Abstract
Pinnipeds (seals, fur seals, sea lions and walrus) form large breeding aggregations with females often remaining faithful to a natal site or area. In these cases, females are philopatric to regional areas on broad geographical scales of hundreds to thousands of kilometres. An investigation of variation in a control region sequence of mtDNA in the Australian sea lion (Neophoca cinerea) has shown a case of extreme female natal site fidelity that has resulted in almost fixed population differentiation across its range (ΦST=0.93). This high level of population subdivision over short geographical distances (approx. 60 km) is unparalleled in any social marine mammal and reflects the unique life-history traits of this rare species. The high level of population subdivision and exclusive female natal site fidelity has important ramifications for conservation management, and poses many interesting questions of both academic and applied interest.
Keywords: natal site fidelity, Australian sea lion, genetic drift
1. Introduction
Pinnipeds are a group of marine mammals that in most cases require terrestrial habitats for mating and parturition. Nearly all species of pinnipeds display annual, synchronous breeding cycles, resulting in the formation of large breeding colonies (Boyd 1991). Breeding colonies are distributed non-uniformly, owing to the availability of suitable habitat, and this may result in a regionally fragmented population structure (e.g. a stepping stone model, see Kimura & Weiss 1964). However, these animals are capable of widespread dispersal and, theoretically, a high degree of gene flow. Countering this potential for high gene flow is the possibility of population subdivision due to female natal site fidelity, which has been demonstrated for several species on broad geographical scales (e.g. Steller sea lions (Eumetopias jubatus) in Bickham et al. 1996, southern elephant seals (Mirounga leonina) in Slade et al. 1998).
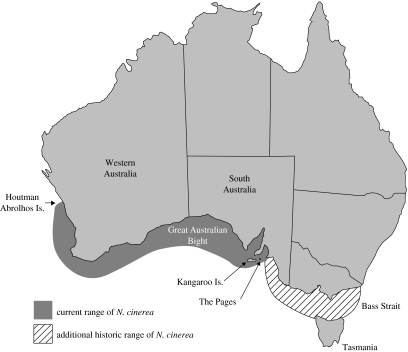
The Australian sea lion is Australia's only endemic otariid (fur seals and sea lions) and is currently listed as Threatened under Federal legislation (‘vulnerable’ category). It inhabits the offshore islands of the southwestern and southern coasts of Australia (figure 1). Its current range is historically contracted, due to the regional extinction in the Bass Strait area as a probable consequence of commercial sealing operations during the eighteenth and nineteenth centuries. A population estimate of 12 000 animals makes this species one of the rarest pinnipeds in the world (Gales et al. 1994). This population is scattered among some 60 individual breeding colonies, most of which produce less than 50 pups in each breeding season (Gales et al. 1994). Most unusually, this species has a unique breeding cycle of 17.5 months, which is asynchronous across its range unlike the annual synchronized breeding cycle of other pinnipeds (Boyd 1991; Higgins 1993; Gales et al. 1994). Small groups of proximate colonies show some synchrony in the timing of breeding, but otherwise the timing of breeding appears random. It was hypothesized that this system of asynchronous breeding was maintained by fine-scale female philopatry to the point where females return exclusively to their natal colony (Gales et al. 1994).
Figure 1.
Distribution of extant Australian sea lion breeding colonies and extinct regional populations due to commercial sealing operations in the eighteenth and nineteenth centuries.
We analysed variation in a control region sequence of mtDNA to determine the genetic structure of populations of Neophoca cinerea and to understand the role of reproductive strategies in regulating female-mediated gene flow.
