Abstract
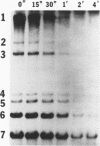
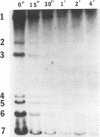
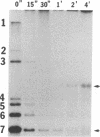
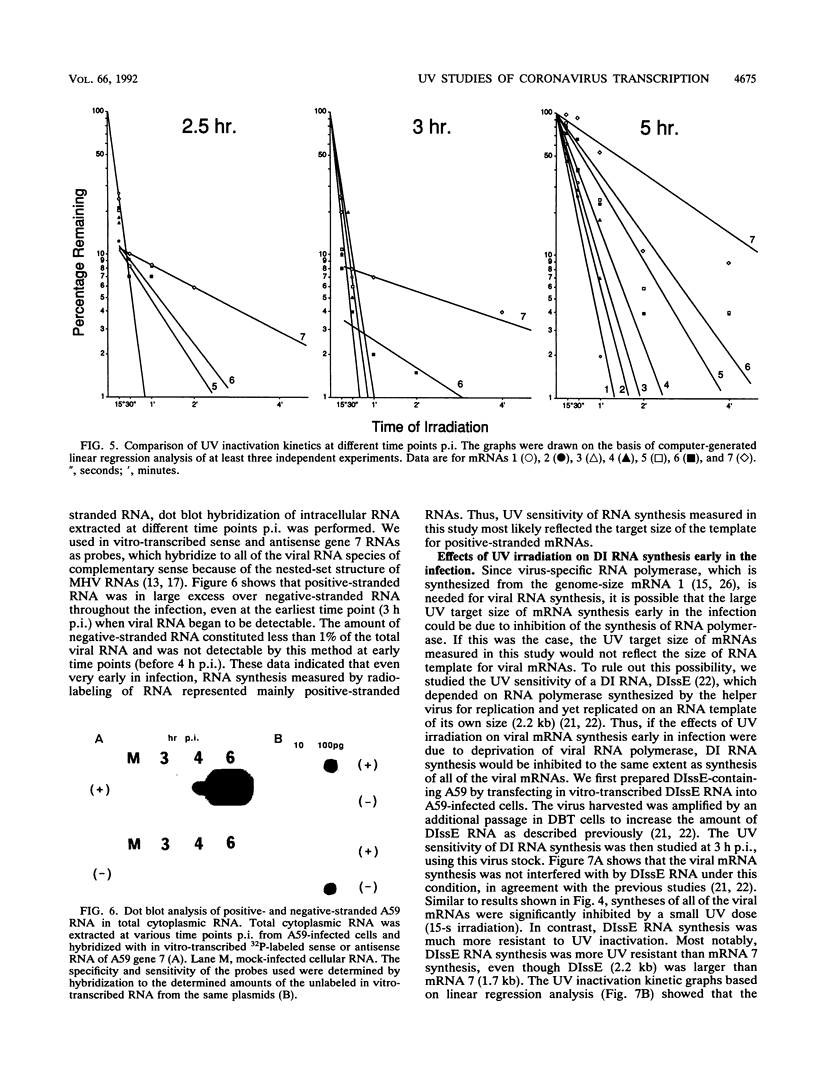
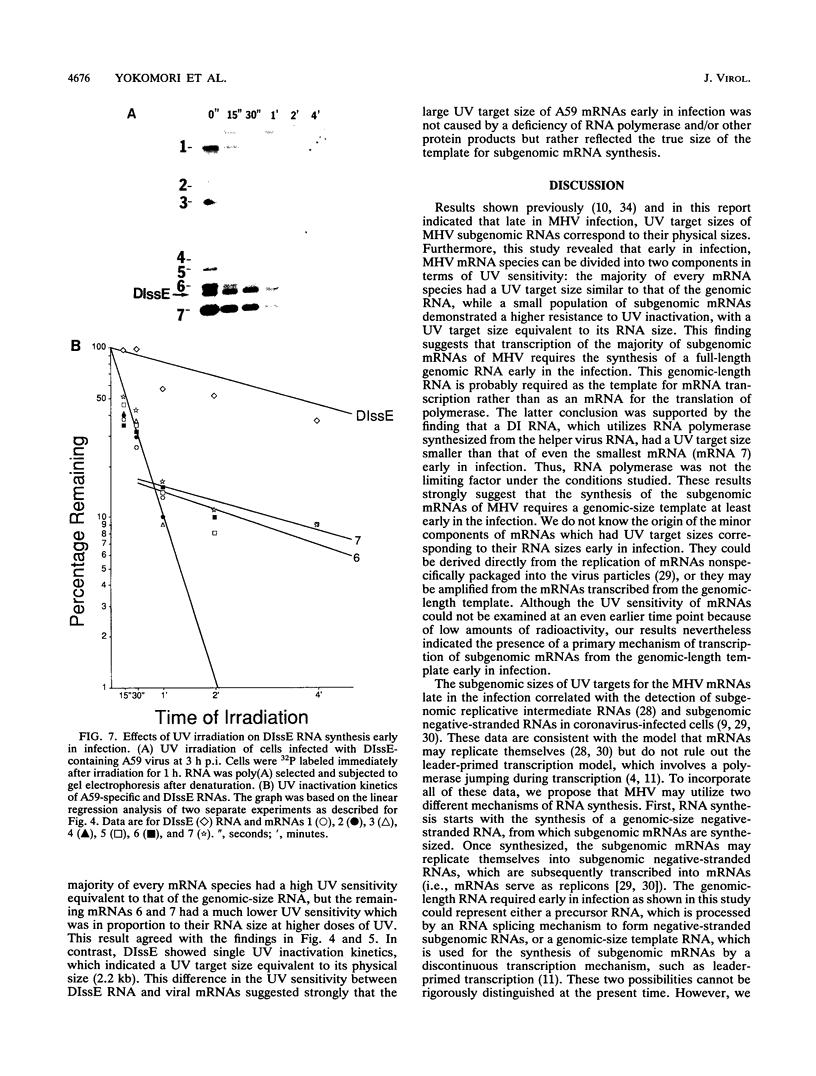
Mouse hepatitis virus (MHV) synthesizes seven to eight mRNAs, each of which contains a leader RNA derived from the 5' end of the genome. To understand the mechanism of synthesis of these mRNAs, we studied how the synthesis of each mRNA was affected by UV irradiation at different time points after infection. When MHV-infected cells were UV irradiated at a late time in infection (5 h postinfection), the syntheses of the various mRNAs were inhibited to different extents in proportion to the sizes of the mRNAs. Analysis of the UV inactivation kinetics revealed that the UV target size of each mRNA was equivalent to its own physical size. In contrast, when cells were irradiated at 2.5 or 3 h postinfection, there appeared to be two different kinetics of inhibition of mRNA synthesis: the synthesis of every mRNA was inhibited to the same extent by a small UV dose, but the remaining mRNA synthesis was inhibited by additional UV doses at different rates for different mRNAs in proportion to RNA size. The analysis of the UV inactivation kinetics indicated that the UV target sizes for the majority of mRNAs were equivalent to that of the genomic-size RNA early in the infection. These results suggest that MHV mRNA synthesis requires the presence of a genomic-length RNA template at least early in the infection. In contrast, later in the infection, the sizes of the templates used for mRNA synthesis were equivalent to the physical sizes of each mRNA. The possibility that the genomic-length RNA required early in the infection was used only for the synthesis of a polymerase rather than as a template for mRNA synthesis was ruled out by examining the UV sensitivity of a defective interfering (DI) RNA. We found that the UV target size for the DI RNA early in infection was much smaller than that for mRNAs 6 and 7, which are approximately equal to or smaller in size than the DI RNA. This result indicates that even though DI RNA and viral mRNAs are synthesized by the same polymerase, mRNAs are synthesized from a larger (genomic-length) template. We conclude that a genomic-length RNA template is required for MHV subgenomic mRNA synthesis at least early in infection. Several transcription models are proposed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham G., Banerjee A. K. