Abstract
Accumulative evidence suggests that more than 20 neuron-specific genes are regulated by a transcriptional cis-regulatory element known as the neural restrictive silencer (NRS). A trans-acting repressor that binds the NRS, NRSF [also designated RE1-silencing transcription factor (REST)] has been cloned, but the mechanism by which it represses transcription is unknown. Here we show evidence that NRSF represses transcription of its target genes by recruiting mSin3 and histone deacetylase. Transfection experiments using a series of NRSF deletion constructs revealed the presence of two repression domains, RD-1 and RD-2, within the N- and C-terminal regions, respectively. A yeast two-hybrid screen using the RD-1 region as a bait identified a short form of mSin3B. In vitro pull-down assays and in vivo immunoprecipitation-Western analyses revealed a specific interaction between NRSF-RD1 and mSin3 PAH1-PAH2 domains. Furthermore, NRSF and mSin3 formed a complex with histone deacetylase 1, suggesting that NRSF-mediated repression involves histone deacetylation. When the deacetylation of histones was inhibited by tricostatin A in non-neuronal cells, mRNAs encoding several neuronal-specific genes such as SCG10, NMDAR1, and choline acetyltransferase became detectable. These results indicate that NRSF recruits mSin3 and histone deacetylase 1 to silence neural-specific genes and suggest further that repression of histone deacetylation is crucial for transcriptional activation of neural-specific genes during neuronal terminal differentiation.
Regulation of gene expression is fundamental to all biological systems, and much of this regulation occurs at the level of transcription (1, 2). Recent evidence suggests that complex regulation of genes during development and in the adult is controlled primarily by the combinatorial action of three classes of transcriptional components: sequence-specific DNA-binding proteins (i.e., activators and repressors), coregulators (i.e., coactivators and corepressors), and components of the basal machinery (including RNA polymerases and core promoter factors) (3). Among these transcriptional components, the specificity of gene regulation is determined primarily by activators and repressors, which bind enhancer- and silencer-DNA elements, respectively. These regulators, in turn, bind specific coregulatory proteins to form complexes that activate or repress the transcription of individual genes (4). Recent evidence suggests that these events take place in concert with the reorganization of chromatin structure (5, 6).
In the mammalian nervous system, cell-specific expression of positive and negative transcriptional regulators containing Pit1-Oct2-Unc86 (POU), helix–loop–helix, or zinc-coordinated finger (Zn-finger) motifs in their DNA-binding domains (DBDs) have been shown to play crucial roles at specific stages of neural cell fate determination and subsequent neuronal differentiation (7, 8). Many of these transcription factors function as activators or repressors for other transcription factors and thus participate in cascades of positive and negative transcriptional regulation. Only a few factors, however, have been shown to be directly involved in establishing the terminally differentiated neuronal phenotype.
Neural restrictive silencer factor (NRSF) (9), also known as RE1-silencing transcription factor (10), functions as a transcriptional repressor of multiple neuron-specific genes in non-neuronal cells and tissues in mammals (8, 11). Many target genes of NRSF (12) encode proteins with neuronal functions, including ion channels [e.g., sodium channel type II (13)], neurotransmitter synthetases [e.g., choline acetyltransferase (14, 15) and dopamine β-hydroxylase (16, 17)], receptors [e.g., muscarinic acetylcholine receptor M4 (18, 19), nicotinic β2 receptor subunit (20), and NMDA receptor type 1 (21)], synaptosomal proteins [e.g., synapsin I (22, 23)], neuronal cell adhesion molecules [e.g., Ng-CAM (24) and L1 (25)], neuronal cytoskeleton [tubulin βIII (see refs. 11 and 12)], neurotrophic factors [e.g., brain-derived neurotrophic factor (BDNF) (26)], and neuronal growth-associated proteins [e.g., SCG10 (27)]. Recent studies of NRSF-deficient mouse by Anderson and his colleagues (11) revealed that the loss of NRSF in vivo promotes the derepression of expression of some of its target genes, including SCG10 and tubulin βIII, in nonneuronal cells. Loss of NRSF thus disrupts neural development and ultimately leads to embryonic lethality, although the actual cause of death is unknown. In spite of its prominent role in neural development, however, the transcriptional mechanisms underlying the neuron-specific gene repression by NRSF are largely unknown.
NRSF is a modular protein that contains an N-terminal repression domain, a DBD with eight consecutive zinc fingers followed by a highly basic region, and a C-terminal repression domain containing a single zinc finger motif (9, 28, 29). Little is known, however, about how the N- and C-terminal repression domains interact with other transcriptional coregulators and basal transcriptional machinery in nucleus. It is also not known whether these two repression domains cooperate with each other or function independently to repress various neuron-specific target genes in vivo. Here, we provide evidence that the N-terminal domain of NRSF represses transcription of NRS-bearing, neuron-specific target genes by binding the transcriptional corepressor mSin3 and thereby recruiting histone deacetylase (HDAC). This work recently has been presented in a preliminary form (30).
Materials and Methods
Plasmids and Expression Vectors.