2. Material and methods
(a) DNA extraction and polymerase chain reaction
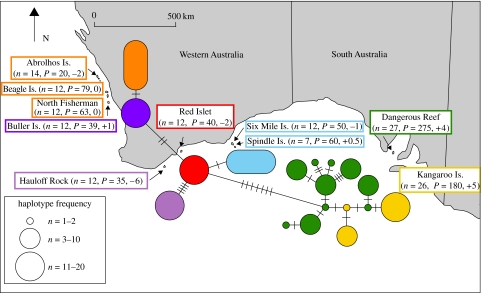
Tissue samples were taken from 149 newborn pups from 10 colonies representative of the spatial and temporal distribution (figure 2). Mean pup production (P) for each colony is displayed in figure 2 and was taken from Gales et al. (1994). Total genomic DNA was extracted from the tissue by the method of Gemmell & Akiyama (1996) and stored in 10 mM Tris, pH 8.0. A fragment of the mitochondrial control region was isolated using polymerase chain reaction (PCR) and the pair of primers, LGL 283 (5′-TACACYRGTCTTCTAAACC-3′) and LGL 1115 (5′-CTGGTTCYTTCAGGGTCAT-3′) designed for Steller sea lions (Bickham et al. 1996). PCR conditions were similar to Bickham et al. (1994) with modification of the annealing temperature to 54°C. This produced a single, approximately 480 bp product which was reduced to a 360 bp sequence for analysis after removing the primer sites and flanking regions.
Figure 2.
Minimum spanning tree of mtDNA haplotypes of Australian sea lions. Individual haplotypes are indicated by circles and extended circles where a haplotype is shared between colonies. Distribution of haplotypes is colour coded with the colony name and the size is related to their frequency of occurrence. Groups of colonies fixed for the same haplotype are given a common colour. Branch lengths are defined by the number of nucleotide changes indicated by the number of cross-hatches on the branch. Sample sizes (n) and mean pup production (P) for individual colonies are listed in parentheses after the colony name. The breeding time of each colony is also given in parentheses in number of months before (−) or after (+) the reference colony of North Fisherman on the west coast of Western Australia.
(b) Sequence and phylogenetic analysis
The sequences were aligned in the program Sequencher (v. 3.1.1, Life Codes, Inc.). Polymorphic sites and haplotypes were defined using the program MacClade v. 3.0.4 (Maddison & Maddison 1992). Population subdivision was measured using an analysis of molecular variance to look at the distribution of genetic variation between and within individual colonies (AMOVA). This was done using a modified measure of the traditional F- and Φ-statistics based on haplotype sequence distances (Tamura & Nei 1993).
Pairwise genetic differences between colonies were calculated by the haplotype distance method (Tamura & Nei 1993) and their significance tested by 1000 permutations (Weir 1996). These calculations were performed in the computer program Arlequin v. 2.000 (Schneider et al. 2000). A minimum spanning tree was created among the haplotypes based on the number of nucleotide changes to infer phylogenetic relationships.
3. Results
Eighteen individual haplotypes were defined by the 21 variable sites of a 360 bp fragment of the control region (accession numbers of sequences are listed below). Only 2 of the 18 haplotypes were shared between colonies, and the remaining 16 haplotypes were unique to a particular colony (figure 2). This resulted in an incredibly high level of population subdivision approaching the theoretical maximum of unity (ΦST=0.93). Nearly, all colony pairwise estimates showed significant differences, again in most cases approaching the theoretical maximum (table 1).
Table 1.
Pairwise comparisons of genetic differentiation (ΦST) among colonies. (Italic indicates pairwise comparisons that are not significantly different from zero at p<0.001. All other comparisons are significantly different. Locations identified in figure 2.)
| Abrolhos Is. | Beagle Is. | North Fisherman | Buller Is. | Hauloff Rock | Red Islet | Six Mile Is. | Spindle Is. | Dangerous Reef | |
|---|---|---|---|---|---|---|---|---|---|
| Beagle Is. | 0.00 | ||||||||
| North Fisherman | 0.00 | 0.00 | |||||||
| Buller Is. | 1.00 | 1.00 | 1.00 | ||||||
| Hauloff Rock | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| Red Islet | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Six Mile Is. | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||
| Spindle Is. | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | ||
| Dangerous Reef | 0.83 | 0.85 | 0.85 | 0.83 | 0.84 | 0.80 | 0.82 | 0.80 | |
| Kangaroo Is. | 0.94 | 0.95 | 0.95 | 0.94 | 0.95 | 0.93 | 0.94 | 0.93 | 0.54 |
The two cases of haplotype sharing occurred between colonies that were near neighbours and bred at approximately the same time (Six Mile Is. & Spindle Is. and North Fisherman Is., Beagle Is. & Houtman Abrolhos Is.; figure 2). Eight of the ten colonies were fixed for a single haplotype, indicating an extremely low level of intra-colony genetic variation. Only two colonies exhibited any considerable haplotype diversity (Kangaroo Is. and Dangerous Reef), though there were no shared haplotypes between these neighbouring colonies.
