Abstract
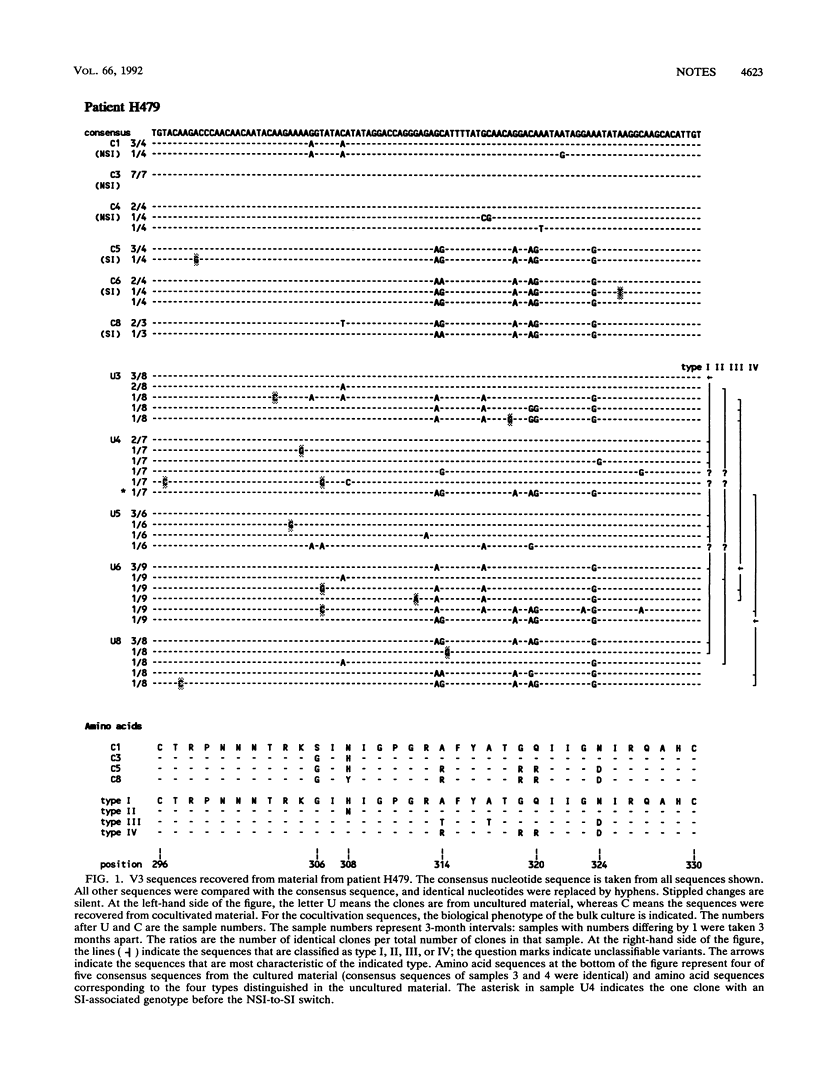
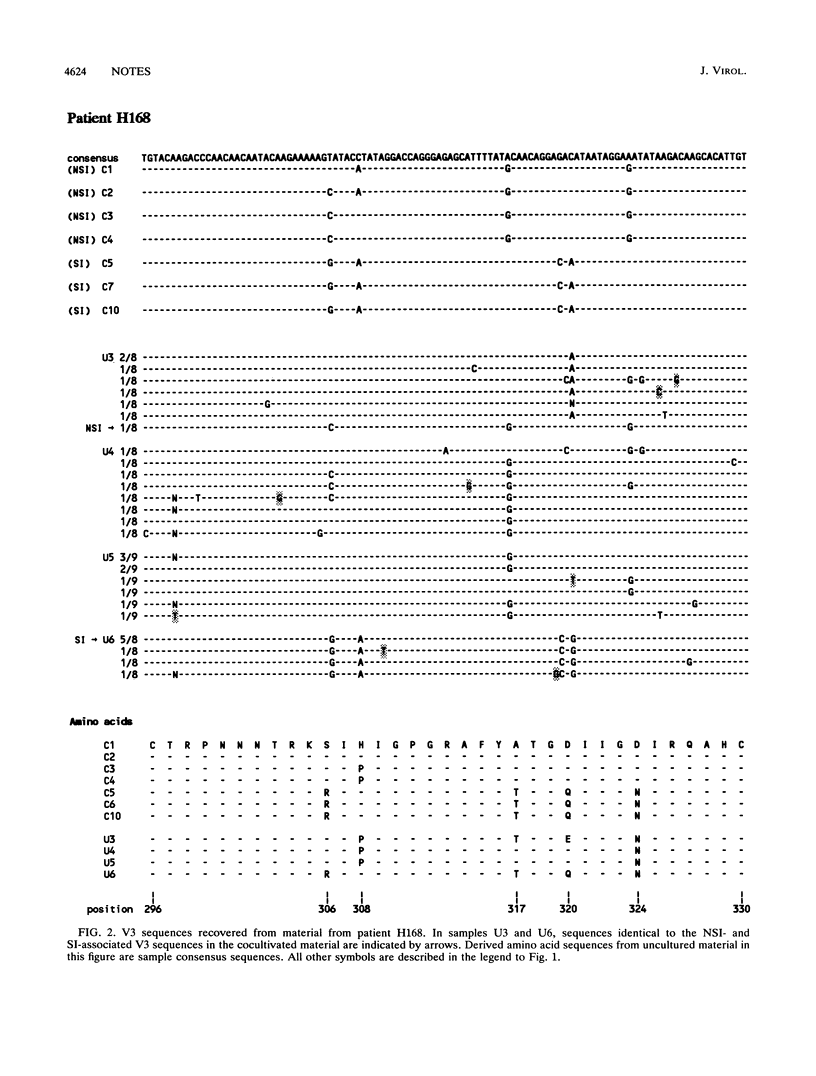
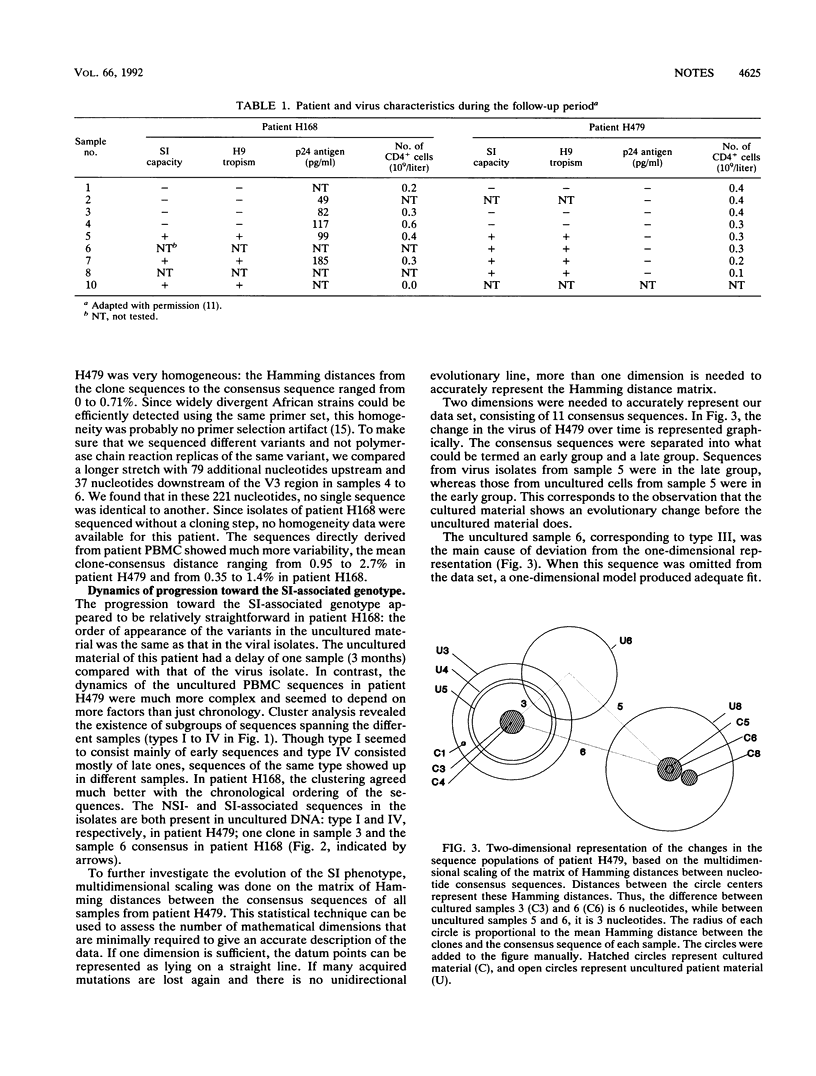
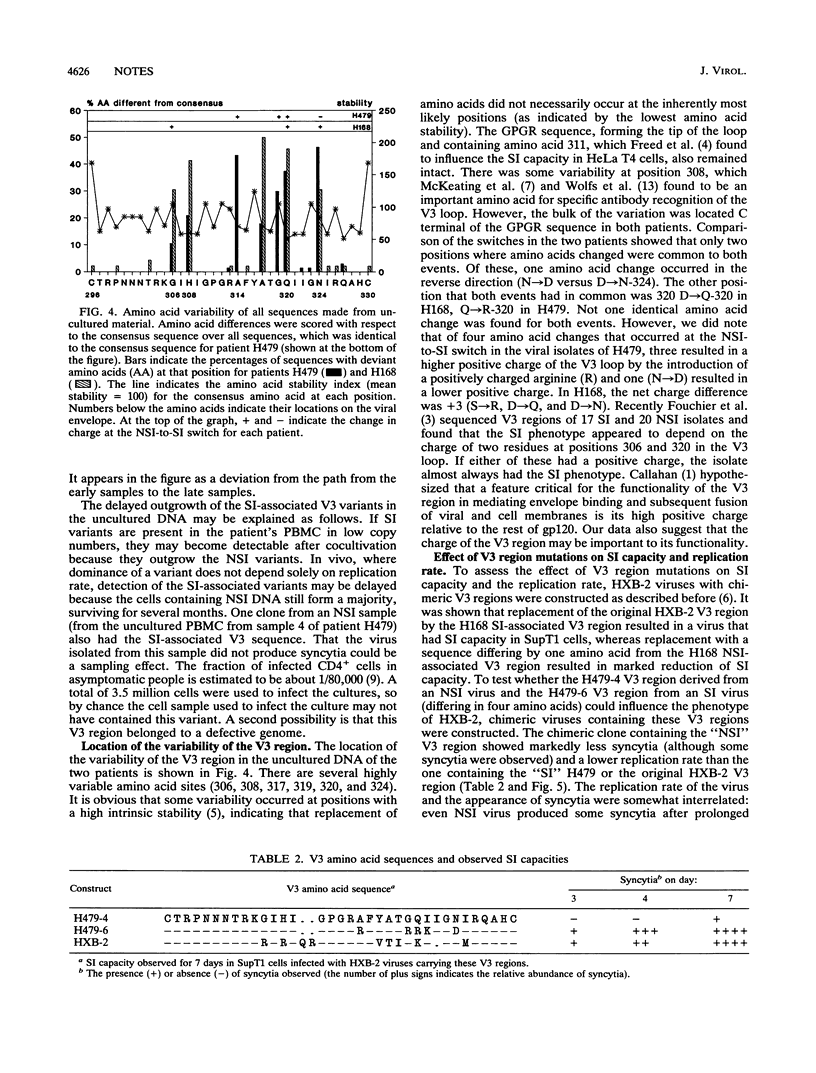
The third variable domain (V3) of the envelope gene of human immunodeficiency virus type 1 contains a major neutralization epitope and determinants of syncytium-inducing (SI) capacity and replication rate (reviewed by J. P. Moore and P. L. Nara, AIDS Suppl. 2:S21-S33, 1991). Sequences were generated from DNA of samples taken 3 months apart over a period of 24 and 30 months from peripheral blood mononuclear cells (PBMC) of two individuals, both before and after cocultivation with uninfected donor PBMC. The isolated virus shifted from the non-syncytium-inducing (NSI) phenotype to the SI phenotype during the study period. This shift was associated with distinct changes in the V3 domain in both patients. The association of the phenotype shift with the V3 sequence changes was confirmed by construction of viruses with chimeric V3 loops. The shift from NSI- to SI-associated V3 variants was also seen in the uncultured PBMC of both patients, but not until 