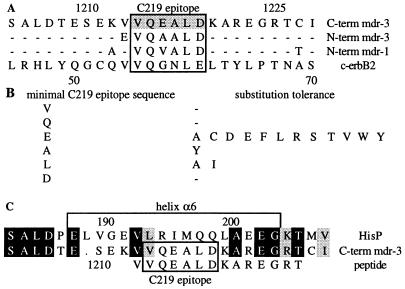
It has been recognized previously that anti-P-glycoprotein antibody C219 shows cross-reactivity with Pgp-related and unrelated proteins (6, 46). The discovery of C219’s cross-reactivity with a 185-kDa transmembrane oncoprotein (p185c-erbB2), which is overexpressed in several carcinomas, imposes complications in the use of C219 for detection of P-glycoprotein and diagnosis of multidrug resistance (14, 15). Liu et al. (14) compared peptide sequences and identified a C219 epitope-like region in p185c-erbB2 (Fig. 5A). The expected cross-reactivity of C219 with this epitope-like sequence VVQGNLE is consistent with our observations and the results from binding studies with amino acid substituted epitope peptides (6, 19). These studies showed that the first (Val 2P), second (Gln 3P), and final (Asp 7P) residues of the epitope are the least tolerant of sequence change. One, two, or multiple substitutions are tolerated at position 4 (Ala 5P), 5 (Leu 6P),and 3 (Glu 4P), as is shown in Fig. 5B.
Figure 5.
(A) Alignment of the human MDR1 and MDR3 sequences of P-glycoprotein with the C219 epitope homologous sequence in c-erbB2. The position of the C219 epitope sequence is indicated. (B) Amino acid positions of the minimal C219 epitope sequence and their tolerance for sequence change (19). (C) Sequence alignment of HisP with the C-terminal ATP-binding domain of P-glycoprotein (MDR3). The position of helix α6 is indicated. Black and gray boxes indicate identical and homologous residues, respectively.
Sequence differences between MDR Pgps and cross-reactive non-MDR Pgps are located at position 3 (Glu 4P) of the minimal epitope. According to the results from binding studies with these epitopes, the binding of C219 is slightly more efficient to the N-terminal half (VQAALD) than to the C-terminal half (VQEALD) of Pgp. The substitution of valine for Glu 4P in the N-terminal half of human class I Pgp (VQVALD) reduces its binding to the antibody (6). As seen in the crystal structure, Glu 4P is not involved in any interaction with the C219 binding site and can be substituted by multiple amino acids. However, substitution of Glu 4P by glycine (as in p185c-erbB2), histidine, lysine, methionine, asparagine, proline, and glutamine shows significant decrease in binding (19). A possible explanation for these results may lie in the disruption of the helical conformation of the peptide by these residues. Residues such as proline and glycine are known to be poor helix formers. The slight differences in binding affinity observed between the NBD epitopes described above are also very likely related to the helix stabilizing/destabilizing properties of the substituted amino acids. The crucial determinants of binding are identical in all cross-reactive Pgps, except for the Asp7Glu substitution in p185c-erbB2. In the crystal structure, Asp 7P appears to play a crucial role in internal hydrogen bond interactions stabilizing the α-helical conformation, thereby emphasizing the importance of the secondary structure of the eptide in the recognition by C219. Substitution of Asp 7P by any other amino acid resulted in a substantial loss of affinity (19). Thus, the p185c-erbB2 sequence predicted to be recognized by C219 should bind with a significantly lower affinity compared with the native epitope sequence. Nevertheless, reports of immunodetection of P-glycoprotein in breast carcinoma, a tumor that can also express p185c-erbB2, could confound the interpretation of P-glycoprotein detection and diagnosis of multidrug resistance
ATPase Inhibition.