The cDNAs of human NRSF, mSin3BLF, mSin3BSF, mSin3A, and MAD were obtained by reverse transcription–PCRs (RT-PCRs) by using sequence-specific oligonucleotide primers based on published sequences. Mouse NRSF cDNAs were obtained by conventional screening and, in part, by RT-PCR. The PCR products were subcloned in a TA cloning vector (Invitrogen), and the DNA sequences were confirmed by conventional sequencing by using an ABI-PE 377 sequencer (Applied Biosystems). Various subdomains of NRSF, mSin3B, or mSin3A were inserted in-frame into pcDNA3-G4DBD(1–147) and/or pcDNA3-FLAG (FLAG epitope tag: MDYKDDDDK) vectors by using standard molecular techniques. The luciferase gene-containing reporter plasmid (pGL3-S10PRS36+) was constructed by subcloning the promoter portion of pCAT3-S36+ (27) into pGL3-Basic (Promega). The GAL4 reporter plasmid pGL3-S10PR5GB was constructed by replacing the S36+ sequence of pGL-S10PRS36+ with five copies of the GAL4-DNA-binding site (excised from pGS-E4CAT; a gift from M. Horikoshi, University of Tokyo, Japan). Glutathione S-transferase (GST) fusion constructs were generated by the ligation of fragments of NRSF-N (1–153), NRSF-C (989–1097), or MAD (1–60) cDNAs in-frame into the pGEX plasmid series (Amersham Pharmacia). Details of these and other constructions are available on request.
Transfections and Reporter Gene Assays.
Neuro2a and PC12 cells were transfected with a series of GAL4DBD-NRSF plasmids by using Lipofectamine Plus (GIBCO/BRL). For luciferase assays, 0.8 × 105 cells in 24-well plates were transfected with 200 ng of luciferase reporter plasmid, 50 ng of effector plasmid, 150 ng of pcDNA3 as a carrier, and 50 ng of control Renilla luciferase vector (pRL-TK) (Promega) as an internal control for transfection efficiency. Cellular extracts were prepared 48 hr after transfection, and dual-luciferase activities were measured with a luminometer (Lummat LB96V) (EG & G, Salem, MA). All the transfection experiments were performed at least three times, and averages and SEs of 3–4 independent experiments with triplicate dishes were shown in the results.
Two-Hybrid Screening and Assays.
The N-terminal portion of NRSF (residues 1–76) was subcloned in-frame into pBTM116, a LexA vector (a gift from S. Hollenberg, Oregon Health Sciences University, Portland), and transfected into the yeast L40 strain (31). An expression library consisting of mouse T lymphoma (V13) cDNA (CLONTECH) then was introduced into the yeast and screened for growth on his−, leu−, trp− plates containing 2.5 mM 3-aminotriazole. His+ candidate clones were tested for β-galactosidase activity, and positive clones’ inserts were sequenced and analyzed by the sequence similarity search program of the National Center for Biotechnology Information.
In Vitro Binding Assays.
FLAG-tagged mSin3 products were labeled with [35S]methionine in vitro by using a single-tube, coupled transcription-translation system (Novagen). Various GST-fusion proteins, including GST-NRSF-N (1–153), GST-NRSF-C (989–1097), GST-Mad (1–60), or GST alone, were expressed in BL21 cells and purified by using glutathione-Sepharose beads (Amersham Pharmacia). The slurry then was incubated with 10 μl of 35S-labeled FLAG-tagged mSin3 products in 1 ml of buffer A, a PBS containing an additional 100 mM KCl and 0.25% NP-40, at 4°C for 2 hr. After the beads had been washed five times with buffer A, the bound proteins were eluted in Laemmli’s loading buffer, fractionated by SDS/PAGE, and detected by autoradiography. For the study of native mSin3 and HDAC1 protein interactions with NRSF-N, GST- and GST-fusion protein-coated beads (20 μg each) were incubated with nuclear extracts of NIH 3T3 cells (100 μg) in buffer B [20 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.5% NP-40/10% glycerol/protease inhibitor complete (Boehringer Mannheim)]. Nuclear extracts were prepared as described (27). The GST beads were washed, and bound proteins were eluted in Laemmli’s loading buffer and analyzed by Western blotting using appropriate antibodies. The primary antibodies used were anti-mSin3B antibodies, i.e., A-20 (against N terminus), Ak-12 [against the paired amphipathic helix 2 (PAH2) domain], E-20 (against carboxyl terminus), anti-mSin3A antibody K-20 (against amino acids 2–21), and anti-HDAC1 antibody C-19 (against amino acids 464–482) (all from Santa Cruz Biotechnology).
Immunoprecipitation and Western Blotting.
Subconfluent NIH 3T3 cells were transfected with 2 μg each of myc-tagged NRSF together with FLAG-tagged mSin3BLF or mSin3BSF by using Lipofectamine Plus reagent (GIBCO/BRL). Immunoprecipitations were performed under low-stringency conditions by using anti-FLAG M2 affinity gel (Kodak). The proteins bound to the gel beads were eluted in Laemmli’s loading buffer and analyzed by SDS/PAGE. Western blotting was performed by standard procedures, and the reacted products were visualized with horseradish peroxidase-conjugated, species-specific secondary antibody and an ECL-Plus chemiluminescence system (Amersham Pharmacia). The primary antibodies used were 9E10 (anti-myc-tag), A-20 (anti-mSin3B), and C-19 (anti-HDAC1).