3 and 9 months after the detection of SI virus in culture. In the samples of uncultured PBMC DNA, several subgroups of sequences were found, indicating that the process of evolution may not be gradual and that several distinct populations can coexist. The paucity of intermediate sequences indicated that strong selection pressure was exerted on this part of the envelope. The early emergence of disease-associated SI variants in cultured material indicates that virus culture may have relevance for the in vivo situation.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Callahan L. N., Phelan M., Mallinson M., Norcross M. A. Dextran sulfate blocks antibody binding to the principal neutralizing domain of human immunodeficiency virus type 1 without interfering with gp120-CD4 interactions. J Virol. 1991 Mar;65(3):1543–1550. doi: 10.1128/jvi.65.3.1543-1550.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R. A., Groenink M., Kootstra N. A., Tersmette M., Huisman H. G., Miedema F., Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992 May;66(5):3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E. O., Myers D. J., Risser R. Identification of the principal neutralizing determinant of human immunodeficiency virus type 1 as a fusion domain. J Virol. 1991 Jan;65(1):190–194. doi: 10.1128/jvi.65.1.190-194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D. Amino acid composition and the evolutionary rates of protein-coding genes. J Mol Evol. 1985;22(1):53–62. doi: 10.1007/BF02105805. [DOI] [PubMed] [Google Scholar]
- McKeating J. A., Gow J., Goudsmit J., Pearl L. H., Mulder C., Weiss R. A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989 Dec;3(12):777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- Moore J. P., Nara P. L. The role of the V3 loop of gp120 in HIV infection. AIDS. 1991;5 (Suppl 2):S21–S33. doi: 10.1097/00002030-199101001-00004. [DOI] [PubMed] [Google Scholar]
- Moore J. P., Nara P. L. The role of the V3 loop of gp120 in HIV infection. AIDS. 1991;5 (Suppl 2):S21–S33. doi: 10.1097/00002030-199101001-00004. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Balfe P., Peutherer J. F., Ludlam C. A., Bishop J. O., Brown A. J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990 Feb;64(2):864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersmette M., Gruters R. A., de Wolf F., de Goede R. E., Lange J. M., Schellekens P. T., Goudsmit J., Huisman H. G., Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989 May;63(5):2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersmette M., de Goede R. E., Al B. J., Winkel I. N., Gruters R. A., Cuypers H. T., Huisman H. G., Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988 Jun;62(6):2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfs T. F., Zwart G., Bakker M., Valk M., Kuiken C. L., Goudsmit J. Naturally occurring mutations within HIV-1 V3 genomic RNA lead to antigenic variation dependent on a single amino acid substitution. Virology. 1991 Nov;185(1):195–205. doi: 10.1016/0042-6822(91)90767-6. [DOI] [PubMed] [Google Scholar]
- Wolfs T. F., de Jong J. J., Van den Berg H., Tijnagel J. M., Krone W. J., Goudsmit J. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J. J., Goudsmit J., Keulen W., Klaver B., Krone W., Tersmette M., de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992 Feb;66(2):757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


