Abstract
Large-scale transitions in genome size from tetraploid to diploid were observed during a previous 1800-generation evolution experiment in Saccharomyces cerevisiae. Whether the transitions occurred via a one-step process (tetraploid to diploid) or through multiple steps (through ploidy intermediates) remained unclear. To provide insight into the mechanism involved, we investigated whether triploid-sized cells sampled from the previous experiment could also undergo ploidy loss. A batch culture experiment was conducted for approximately 200 generations, starting from four triploid-sized colonies and one contemporaneous tetraploid-sized colony. Ploidy reduction towards diploidy was observed in both triploid and tetraploid lines. Comparative genomic hybridization indicated the presence of aneuploidy in both the founder and the evolved colonies. The specific aneuploidies involved suggest that chromosome loss was not haphazard but that nearly full sets of chromosomes were lost at once, with some additional chromosome mis-segregation events. These results suggest the existence of a mitotic mechanism allowing the elimination of an entire set of chromosomes in S. cerevisiae, thereby reducing the ploidy level.
Keywords: yeast, ploidy, aneuploidy
1. Introduction
The range and the scale of viable mutational events that affect genome size remain largely unknown. By tracking genome size of Saccharomyces cerevisiae lines for approximately 1800 generations in rich medium (standard laboratory YPD), we previously found that replicate tetraploid lines rapidly decreased in genome size towards a diploid genomic content (Gerstein et al. 2006). Using comparative genomic hybridization (CGH), we found that the evolved diploids were euploid (or nearly euploid). As all lines were maintained asexually, this suggests a mechanism whereby S. cerevisiae cells rapidly and mitotically decrease ploidy levels.
One possible mechanism for the halving of DNA content is a rare meiotic event, which is unlikely in this study because the strain used was homozygous for MATa-a1 and should not undergo meiosis (for further explanation, see Gerstein et al. (2006)). Another possible mechanism is an accidental error in the mitotic cell cycle, causing a reductive cell division without an intervening round of DNA replication (the reductive division would be irregular, involving the separation of homologues). These mechanisms cannot, however, account for a genomic reduction from near tetraploid to near triploid levels. In our previous experiment, triploid-sized colonies arose within 200 generations in several lines and were sampled at multiple time points throughout the approximately 1800 generations (Gerstein et al. 2006). However, whether these triploids were intermediate in the process of genomic reduction to diploidy or were evolutionary dead ends remains unclear from the sampling we had done.
The transition from tetraploidy to triploidy suggests an alternative mechanism, whereby unequal mitosis occurs in a manner that ensures a balanced set of chromosomes in each daughter cell. Meiotic distributive disjunction, the proper segregation of chromosomes during meiosis I in the absence of recombination, was shown to occur for non-recombining artificial chromosomes in S. cerevisiae (Dawson et al. 1986). The exact mechanism and the role of distributive disjunction remain unknown in S. cerevisiae, nor is it known whether such a system could operate during mitosis. A multipolar mitotic division (Storchová & Pellman 2004), with three spindle pole bodies on one side of the dividing cell and one on the other, is one possibility. Here, we ask whether the elimination of a haploid set of chromosomes is unique to tetraploid cells or is also possible in triploid cells. Rapid transitions from triploid to diploid cells would provide further support for the existence and generality of a mitotic distributive disjunction system.
2. Material and methods
Four triploid-sized colonies and one tetraploid-sized colony were isolated from three independently evolved lines (denoted P, R and S) in our previous experiment; these three lines were founded from the same ancestral tetraploid and propagated in YPD by batch culture for approximately 1800 generations (Gerstein et al. 2006). One triploid-sized colony was isolated from each of these three lines at generation 300 (, , ), one triploid-sized colony was isolated from line R at generation 500 () and one tetraploid-sized colony was isolated from line R at generation 300 (). We refer to these five colonies as the ‘founder colonies’. The tetraploid colony () served as a control to ensure that ploidy loss again occurred in the current experiment and to assess the effect of initial ploidy level on the rate of genomic reduction.
Each founder colony was grown up overnight in rich medium (YPD) and the resultant culture was used to initiate six replicate lines (; ; ; ; ) for a total of 30 lines. These lines were evolved asexually in separate test tubes for 28 days (t=186 generations) using the same batch culture techniques as in Gerstein et al. (2006). Specifically, 1 : 100 dilutions of 100 μl of stationary-phase culture into 10 ml of YPD were performed every 24 hours (±1 hour). Cultures from each population were frozen down weekly in 15% glycerol.