Tricostatin A (TSA) Experiments, RT-PCR Analysis, and Immunocytochemistry.
Levels of neuronal and non-neuronal mRNAs were determined by semiquantitative RT-PCR. Total cellular RNA was isolated from NIH 3T3 cells treated or not with TSA (100 ng/ml), and 1-μg aliquots were reverse-transcribed with RNase H− reverse transcriptase (GIBCO). PCRs were performed with the following sequence-specific primer sets for detecting each mRNA: SCG10, (5′ primer) CGTGCACATCCCTACAAT, (3′ primer) CTTCAGCCAGACAGTTC; NMDA receptor type 1, (5′ primer) TACACTGCCAACTTGGCAGCTTTC, (3′ primer) CATGAAGACCCCTGCCATGTT; choline acetyltransferase, (5′ primer) GCTTACTACAGGCTTTAC, (3′ primer) GACAAACCGGTTGCTCAT; glial fibrillary acidic protein, (5′ primer) AAGCTCCAAGATGAAACCAACCTGA, (3′ primer) GCGATCTCGATGTCCAGGGC; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), (5′ primer) ACCACAGTCCATGCCATCAC, (3′ primer) TCCACCACCCTGTTGCTGTA. In most cases, PCR was performed for 30–35 cycles with an annealing temperature at 60°C. The control PCR for GAPDH was terminated at 20 cycles. The PCR products were resolved on 2% agarose gels and detected by ethidium bromide staining. Immunofluorescent staining of the TSA-treated and untreated cells grown on coverslips was performed with mAb TuJ1 (Babco, Richmond, CA) at a 1:200 dilution. Cellular images were captured by a BX60 microscope (Olympus) equipped with the MicroRadience confocal scanning system (Bio-Rad) and were reconstructed by use of Adobe photoshop software.
Results
Delineation of Repression Domains of NRSF.
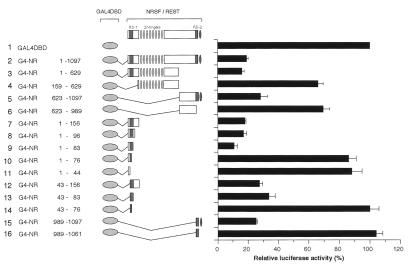
To analyze the mechanism of the NRSF-mediated transcriptional silencing, we first determined minimal regions required for the repression activity of NRSF. A series of GAL4-fused NRSF mutants were generated by using both human and mouse clones, and their repression activities were analyzed in transient cotransfection assays in Neuro2a cells using a luciferase reporter plasmid containing five GAL4-binding sites in the promoter-proximal region (Fig. 1). The full-length NRSF (Fig. 1, lane 2) and the N- and C-terminal halves, i.e., NRSF(1–629) and NRSF(623–1097) (Fig. 1, lanes 3 and 5), significantly repressed transcription (6- and 4-fold, respectively), whereas the internal domains, e.g., NRSF(159–629) and NRSF(623–989), caused only modest or no repression of transcription (Fig. 1, lanes 4 and 6), compared with levels of transcription obtained with a control plasmid containing the GAL4-DBD alone (Fig. 1, lane 1). Additional deletion experiments revealed that the N-terminal 83 aa residues contributed to the maximum transcriptional repression (Fig. 1, lanes 7–9). Surprisingly, however, deletion to residues 76 or 44 totally abolished repression (Fig. 1, lanes 10 and 11), suggesting that a repression domain is located between residues 44 and 83. When the NRSF fragment between amino acids 43 and 83 was tested for repression, it was found to strongly repress transcription (Fig. 1, lane 13), whereas deletion to residue 76 abolished the repression (Fig. 1, lane 14). Essentially the same results were obtained when HeLa and PC12 cells were used (data not shown). These results indicate that amino acid residues 76–83 are necessary and 43–83 are sufficient for the repression activity of the N-terminally located repressing domain (RD-1) of NRSF. On the other hand, we also found that the C terminus of NRSF (989–1097) significantly repressed transcription (Fig. 1, lane 15) compared with the control. Amino acid residues 1061–1097 containing the C-terminally located zinc finger are essential for this repression activity (Fig. 1, compare lanes 15 and 16). These results define amino acids 989–1097 as a second repression domain (RD-2) in NRSF. RD-1 and RD-2 can function independently and do not seem to have additive or synergistic effects on transcriptional repression. These results confirm and extend previous observations by others (29, 30) and confirm that both human and mouse NRSF have the same functional domains. Having shown that the RD-1 and RD-2 domains are evolutionally conserved, we proceeded to use these regions as bait for screening for molecules that interact with NRSF in the yeast two-hybrid screen (see below).
Figure 1.
The amino-terminal and carboxyl-terminal domains of NRSF are sufficient to mediate transcriptional repression. Various regions of NRSF, depicted schematically on the left with amino acid residue numbers, were fused in-frame to GAL4-DBD. The resultant series of GAL4 DBD-NRSF (G4-NR) constructs was cotransfected into Neuro2a cells with a luciferase reporter plasmid, pGL-S10PR5GB, containing the SCG10 promoter flanked by five copies of the GAL4-binding site linked to a luciferase gene, together with pRL-TK as an internal control.
Selective Interaction of mSin3BSF with NRSF-N-Terminal Domain in Yeast.