The minimum spanning tree shows the moderate phylogeographic signal which indicates relatively long-term population structure (figure 2). There appears to be a moderate division across the Great Australian Bight.
4. Discussion
The evolution of the unique Australian sea lion breeding system has resulted in a highly subdivided population. The level of female philopatry is on a far finer scale than previously seen in any other marine mammal (Maldonado et al. 1995; Bickham et al. 1996; Lamont et al. 1996; Slade et al. 1998), more akin to isolated populations of terrestrial animals (e.g. white-tailed deer in Purdue et al. 2000). Haplotype diversity was very low within colonies, with all but two colonies displaying fixation of the control region haplotype. Fixation was observed at the relatively small Western Australian, where mean pup production was usually less than 50. The two colonies that displayed any degree of haplotype diversity, Kangaroo Island and Dangerous Reef, were three- to fivefold larger and presumably have maintained this diversity despite the effects of genetic drift.
It is possible that commercial exploitation and potential bottleneck events within the last 200 years could have played a role in the genetic structure of the Australian sea lion population presented here. However, it was recorded that the majority of Australian sea lion harvesting occurred around Kangaroo Island and that limited commercial harvesting occurred in Western Australia (Ling 1999).
The phylogeographic patterns and results of the AMOVA suggest that the regional population structure is more probably due to population discontinuity and the subsequent role of genetic drift, rather than the extreme female natal site fidelity alone. These results relate strictly to maternal lineages and it is quite probable that the investigation of nuclear genes (biparentally inherited DNA) will show a lower level of population subdivision.
The extremely high level of female natal site fidelity means that female recruitment is only from within the colony. This has important implications for the conservation of many of the small colonies of N. cinerea, which due to their size are at risk of localized extinction due to demographic and environmental stochasticity and human perturbations (Goldsworthy & Page 2007). Recolonization of previous breeding areas is highly unlikely due to the low level of female dispersal.
The evolution of such fine-scale population structure may be linked to the non-seasonal, supra-annual breeding cycle. This breeding cycle is postulated to be a response to a nutrient-poor environment (Gales et al. 1994). The coastal marine environment of western and southern Australia experiences low nutrient levels due to the combination of a lack of seasonal upwelling events and a warm-water, nutrient-poor current (the Leeuwin current) which extends down the west coast and along the southern coast of Australia (Pearce 1991).
The evolution of a non-seasonal breeding cycle has relaxed the stabilizing selection pressure evident in the temporal patterns of breeding seen in other pinniped species (Boyd 1991). As temporal variation in breeding times between colonies developed, strong selection against female dispersal would occur as individuals would be cued into the breeding cycle of their natal colony. Dispersal from the natal colony would result in entering a breeding system possibly out of synchrony with conspecifics, and limit the chances of finding a mate. This would result in reinforcing selection against dispersal and the development of isolated colonies of philopatric females, conditions suitable for the operation of genetic drift (Wright 1931). Other behavioural mechanisms, such as foraging site fidelity may also have evolved in concert with the temporal patterns in breeding, further reinforcing the selection pressure for natal site fidelity among females.
These results represent a fascinating example of the interplay between environment, ecology and the evolution of constraints on population genetic structure. In effect each individual colony that is represented by a novel set of mtDNA haplotypes, most often a single matriline, becomes a separate management unit (Moritz 1994). Conservation management strategies to ensure the continued survival of the Australian sea lion should reflect this unique population structure as once a breeding colony is lost there appears to be a low probability of recolonization.
Acknowledgments
Fieldwork practices were conducted under the auspices of the ethics permit issued to R.A.C. by the University of Western Australia.