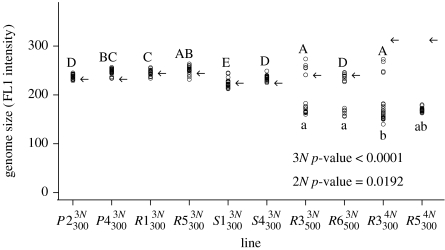
Flow cytometry was performed weekly on approximately 30 000 cells from each line to scan populations for changes in total genome size (electronic supplementary material S1); more fine-scale within-population variation after 186 generations was subsequently assessed by flow cytometry of 20 colonies isolated from lines of interest. The genome size of all colonies fell into three discrete size clusters (figure 1); we categorized fluorescence readings (FL1 intensity) between 100 and 200 as diploid, 200 and 300 as triploid and more than 300 as tetraploid.
Figure 1.
Variation in genome size measured by flow cytometry. Twenty colonies (circles) were analysed from each line at t=186. Arrows indicate the initial genome sizes of tetraploid- and triploid-sized founder colonies at t=0 (averaged across three colonies). Letters denote significant (p<0.05) genome size differences among evolved triploid-sized lines (uppercase letters) and among the diploid-sized lines (lowercase letters; analysed separately) according to post hoc Tukey tests following significant ANOVA results (p-values in figure).
To detect aneuploidy, CGH was performed (Hughes et al. 2000) on the five founder colonies used in this experiment (, , , , ), on four t=186 colonies where ploidy reduction was observed (choosing the two diploid-sized colonies that had the highest and lowest FL1 intensities from lines and ; labelled ‘high’ and ‘low’ in figure 2) and on the haploid and tetraploid ancestral colonies from Gerstein et al. (2006). In each case, the control colony was a distantly related haploid strain (YPH499). DNA was labelled to S. cerevisiae microarrays obained from the University Health Network Microarray Centre (Toronto), with each analysis performed twice with dye swapping (electronic supplementary material S2).
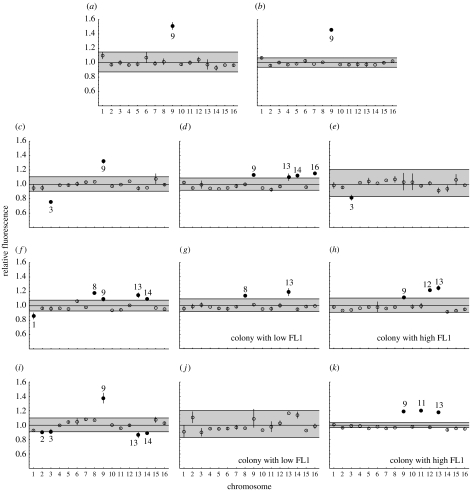
Figure 2.
Fluorescence ratio by chromosome using CGH. Each panel presents the results of two microarrays (dots, mean; bars, range). Euploid lines are expected to have fluorescence ratios (experimental/control) of one. Putative aneuploid chromosomes (solid dots with chromosome numbers) exhibited fluorescence ratios substantially different from one (more than 10%) and fell outside of the rejection region (grey shading). (a) Approximately 1N ancestor, (b) approximately 4N ancestor, (c) approximately 3N at t=0 (), (d) approximately 3N at t=0 (), (e) approximately 3N at t=0 (), (f) approximately 3N at t=0 (), (g) approximately 2N at t=186 (), (h) approximately 2N at t=186 (), (i) approximately 4N at t=0 (), (j) approximately 2N at t=186 () and (k) approximately 2N at t=186 ().
Data were ordered according to chromosomal location using Excel (Microsoft Corporation) and analysed using Mathematica (Wolfram Research). All analyses were based on log-fluorescence ratios, with back-transformed ratios presented in the figures. In a CGH comparison of euploid colonies, fluorescence ratios should equal one for each chromosome. Aneuploidy can be detected as a marked departure from this expectation across part or all of a chromosome. To be considered aneuploid, a chromosome had to depart by more than 10% from this expected ratio and had to lie outside a rejection region (electronic supplementary material S2).