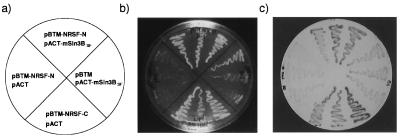
By screening 1.52 × 107 independent clones of a mouse lymphoma cDNA library with a LexA-fused RD-1 region (residues 1–76 of NRSF) used as a bait we obtained 32 positive colonies, of which 21 comprised overlapping clones. The DNA sequences of the overlapping clones predicted an ORF of 293 aa that was identical to the short isoform of mouse Sin3B (mSin3BSF) (32). The first 274 aa of this sequence are identical to the N-terminal sequence of the long form of mSin3B protein (mSin3BLF), although the remaining 19 C-terminal amino acid residues were unique. The mSin3A and B proteins are known to contain four PAHs (PAH1–4), which are thought to mediate protein–protein interactions (33). The mSin3BSF includes the N-terminal portion of mSin3BLF and contains PAH1 and PAH2 only. The screening results suggest that mSin3BSF or a related molecule may be a potential corepressor of NRSF-mediated transcriptional repression of neuronal-specific genes. We also used the RD-2 region of NRSF in the LexA and GAL4 systems to screen for additional corepressor(s). However, these screenings were unsuccessful because the NRSF-C-terminal RD-2 region alone activated reporter genes in both the LexA- and the GAL4-based two-hybrid systems (data not shown; see Fig. 2). We do not understood why the RD-2 domain functions as an activator in yeast, considering that RD-2 linked to the GAL4-DNA-binding site is a strong repressor in mammalian cells (e.g., Neuro2a and PC12) (see Fig. 1, lanes 5 and 15). In the in vivo interaction assay in yeast, mSin3BSF did show a strong association with NRSF-N (Fig. 2c Upper) and had only a weak interaction with an empty vector pBTM (Fig. 2c Right). On the other hand, the plasmid expressing the RD-2-containing C-terminal fragment of NRSF (pBTM-NRSF-C) showed activation in both LexA (Fig. 2c Lower) and GAL4 (not shown) systems. Thus, our yeast two-hybrid screen and assays revealed that mSin3BSF functions as a major partner of the NRSF-N-terminal RD-1 domain. However, the identity of the molecule(s) that interact with the NRSF-C-terminal RD-2 remains uncertain.
Figure 2.
Selective interaction of mSin3BSF with NRSF N-terminal domain in yeast. Interaction of NRSF subdomains and mSin3BSF was examined by using a two-hybrid assay in yeast. (a) Combinations of “bait” and “prey” constructs introduced into the yeast L40 strain. (b) Growth of yeast strains carrying each construct on a his−, leu−, trp− plate. (c) A filter assay for the β-galactosidase activity on a leu−, trp−, his+ plate. Note that β-galactosidase activity was induced in yeast cells expressing both NRSF-N and mSin3BSF, but not in the cells expressing mSin3BSF alone.
NRSF-N Forms a Complex with mSin3 and HDAC in Mammalian Cells.
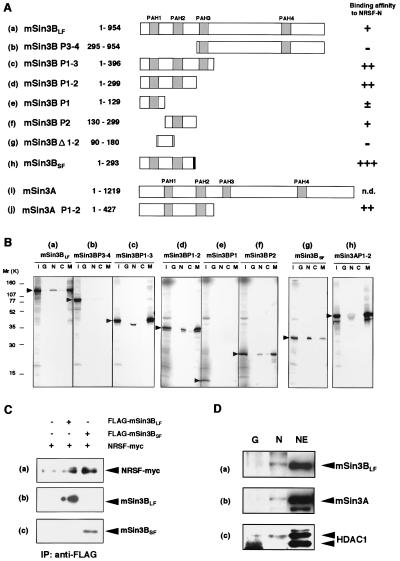
We next examined whether the N- and C-terminal domains physically interact with mSin3B and/or its structurally and functionally related homologue mSin3A in in vitro pull-down assays by using GST-fused proteins. Because the specific interaction of MAD with mSin3B is well characterized (34, 35), GST-MAD and GST alone were used as positive and negative controls, respectively. Among the series of mSin3B deletion constructs examined (see Fig. 3A), mSin3BSF and its equivalent (i.e., mSin3B P1–2) had strong affinity for NRSF-N (Fig. 3 Bg and Bd and Ah and Ad). A construct containing the PAH2 region (mSin3B P2) represented significant binding to NRSF-N (Fig. 3 Bf and Af), whereas the adjacent PAH1 region (i.e., mSin3B P1) was less efficient (Fig. 3 Be and Ae). In contrast, constructs containing the C-terminal half of mSin3B, encompassing PAH3 through PAH4 domains (mSin3B P3–4), as well as the intervening regions between PAH1 and PAH2 (mSin3B Δ1–2) showed very little or no interaction with NRSF-N [Fig. 3 Bb and Ab and Ag). Mammalian Sin3A (PAH1-PAH2) also bound to NRSF-N with similar or a little weaker affinity compared with mSin3B (see Fig. 3 Ad and Aj and Bd and Bh). Thus, mSin3B and mSin3A interacted with the NRSF-N-terminal domain through their PAH1-PAH2 (or PAH2) domain(s). Mammalian Sin3B, however, does not seem to interact with NRSF-C (RD-2), because a little or no association was observed in these in vitro assays (see Fig. 3B, lane C).