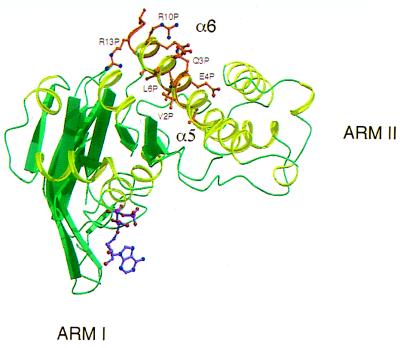
The NBD epitope sequence aligns with an α-helical segment (residues 192–205; Fig. 5C) in a recent crystal structure of HisP (47) corresponding to helix α6 (Fig. 6). Helix α6, located in arm II of the HisP structure, is surface-exposed and has continuous amphipathic characteristics similar to the NBD-epitope peptide in the C219 binding site. However, in the HisP structure, the hydrophobic side of the amphipathic α6 helix is partially buried. A superposition of the helical NBD-epitope peptide on its homologous sequence in HisP is shown in Fig. 6. If the structure of Pgp’s nucleotide binding domain is indeed similar to HisP and the sequence alignment is representative of the structural superposition, this implies either that the C219 epitope is flexible or that binding of C219 to helix α6 disrupts the NBD structure by its interaction with this partially buried epitope. In fact, the accessibility of the C219 epitope might be related to conformational differences between the apo and ATP-bound state of the NBD. Strong evidence supporting this idea is provided by the results from studies by Georges et al. (7) and Kokubu et al. (8) showing inhibition of P-glycoprotein’s ATP-binding capacity caused by the binding of C219. Other evidence for a possible connection between conformational changes in helix α6 and the ATP-binding site is found in G proteins and molecular motors, such as myosin and kinesin. The analogous helix in G proteins appears to be linked to nucleotide-induced conformational changes (48, 49). In NBDs, this conformational switching might involve the highly conserved LSGGQ motif C sequence located in helix α5, which is partially buried by helix α6 in HisP (Fig. 6). Welsh et al. (50) postulate that ATP-induced conformational changes in helix α6 might expose the LSGGQ motif, allowing interactions with the membrane-spanning domains. The possible shielding of this motif on binding of C219 to its epitope in helix α6 might explain its capacity to inhibit the binding of calcium channel blocker azidopine to Pgp (7). In a reverse manner, the results from a surface plasmon resonance study on the recognition of Pgp by C219 show an increased binding of Pgp by the antibody after preincubation with cyclosporin A (CsA), suggesting that binding of CsA, a competitive inhibitor of drug efflux, to Pgp could bring about a conformational change in the Pgp molecule such that its C219 epitope would become more readily available for interacting with the antibody (51). The binding sites of azidopine and CsA have both been located on predicted transmembrane regions 11 and 12 (52, 53), suggesting that ATP-induced conformational changes in NBD’s arm II, drug binding, and transport occur in concert with these transmembrane domains.
Figure 6.
Superposition of NBD helix onto HisP helix α6. Only a monomer of HisP is shown. The figure was produced with molscript (34) and raster3d (35).
Our results interpreted in the framework of the HisP structure permit us to postulate that the C219 epitope, the α6 helix in HisP nomenclature, is an indicator of the linkage between ATPase activity and drug binding. When the conformation of this region is fixed—for example, by C219 interaction—this linkage is broken and drug efflux is inhibited.
Conclusions
The structure of the scFv C219 in complex with its P-glycoprotein epitope shows that continuous α-helical epitopes can give rise to specific recognition of linear peptides in a helical conformation. In the structure reported here, the recognition is mostly driven by tight shape complementarity of nonpolar interactions between aromatic and long aliphatic side chains. Only a small number of electrostatic contacts are involved. Furthermore, binding affinity depends on the stability of the helical secondary structure; introduction of helix destabilizing residues can weaken the interaction.
When a peptide is bound in a helical conformation, its backbone cannot easily engage in hydrogen bonding to its binding protein, in contrast to peptides in an extended or β-turn conformation, because of the lateral intrapeptide hydrogen bonding along the helix. Because the antibody fold does not permit dramatic conformational transformations, as seen in calmodulin-target complexes (42), the binding of the helical NBD epitope is very superficial, covering only 30% of the peptide’s surface. As 70% of the bound peptide is exposed, considerable variation in sequence can be tolerated without disrupting C219 binding, thus explaining the variability of the context of the C219 epitope in crossreacting molecules.
Nevertheless, C219 is an important tool for studying the structural aspects of NBDs and their role in ATP-hydrolysis and drug transport. In the future, our results, together with the availability of the recombinant scFv, will be used to investigate the potential of improving the affinity and specificity of the interaction between C219 and its NBD epitope.