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci U S A. 1976 May;73(5):1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Lai M. M. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990 Dec;9(12):4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L. A., White C. N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Lai M. M. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983 Dec;48(3):633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Razavi M. K., Lai M. M. Characterization of leader-related small RNAs in coronavirus-infected cells: further evidence for leader-primed mechanism of transcription. Virus Res. 1985 Jul;3(1):19–33. doi: 10.1016/0168-1702(85)90038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton P. R., Stohlman S. A., Lai M. M. Further characterization of mouse hepatitis virus RNA-dependent RNA polymerases. Virology. 1984 Feb;133(1):197–201. doi: 10.1016/0042-6822(84)90439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier K., Raghow R., Kingsbury D. W. Regulation of Sendai virus transcription: evidence for a single promoter in vivo. J Virol. 1977 Mar;21(3):863–871. doi: 10.1128/jvi.21.3.863-871.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44(3):298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Hofmann M. A., Sethna P. B., Brian D. A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990 Sep;64(9):4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs L., Spaan W. J., Horzinek M. C., van der Zeijst B. A. Synthesis of subgenomic mRNA's of mouse hepatitis virus is initiated independently: evidence from UV transcription mapping. J Virol. 1981 Aug;39(2):401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Monica N., Yokomori K., Lai M. M. Coronavirus mRNA synthesis: identification of novel transcription initiation signals which are differentially regulated by different leader sequences. Virology. 1992 May;188(1):402–407. doi: 10.1016/0042-6822(92)90774-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Brayton P. R., Armen R. C., Patton C. D., Pugh C., Stohlman S. A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981 Sep;39(3):823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Leibowitz J. L., Weiss S. R., Paavola E., Bond C. W. Cell-free translation of murine coronavirus RNA. J Virol. 1982 Sep;43(3):905–913. doi: 10.1128/jvi.43.3.905-913.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J. L., Wilhelmsen K. C., Bond C. W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology. 