Figure 3.
NRSF-N forms a complex with mSin3 and HDAC in mammalian cells. (A) Regions of mSin3 involved in the interaction with NRSF-N. Relative strengths of NRSF-N and mSin3 interactions were assessed by quantification of the autoradiograms and scored from background level (−) to strong interaction (+++); n.d., not determined. PAH domains and a unique region in mSin3BSF are distinguished by hatched and shaded boxes, respectively. Numbers correspond to amino acid positions in the protein. (B) Specific binding of NRSF-N and mSin3 in vitro. GST-NRSF fusion proteins, i.e., GST-NRSF-N (N), GST-NRSF-C (C), GST alone (G), or GST-MAD (M), were incubated with 35S-labeled FLAG-tagged mSin3 deletion constructs. Specifically bound mSin3 subforms then were resolved on an SDS/PAGE gel and detected by autoradiography. An aliquot comprising one-fifth of the total input protein was loaded in lane I as a control. (C) Interaction of NRSFMyc and mSin3BFLAG in NIH 3T3 cells. Expression plasmids encoding Myc-tagged NRSF and the long or short isoform of FLAG-tagged mSin3B were cotransfected into NIH 3T3 cells, and the expressed mSin3BFLAG was immunoprecipitated with anti-FLAG M2 antibody. Proteins that interacted with mSin3BFLAG were resolved by SDS/PAGE and visualized by Western blotting. Shown are fluorograms of NRSF, visualized by using the anti-myc antibody 9E10 (a), and mSin3BLF (b) and mSin3BSF (c), visualized by using the anti-mSin3B antibody A-20. (D) Association of NRSF-N with mSin3A, mSin3B, and HDAC1 in NIH 3T3 cells. GST pull-down experiments show that NRSF-N interacts with mSin3A, mSin3BLF, and HDAC1. Nuclear extracts of NIH 3T3 cells were incubated with either immobilized GST alone (lane G) or GST-NRSF-N fusion protein (lane N), precipitated, and visualized by using anti-mSin3B (A-20) (a), anti-mSin3A (K-20) (b), or anti-HDAC1 (C-19) (c) antibody. Nuclear extracts were loaded into lane NE as a control.
We next asked whether the interaction between NRSF and mSin3B also occurs in vivo. In cotransfection experiments using expression plasmids encoding Myc-tagged NRSF and FLAG-tagged mSin3B isoforms in NIH 3T3 cells, Western blot analysis showed that immunoprecipitation with a FLAG antibody caused the coprecipitation of NRSF with mSin3B (Fig. 3C). A similar interaction also was demonstrated with anti-Myc antibody immunoprecipitation followed by FLAG antibody detection (data not shown).
Mammalian Sin3 and its yeast homologue are known to exert their effects through the interaction with HDACs (32, 36–42), which silence gene expression in vivo by altering chromatin structure (43). Therefore, we tested whether the interaction of NRSF with mSin3B involves HDACs. For this purpose, we performed in vitro pull-down assays by using nuclear extracts of NIH 3T3 cells expressing the GST-fused N-terminal domain of NRSF or GST alone, and probed proteins bound to these complexes with antibodies against mSin3B, mSin3A, and HDAC1 (Fig. 3D). These experiments showed that GST-NRSF pulled down mSin3BLF, mSin3A, and HDAC1 (Fig. 3D, lane N), whereas GST alone failed to pull down mSin3A, mSin3B, or HDAC (Fig. 3D, lane G). A similar interaction also was demonstrated to take place by using nuclear extracts of Neuro2a cells (data not shown). These results indicate that the NRSF-mSin3 complex recruits HDAC1 and suggest the involvement of histone deacetylation in transcriptional repression by the NRS-NRSF system.
Inhibition of HDAC Relieves Transcriptional Repression by NRSF.
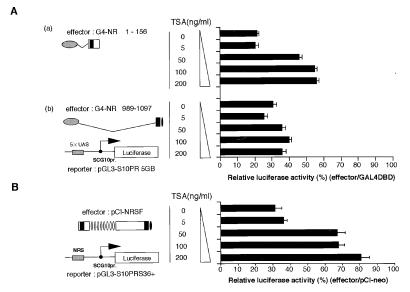
To determine whether histone deacetylation is essential for transcriptional silencing via NRSF in vivo, we attempted to block transcriptional repression with TSA, a specific inhibitor of HDACs (44). Neuro-2a cells were transfected with GAL4-fused NRSF expression plasmids (either N- or C-terminal domains) together with a GAL4-binding, site-containing, luciferase-expressing reporter plasmid and treated with various concentrations of TSA. Twenty-four hours after the TSA treatment, the cells were lysed and the luciferase activities were measured. Transcriptional repression by the N-terminal portion of NRSF (GAL4 DBD-NRSF 1–156) was derepressed at least 2.5-fold by treatment with 50–100 ng/ml TSA (Fig. 4Aa). In contrast, transcriptional repression by the C-terminal domain of NRSF (GAL4 DBD-NRSF 989-1097) was not affected, even at 200 ng/ml TSA (Fig. 4A). Repression by the full-length NRSF via NRS also was inhibited 2.6-fold by 200-ng/ml treatment (Fig. 4B). These results strongly suggest that histone deacetylation is involved in the transcriptional silencing via RD-1 in the N-terminal domain of NRSF. In contrast, repression via RD-2 at the C terminus of NRSF apparently uses a mechanism other than histone deacetylation to repress transcription.