Acknowledgments
T. Signorelli participated in constructing, purifying, and crystal studies of the C219 single chain Fv. We thank Drs. M. Ikura and J. Forman-Kay for their helpful discussions. Collaboration with Dr. V. Ling was essential in the early stages of this work. Coordinates for the HisP structure were kindly provided by Dr. S.-H. Kim. J.vdE. is supported by an Amgen/OCI Fellowship, and the work was funded by the National Cancer Institute of Canada with funds from the Canadian Cancer Society.
Abbreviations
- Pgp
P-glycoprotein
- MDR
multidrug resistance
- NBD
nucleotide-binding domain
- Fv
variable fragment
- scFv
single-chain Fv
- ABC
ATP-binding cassette
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and diffraction data have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 2AP2).
References
- 1.Kartner N, Shales M, Riordan J R, Ling V. Cancer Res. 1983;43:4413–4419. [PubMed] [Google Scholar]
- 2.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 3.Kim R B, Fromm M F, Wandel C, Leake B, Wood A J, Roden D M, Wilkinson G R. J Clin Invest. 1998;101:289–294. doi: 10.1172/JCI1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg M F, Callaghan R, Ford R C, Higgins C F. J Biol Chem. 1997;272:10685–10694. doi: 10.1074/jbc.272.16.10685. [DOI] [PubMed] [Google Scholar]
- 5.Kartner N, Evernden-Porelle D, Bradley G, Ling V. Nature (London) 1985;316:820–823. doi: 10.1038/316820a0. [DOI] [PubMed] [Google Scholar]
- 6.Georges E, Bradley G, Gariepy J, Ling V. Proc Natl Acad Sci USA. 1990;87:152–156. doi: 10.1073/pnas.87.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georges E, Zhang J T, Ling V. J Cell Physiol. 1991;148:479–484. doi: 10.1002/jcp.1041480321. [DOI] [PubMed] [Google Scholar]
- 8.Kokubu N, Cohen D, Watanabe T. Biochem Biophys Res Commun. 1997;230:398–401. doi: 10.1006/bbrc.1996.5970. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein R S, Grogan T M, Kuszak J R, Jakate S M, Kluskens L F, Coon J S. Adv Pathol Lab Med. 1991;4:207–234. [Google Scholar]
- 10.Barrand M A, Twentyman P R. Br J Cancer. 1992;65:239–245. doi: 10.1038/bjc.1992.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J J, Hughes C S, Fine R L, Page R L. Cancer. 1996;77:1892–1898. doi: 10.1002/(SICI)1097-0142(19960501)77:9<1892::AID-CNCR20>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 12.Krishan A, Sauerteig A, Stein J H. Cytometry. 1991;12:731–742. doi: 10.1002/cyto.990120807. [DOI] [PubMed] [Google Scholar]
- 13.Norris M D, Bordow S B, Marshall G M, Haber P S, Cohn S L, Haber M. N Engl J Med. 1996;334:231–238. doi: 10.1056/NEJM199601253340405. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Sun D, Xia W, Hung M C, Yu D. J Natl Cancer Inst. 1997;89:1524–1529. doi: 10.1093/jnci/89.20.1524. [DOI] [PubMed] [Google Scholar]
- 15.Chan H S, Ling V. J Natl Cancer Inst. 1997;89:1473–1476. doi: 10.1093/jnci/89.20.1473. [DOI] [PubMed] [Google Scholar]
- 16.Jette L, Pouliot J F, Murphy G F, Beliveau R. Biochem J. 1995;305:761–766. doi: 10.1042/bj3050761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaulieu E, Demeule M, Pouliot J F, Averill-Bates D A, Murphy G F, Beliveau R. Biochim Biophys Acta. 1995;1233:27–32. doi: 10.1016/0005-2736(94)00239-l. [DOI] [PubMed] [Google Scholar]
- 18.Schinkel A H, Roelofs E M, Borst P. Cancer Res. 1991;51:2628–2635. [PubMed] [Google Scholar]
- 19.Childs S, Yeh R L, Georges E, Ling V. Cancer Res. 1995;55:2029–2034. [PubMed] [Google Scholar]
- 20.Hoedemaeker F J, Signorelli T, Johns K, Kuntz D A, Rose D R. J Biol Chem. 1997;272:29784–29789. doi: 10.1074/jbc.272.47.29784. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Navaza J. Acta Crystallogr A. 1987;43:645–653. [Google Scholar]