3. Results
According to population-wide flow cytometry, diploid-sized cells arose and spread to high frequency within two of the six initially tetraploid lines (figure S3 in the electronic supplementary material (e) , (f) ) by generation 186. The remaining initially tetraploid lines either evolved towards a triploid genomic content (figure S3 in the electronic supplementary material (a) , (b) ) or exhibited a polymorphism for triploid- and diploid-sized cells (figure S3 in the electronic supplementary material (c) , (d) ) by this time point. A polymorphism for triploid- and diploid-sized cells was also observed in three of the initially triploid-sized lines (figure S4 in the electronic supplementary material (s) , (u) , (x) ), demonstrating that mutations reducing genome size from roughly triploid to diploid levels were possible. The remaining 21 initially triploid lines appear to have maintained a roughly triploid genome size (figure S4 in the electronic supplementary material). Genome size was thus more likely to decrease in the tetraploid lines than in the triploid lines (two-tailed Fisher's exact test, p=0.00003). The results also suggest that not all triploids were equally likely to undergo a reduction in genome size, as only lines initiated from the founder colony exhibited ploidy reduction (p=0.04 for the probability that the second and third case of ploidy reduction would occur in the same triploid line as the first case).
Two lines evolved from each of the five founder colonies were chosen to examine within-line variation in genome size at generation 186, focusing on those lines that had undergone ploidy transitions (figure 1). Results were consistent with the population-level data. Lines polymorphic in the population-wide scan showed distinct clusters of cells with approximately triploid and diploid contents (percentage of diploid-sized cells: 70% in , 30% in and 80% in). In addition, significant variation in genome size was observed within a ploidy level (figure 1) among the ancestral triploid lines (F3,30=13.94, p<0.0001), among triploids isolated from evolved triploid and tetraploid lines (F8,134=29.45, p<0.0001) and among diploids isolated from evolved triploid and tetraploid lines (F3,56=7.989, p=0.0002). Such variation suggests the presence of aneuploidy.
The CGH experiments confirmed that aneuploidy was common (figure 2; see also sliding window analyses in electronic supplementary material S3). Importantly, the ancestral haploid used to construct the diploid and tetraploid isogenic lines in the first series of experiments (Gerstein et al. 2006) was itself aneuploid (figure 2a,b), carrying an extra copy of chromosome 9 (our previous CGH analyses had not compared the haploid ancestor to an external line). The additional copy of chromosome 9 might have increased the mutation rate or selection pressure for genome-size change in our previous study, but it does not explain why both haploid and tetraploid lines converged towards diploidy. The fluorescence ratios of the ancestral tetraploid and haploid lines used in Gerstein et al. (2006) were similar (figure 2a,b), suggesting that the ancestral tetraploid was 4N+4. Among the founder colonies used here (isolated after 300 or 500 generations), however, the degree of chromosome 9 aneuploidy was reduced or eliminated (figure 2c–k). Nevertheless, the triploid founder colonies exhibited varying degrees of aneuploidy (figure 2c–f), in a manner consistent with flow cytometry (figure 1). Interestingly, lines with reduced ploidy levels were often aneuploid in similar ways to their founder colony (figure 2; compare diploid-sized colonies at t=186 (panels g,h) to their triploid-sized founder colony at t=0 (panel f)).
4. Discussion
We observed ploidy reduction, often coupled with aneuploidy, over a short time period (186 generations) in all six of our tetraploid replicates and in three out of 24 triploid replicate lines. These results demonstrate that ploidy reduction is not restricted to tetraploids. As pointed out by Gerstein et al. (2006), the short time frame over which ploidy reduction is observed suggests a concerted mechanism for chromosome loss across the genome. Without a concerted mechanism of loss, it is difficult to explain the production of nearly euploid diploids from tetraploids within 200 generations unless selection coefficients favouring aneuploidy are very high (on the order of a doubling of fitness every time a chromosome is lost; Gerstein et al. 2006). Such strong selection is inconsistent with the relatively minor differences in growth rate observed among lines (Gerstein 2006). The same aneuploid chromosomes were often maintained in transitions from tetraploid- and triploid-sized to diploid-sized colonies (figure 2), which is further evidence for concerted chromosome loss. Concerted chromosome loss leading to ploidy reduction has also been inferred in the related yeast species, Candida albicans (Bennett & Johnson 2003).