Figure 4.
Inhibition of HDAC activity by TSA relieves transcriptional repression by RD-1 but not that by RD-2. (A) TSA relieves transcriptional repression mediated by the N-terminal (a) but not by the C-terminal (b) domain. Neuro2a cells were transfected with plasmids expressing either GAL4DBD-NRSF-N-terminal (G4-NR 1–156) or GAL4DBD-NRSF-C-terminal (G4-NR 989-1097) together with a luciferase reporter plasmid pGL-S10PR5GB. After 24 hr, the cells were treated with various concentrations of TSA (0–200 ng/ml) for an additional 24 hr. Relative luciferase activities are calculated relative to the activity obtained by using the GAL4DBD plasmid as an effector. (B) TSA relieves NRS-mediated transcriptional repression by NRSF. Neuro2a cells were transfected with the NRSF expression plasmid pCI-NRSF and the NRS-containing reporter plasmid pGL-S10PRS36+ and treated as described above.
Ectopic Activation of Neuron-Specific Genes by TSA in Non-Neuronal Cells.
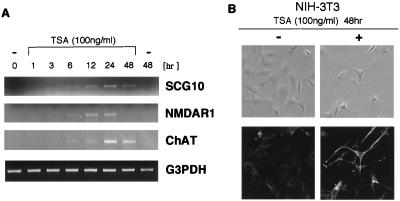
Do neuron-specific genes of NRSF target become activated if the histone deacetylation is blocked? To answer this question we treated NIH 3T3 cells with TSA and determined the levels of mRNAs encoding neuron-specific genes by using PCR. The genes we examined included growth-associated gene SCG10, NMDA receptor type 1 (NMDAR1), and choline acetyltransferase. When NIH 3T3 cells were treated with TSA, each of the NRS-regulated neuronal mRNAs examined became detectable, whereas the levels of constitutively expressed mRNAs, e.g., glyceraldehyde-3-phosphate dehydrogenase, remained unchanged (Fig. 5A). In contrast, mRNA encoding glial fibrillary acidic protein, a glial marker, also was not affected by TSA (not shown). Transcripts encoding SCG10 and other neuron-specific genes were up-regulated in TSA-treated C6 glioma as well (data not shown). We also observed that neuron-specific tubulin βIII gene was induced in vivo as shown by the appearance of immunostaining using TuJ1 antibody (Fig. 5B). These results are consistent with the idea that neural-specific gene regulation by NRSF involves histone acetylation and deacetylation.
Figure 5.
Ectopic activation of neuron-specific genes in TSA-treated fibroblasts. NIH 3T3 cells were incubated with TSA (100 ng/ml) for the times indicated, and the expression levels of various mRNAs were determined by RT-PCR. (A) Three neuron-specific genes: SCG10, NMDA-receptor type 1 (NMDAR1), and choline acetyltransferase (ChAT), as well as a constitutively expressed control gene glyceraldehyde-3-phosphate dehydrogenase (G3PDH). (B) Immunohistochemical staining using TuJ1 antibody reveals that neuron-specific βIII tubulin also is induced in the TSA-treated NIH 3T3 cells (Lower). Note that cell morphology is changed in the TSA-treated cells (Upper, phase contrast).
Discussion
Our experiments demonstrate that NRSF recruits mSin3 via the RD-1 domain and forms a complex with HDAC. These results suggest that NRSF represses transcription by facilitating the deacetylation of nearby histones. Consistent with this model, inhibition of HDAC caused the derepression of NRS-regulated, neuron-specific genes in nonneuronal cells. In contrast, the C-terminal repression domain represses target gene transcription independently of the action of HDAC. NRSF thus is a bipartite repressor of gene expression.
Transcriptional repression via the recruitment of HDAC has been characterized for many transcription factors including UME6, Mad, Mxi1, nuclear hormone receptors, Rb, YY1, CBF1, and Ikaros (refs. 32, 37–40, 43, and 45–48; for reviews see refs. 49–52). In most cases, these repressors recruit HDAC by binding the corepressor mSin3 or NCoR/SMRT (32, 37–40, 48), although in the case of Rb and YY1, the interaction with HDAC could be direct (45, 46). There are also cases in which some corepressor complexes targeted by sequence-specific transcription factors do not contain Sin3 although they contain HDAC (53). We have demonstrated that NRSF directly associates with the PAH1 through PAH2 regions of mSin3A and mSin3B. Although early studies on mSin3-mediated transcriptional repression (32, 38) identified NCoR in the complex containing mSin3, HDAC, and MAD, our preliminary Western blot analysis indicated that neither NCoR nor SMRT was included in the complex containing NRSF and mSin3 (T.A. and N.M., unpublished observations). Because the site of HDAC interaction on mSin3 is known to depend on the PAH3–4 region (32), a reasonable scenario would be that NRSF directly binds mSin3 through PAH1–2, thereby recruiting HDAC indirectly through the mSin3 PAH3 region to the complex containing NRSF. Although the complex may contain additional corepressors, mSin3 binding apparently is sufficient to recruit HDAC. Our finding that incubation of NIH 3T3 cells with the HDAC inhibitor TSA relieves NRS-mediated repression of transcription of SCG10 and other neuron-specific genes also supports the model that transcriptional repression by NRSF is mediated by HDAC. The tissue-specific formation of a DNase I-hypersensitive site in the SCG10 proximal promoter in the presence of NRS, but not in the absence of NRS (54), also is consistent with the idea that alternating chromatin structure triggered by the HDAC activity is involved in the mechanisms of NRS-mediated gene silencing.