1981 Oct 15;114(1):39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J. K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991 Nov;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. Evolution of the 5'-end of genomic RNA of murine coronaviruses during passages in vitro. Virology. 1989 Mar;169(1):227–232. doi: 10.1016/0042-6822(89)90060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J Virol. 1989 Dec;63(12):5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Keck J. G., Lai M. M. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988 Mar;163(1):104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Soe L. H., Baker S. C., Lai M. M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988 Oct;166(2):550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Soe L. H., Shieh C. K., Lai M. M. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J Virol. 1988 Oct;62(10):3870–3873. doi: 10.1128/jvi.62.10.3870-3873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Stohlman S. A., Lai M. M. Leader sequences of murine coronavirus mRNAs can be freely reassorted: evidence for the role of free leader RNA in transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4204–4208. doi: 10.1073/pnas.83.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Hirano N., Fujiwara K. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984 Nov;139(1):138–151. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986 Jan;57(1):328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990 Mar;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hofmann M. A., Brian D. A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991 Jan;65(1):320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hung S. L., Brian D. A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C. K., Lee H. J., Yokomori K., La Monica N., Makino S., Lai M. M. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol. 1989 Sep;63(9):3729–3736. doi: 10.1128/jvi.63.9.3729-3736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C. K., Soe L. H., Makino S., Chang M. F., Stohlman S. A., Lai M. M. The 5'-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987 Feb;156(2):321–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B. A., Siddell S. G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. F., Sefton B. M. Synthesis of coronavirus mRNAs: kinetics of inactivation of infectious bronchitis virus RNA synthesis by UV light. J Virol. 1982 May;42(2):755–759. doi: 10.1128/jvi.42.2.755-759.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L. S., Takemoto K. K. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect Immun. 1972 Oct;6(4):501–507. doi: 10.1128/iai.6.4.501-507.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]