- 23.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszweski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Rice L M, Brünger A T. Proteins Struct Funct Genet. 1994;19:277–290. doi: 10.1002/prot.340190403. [DOI] [PubMed] [Google Scholar]
- 26.Pannu N S, Read R J. Acta Crystallogr A. 1996;52:659–668. [Google Scholar]
- 27.Adams P D, Pannu N S, Read R J, Brünger A T. Proc Natl Acad Sci USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M A. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Connolly M L. J Appl Crystallogr. 1988;16:548–558. [Google Scholar]
- 30.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 31.Vriend G. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 32.Nicholls A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 33.Wallace A C, Laskowski R A, Thornton J M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 35.Merrit E A, Murphy M E. Acta Crystallogr D. 1994;50:219–220. [Google Scholar]
- 36.Stanfield R L, Wilson I A. Curr Opin Struct Biol. 1995;5:103–113. doi: 10.1016/0959-440x(95)80015-s. [DOI] [PubMed] [Google Scholar]
- 37.Padlan E A, Silverton E W, Sheriff S, Cohen G H, Smith-Gill S J, Davies D R. Proc Natl Acad Sci USA. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lescar J, Pellegrini M, Souchon H, Tello D, Poljak R J, Peterson N, Greene M, Alzari P M. J Biol Chem. 1995;270:18067–18076. doi: 10.1074/jbc.270.30.18067. [DOI] [PubMed] [Google Scholar]
- 39.Murali R, Sharkey D J, Daiss J L, Murthy H M. Proc Natl Acad Sci USA. 1998;95:12562–12567. doi: 10.1073/pnas.95.21.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanfield R L, Fieser T M, Lerner R A, Wilson I A. Science. 1990;248:712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- 41.Tsang P, Rance M, Fieser T M, Ostresh J M, Houghten R A, Lerner R A, Wright P E. Biochemistry. 1992;31:3862–3871. doi: 10.1021/bi00130a018. [DOI] [PubMed] [Google Scholar]
- 42.Meador W E, Means A R, Quiocho F A. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 43.Rini J M, Schulze-Gahmen U, Wilson I A. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 44.Wilson I A, Stanfield R L. Curr Opin Struct Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 45.van den Elsen J, Vandeputte-Rutten L, Kroon J, Gros P. J Biol Chem. 1999;274:1495–1501. doi: 10.1074/jbc.274.3.1495. [DOI] [PubMed] [Google Scholar]
- 46.Thiebaut F, Tsuruo T, Hamada H, Gottesman M M, Pastan I, Willingham M C. J Histochem Cytochem. 1989;37:159–164. doi: 10.1177/37.2.2463300. [DOI] [PubMed] [Google Scholar]
- 47.Hung L W, Wang I X, Nikaido K, Liu P Q, Ferro-Luzzi Ames G F, Kim S H. Nature (London) 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 48.Smith C A, Rayment I. Biophys J. 1996;70:1590–1602. doi: 10.1016/S0006-3495(96)79745-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vale R D. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welsh M J, Robertson A D, Ostedgaard L S. Nature (London) 1998;396:623–624. doi: 10.1038/25234. [DOI] [PubMed] [Google Scholar]
- 51.Demeule M, Vachon V, Delisle M C, Beaulieu E, Averill-Bates D, Murphy G F, Beliveau R. Anal Biochem. 1995;230:239–247. doi: 10.1006/abio.1995.1469. [DOI] [PubMed] [Google Scholar]
- 52.Bruggemann E P, Currier S J, Gottesman M M, Pastan I. J Biol Chem. 1992;267:21020–21026. [PubMed] [Google Scholar]
- 53.Demeule M, Laplante A, Murphy G F, Wenger R M, Beliveau R. Biochemistry. 1998;37:18110–18118. doi: 10.1021/bi981992c. [DOI] [PubMed] [Google Scholar]