The possibility remains, however, that the recruitment of HDAC via mSin3 by itself may not be sufficient for repressing NRS-mediated neuronal genes in vivo; HDAC-mediated transcriptional repression might be only one aspect of this bipartite repressor. We have shown that RD-2 in the C terminus represses transcription in a manner independent of TSA, suggesting that RD-2 inhibits transcription by a mechanism unrelated to histone deacetylation. One possibility is that RD-2 interacts directly with core promoter factors. Recent studies by others have shown that the transcription factors RBP/CBF1 and Eve bind TFIID, whereas the HMGP1-SMRT-mSin3 complex binds TFIIB (55–58). Thus, the recruitment of such core promoter factors may be necessary for NRSF to be recruited to the promoter region. Because HDAC probably affects only one or two histone octamers in chromatin (36), relocation of the NRSF-mSin3-HDAC complex to the core promoter region would be essential for gene silencing by NRSF. Furthermore, transcription factors other than the histones also might be targets for deacetylation (59). It, therefore, will be of great interest to identify and characterize proteins that interact with RD-2 to explain completely the mechanisms by which NRSF blocks the expression of neuron-specific genes in non-neuronal cells.
Identification of RD-1 at the N terminus of NRSF and the demonstration that it associates with mSin3 support the idea that a recently identified neural-specific splicing variant of NRSF (ref. 60; Y.N. and N.M., unpublished results) functions as a transcriptional repressor. This neuronal variant of NRSF contains the entire RD-1 region and five consecutive zinc fingers, but lacks the C-terminal half of authentic NRSF. The neuronal variant is a major isoform of NRSF expressed in neurons (Y.N. and N.M., unpublished observations) and is induced after neuronal stimulation (60). Its physiological function, however, currently is unknown. Our preliminary results suggest that the neuronal variant has repressing activity in a heterologous expression system (Y.N. and N.M., unpublished data). However, the expression of both the authentic long form and the short isoforms of neuronal variants also may contribute to the fine tuning of the expression of a variety of neuron-specific genes in individual subsets of neurons.
Finally, the functional cooperation between NRSF and HDAC may explain the developmental defects observed in NRSF-deficient mice and chick embryos expressing dominant-negative NRSF (dnNRSF) (11). In both NRSF-deficient mice and dnNRSF-overexpressing chick embryos, a subset of neuron-specific genes was ectopically expressed in non-neuronal cells (11). Given our results, this finding can be interpreted as being caused by the inability of these organisms to recruit the mSin3-HDAC complex to the NRS sites (or further to the appropriate promoter regions). Interestingly, the NRSF-deficient mice apparently underwent normal induction and patterning of the nervous system, even though the mice eventually died between embryonic day 9.5 (E9.5) and E10. That the mice die after neuronal initial differentiation (E9.5–10), and not before neuronal commitment (E8), suggests that NRSF-mediated repression is not essential for early development of neuronal progenitors, but is essential during neuronal terminal differentiation stages. NRS-mediated gene regulation seems to be initiated at least around E9-E9.5, as evidenced by the pattern of TuJ1 staining (tubulin βIII expression) at E9.25. It is probably at this stage that the configuration of chromatin changes in and around the NRS-bearing neuronal genes because of histone deacetylation resulting from the recruitment of HDAC via mSin3 by NRSF. These considerations favor the idea that the fundamental role of NRSF is to repress NRS-bearing neuronal genes in non-neuronal tissues and differentiated neurons; it may not be needed to repress neuronal genes before specification stage of neural development. Derepression of neuron-specific genes resulting from reduced transcription of the NRSF gene or from the inhibition of NRSF by other factors may be crucial for establishing neuronal terminal differentiation as well as for maintaining neural functions in adulthood.
Acknowledgments
We thank S. M. Hollenberg for plasmids pBTM116 and pVP16 and yeast strains L40 and AMR70, M. Horikoshi and M. Fujii for pGS-E4CAT, M. Sato for help with immunohistochemical staining, and T. Okazaki for recombinant protein production. We thank D. J. Anderson for helpful discussions and encouragement and D. Saffen for critical reading of the manuscript. We also thank Y. Ono and N. Takahashi for continuous support and encouragement to Y.N. and T.K., respectively. Technical assistance of S. Muraoka, I. Nakano, and T. Miura are greatly acknowledged. The early phase of this work was performed in the laboratory of “Inheritance and Variation” at Keihanna Plaza and was supported, in part, by a grant from Precursory Research for Embryonic Science and Technology (PRESTO), Science and Technology Corporation of Japan (to N.M.).