Polyploid genomes are known to be unstable in S. cerevisiae (Mayer & Aguilera 1990; Andalis et al. 2004; Storchová & Pellman 2004; Storchová et al. 2006). Storchová et al. (2006) suggested a possible physiological basis, finding that wild-type tetraploid individuals have a 3.6-fold increased frequency of syntelic/monopolar microtubule–kinetochore attachments (rather than biorientation, where kinetochores attach to microtubules from both poles) compared with diploids. If uncorrected, these syntelic attachments lead to chromosome non-disjunction. They suggest that scaling restrictions occur in polyploid yeast, creating mismatches between the size of spindle pole bodies, the length of spindle, and size and geometry of kinetochores, potentially explaining the increased frequency of syntelic attachments.
Although triploid lines also underwent ploidy reduction, they did so at a lower frequency (3 out of 24 replicates compared with 6 out of 6 of the tetraploid replicates). It may be that scaling mismatches are not as problematic in triploids as in tetraploids, causing ploidy-reducing mutations to be less common. Alternatively, the triploid lines might have been fitter than the tetraploid lines, decreasing the relative advantage of any diploid-sized cells that might have arisen.
Aneuploidy might also affect the rate of ploidy evolution. As the haploid progenitor used to create the ancestral series of isogenic haploid, diploid and tetraploid lines was itself aneuploid for chromosome 9, rates of chromosome instability might have been elevated throughout these experiments. Furthermore, the only triploid line that gave rise to diploid cells, , was one of the more aneuploid lines examined (figure 2), although an alternative explanation for this observation is that diploid cells arose in during founder colony formation or the initial overnight inoculation.
Aneuploidy can reduce fitness by altering gene dosage and gene expression, and it is likely that selection for euploidy acted during our experiments. In our previous experiment, lines were nearly euploid after approximately 1800 generations (Gerstein et al. 2006). Aneuploidy is more common at earlier time points (after only 300 or 500 generations in the previous experiment; figure 2c–f,i), and it remained common after the 186 generations of batch culture evolution reported here (figure 2g,h,j,k). Yet even over these shorter time frames, the replicates analysed by CGH appear to have lost many if not all of the additional copies of chromosome 9 present in the ancestor.
Our results paint a picture of a dynamic process of ploidy change in mitotically reproducing S. cerevisiae, which tends towards genomic stabilization at diploid levels. Cells with approximately tetraploid and triploid genomic contents were observed to undergo ploidy reduction, providing evidence for a mitotic distributive disjunction process in yeast.
Acknowledgments
We thank L. Glaubach, M. Mandegar and L. Cleathero for technical assistance and three anonymous reviewers for helpful comments. We are grateful to Michael Whitlock for statistical advice. Funding was provided by a Natural Sciences and Engineering Research Council (Canada) post-graduate scholarship (A.C.G.), a discovery grant (S.P.O.) and by a sabbatical grant from the National Evolutionary Synthesis Center (NSF #EF-0423641; S.P.O.).
Supplementary Material
S1: flow cytometry
S2: aneuploid detection
S3: sliding window analyses
References
- Andalis A.A, Storchová Z, Styles C, Galitski T, Pellman D, Fink G.R. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics. 2004;167:1109–1121. doi: 10.1534/genetics.104.029256. doi:10.1534/genetics.104.029256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.J, Johnson A.D. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. doi:10.1093/emboj/cdg235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson D.S, Murray A.W, Szostak J.W. An alternative pathway for meiotic chromosome segregation in yeast. Science. 1986;234:713–717. doi: 10.1126/science.3535068. doi:10.1126/science.3535068 [DOI] [PubMed] [Google Scholar]
- Gerstein, A. C. 2006 Genomic convergence towards diploidy in Saccharomyces cerevisiae: patterns and mechanisms. MSc thesis, University of British Columbia, Vancouver, Canada.
- Gerstein A.C, Chun H.E, Grant A, Otto S.P. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. doi:10.1371/journal.pgen.0020145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.R, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 2000;25:333–337. doi: 10.1038/77116. doi:10.1038/77116 [DOI] [PubMed] [Google Scholar]
- Mayer V.W, Aguilera A. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat. Res. 1990;231:177–186. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- Storchová Z, Pellman D. From polyploidy to aneuploidy, genome stability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. doi:10.1038/nrm1276 [DOI] [PubMed] [Google Scholar]
- Storchová Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. doi:10.1038/nature05178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1: flow cytometry
S2: aneuploid detection
S3: sliding window analyses