Abbreviations
- NRS
neural restrictive silencer
- NRSF
NRS factor
- RD
repression domain
- HDAC
histone deacetylase
- TSA
tricostatin A
- GST
glutathione S-transferase
- RT-PCR
reverse transcription–PCR
- DBD
DNA-binding domain
- PAH
paired amphipathic helix
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McKnight S, Yamamoto Y, editors. Transcriptional Regulation. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 2.Holstege F C P, Young R A. Proc Natl Acad Sci USA. 1999;96:2–4. doi: 10.1073/pnas.96.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodrich J A, Cutler G, Tjian R. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 4.Chi T, Lieberman P, Ellwood K, Carey M. Nature (London) 1999;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 5.Struhl K. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 6.Wolffe A P. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 7.Simpson P. Neuron. 1995;15:739–742. doi: 10.1016/0896-6273(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 8.Schoenherr C J, Anderson D J. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 9.Schoenherr C J, Anderson D J. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 10.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z F, Paquette A J, Anderson D J. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 12.Schoenherr C J, Paquette A J, Anderson D J. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraner S D, Chong J A, Tsay H J, Mandel G. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 14.Lonnerberg P, Schoenherr C J, Anderson D J, Ibanez C F. J Biol Chem. 1996;271:33358–33365. doi: 10.1074/jbc.271.52.33358. [DOI] [PubMed] [Google Scholar]
- 15.Li Y P, Baskin F, Davis R, Hersh L B. J Neurochem. 1993;61:748–751. doi: 10.1111/j.1471-4159.1993.tb02181.x. [DOI] [PubMed] [Google Scholar]
- 16.Ishiguro H, Kim K T, Joh T H, Kim K S. J Biol Chem. 1993;268:17987–17994. [PubMed] [Google Scholar]
- 17.Ishiguro H, Kim K S, Joh T H. Brain Res Mol Brain Res. 1995;34:251–261. doi: 10.1016/0169-328x(95)00170-w. [DOI] [PubMed] [Google Scholar]
- 18.Mieda M, Haga T, Saffen D W. J Biol Chem. 1997;272:5854–5860. doi: 10.1074/jbc.272.9.5854. [DOI] [PubMed] [Google Scholar]
- 19.Wood I C, Roopra A, Buckley N J. J Biol Chem. 1996;271:14221–14225. doi: 10.1074/jbc.271.24.14221. [DOI] [PubMed] [Google Scholar]
- 20.Bessis A, Champtiaux N, Chatelin L, Changeux J P. Proc Natl Acad Sci USA. 1997;94:5906–5911. doi: 10.1073/pnas.94.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai G, Norton D D, Prenger M S, Kusiak J W. J Biol Chem. 1998;273:1086–1091. doi: 10.1074/jbc.273.2.1086. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Suzuki T, Mori N, Greengard P. Proc Natl Acad Sci USA. 1993;90:1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoch S, Cibelli G, Thiel G. J Biol Chem. 1996;271:3317–3323. doi: 10.1074/jbc.271.6.3317. [DOI] [PubMed] [Google Scholar]
- 24.Kallunki P, Jenkinson S, Edelman G M, Jones F S. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 25.Kallunki P, Edelman G M, Jones F S. J Cell Biol. 1997;138:1343–1354. doi: 10.1083/jcb.138.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmusk T, Palm K, Lendahl U, Metsis M. J Biol Chem. 1999;274:1078–1084. [PubMed] [Google Scholar]
- 27.Mori N, Schoenherr C, Vandenbergh D J, Anderson D J. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 28.Tapia-Ramirez J, Eggen B J, Peral-Rubio M J, Toledo-Aral J J, Mandel G. Proc Natl Acad Sci USA. 1997;94:1177–1182. doi: 10.1073/pnas.94.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiel G, Lietz M, Cramer M. J Biol Chem. 1998;273:26891–26899. doi: 10.1074/jbc.273.41.26891. [DOI] [PubMed] [Google Scholar]
- 30.Mori N, Naruse Y, Aoki T, Kojima T. FASEB J. 1999;13:A1333. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Clark I, Nicholson P R, Herskowitz I, Stillman D J. Mol Cell Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper S E, Qiu Y, Sharp P A. Proc Natl Acad Sci USA. 1996;93:8536–8540. doi: 10.1073/pnas.93.16.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 38.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 39.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 40.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 41.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 43.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 45.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 46.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsieh J J, Zhou S, Chen L, Young D B, Hayward S D. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koipally J, Renold A, Kim J, Georgopoulos K. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe A P. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 50.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 51.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 52.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 53.Wade P A, Jones P L, Vermaak D, Wolffe A P. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 54.Vandenbergh D J, Wuenschell C W, Mori N, Anderson D J. Neuron. 1989;3:507–518. doi: 10.1016/0896-6273(89)90209-2. [DOI] [PubMed] [Google Scholar]
- 55.Olave I, Reinberg D, Vales L D. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C, Manley J L. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sutras-Grau M, Bianchi M E, Bernues J. J Biol Chem. 1999;274:1628–1634. doi: 10.1074/jbc.274.3.1628. [DOI] [PubMed] [Google Scholar]
- 58.Wong C W, Privalsky M L. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1998;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 60.Palm K, Belluardo N, Metsis M, Timmusk T. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]