We thank M. S. Johnson for draft reviews, and also B. Haberley, P. Collins, P. D. Shaughnessy, T. E. Dennis, M. Berriss and all staff at S. A. Parks and Wildlife and K. Brooks for their assistance in the field. We also thank K. Boxen and all members of the School of Biological Sciences, University of Auckland for their advice and help in the laboratory. R.A.C. was supported by Environment Australia, Department of Conservation and Land Management, University of Western Australia, University of Auckland and Australian Geographic for this work. The sequences determined for this study are available in Genbank under the accession numbers AF522370–AF522383 and AF522386–AF522389.
References
- Bickham J.W, Patton J.C, Loughlin T.R. High variability for control-region sequences in a marine mammal: implications for conservation and maternal phylogeny of Steller sea lions (Eumetopias jubatus) J. Mamm. 1996;77:95–108. doi:10.2307/1382712 [Google Scholar]
- Boyd I.L. Environmental and physiological factors controlling the reproductive cycles of pinnipeds. Can. J. Zool. 1991;69:1135–1148. [Google Scholar]
- Gales N, Shaughnessy P.D, Dennis T.E. Distribution, abundance and breeding cycle of the Australian sea lion, Neophoca cinerea. J. Zool. 1994;234:353–370. [Google Scholar]
- Gemmell N.J, Akiyama S. A simple and efficient method for the extraction of DNA. Trends Genet. 1996;12:338–339. doi: 10.1016/s0168-9525(96)80005-9. doi:10.1016/S0168-9525(96)80005-9 [DOI] [PubMed] [Google Scholar]
- Goldsworthy S.D, Page B. A risk-assessment approach to evaluating the significance of seal bycatch in two Australian fisheries. Biol. Conserv. 2007;139:269–285. doi:10.1016/j.biocon.2007.07.010 [Google Scholar]
- Higgins L. The non-annual, nonseasonal breeding cycle of the Australian sea lion, Neophoca cinerea. J. Mamm. 1993;74:270–274. doi:10.2307/1382381 [Google Scholar]
- Kimura M, Weiss G.H. The stepping-stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont M.M, Vida J.T, Harvey J.T, Jeffries S, Brown R, Huber H.H, Delong R, Thomas W.K. Genetic substructure of the Pacific harbour seal (Phoca vitulina richardsi) off Washington, Oregon and California. Mar. Mamm. Sci. 1996;12:402–413. doi:10.1111/j.1748-7692.1996.tb00592.x [Google Scholar]
- Ling J.K. Exploitation of fur seals and sea lions from Australian, New Zealand and adjacent subantarctic islands during the eighteenth, nineteenth and twentieth centuries. Aust. Zool. 1999;31:323–350. [Google Scholar]
- Maddison, W. P. & Maddison, D. R. MacClade: 1992 analysis of phylogeny and character evolution, v. 3, Sunderland, MA: Sinauer Associates.
- Maldonado J.E, Davila F.O, Stewart B.S, Geffen E, Wayne R.K. Intraspecific genetic differentiation in California sea lions (Zalophus californianus) from southern California and the Gulf of California. Mar. Mamm. Sci. 1995;11:46–58. doi:10.1111/j.1748-7692.1995.tb00273.x [Google Scholar]
- Moritz C. Applications of mitochondrial DNA analysis in conservation: a critical review. Mol. Ecol. 1994;3:401–411. [Google Scholar]
- Pearce A.F. Eastern boundary currents of the Southern Hemisphere. J. R. Soc. West. Aust. 1991;74:35–45. [Google Scholar]
- Purdue J.R, Smith M.H, Patton J.C. Female philopatry and extreme spatial genetic heterogeneity in white-tailed deer. J. Mamm. 2000;81:179–185. doi:10.1644/1545-1542(2000)081<0179:FPAESG>2.0.CO;2 [Google Scholar]
- Schneider, S., Roessli, D. & Excoffier, L. 2000 Arlequin: a software for population genetic analysis, v. 2.000, Geneva, Switzerland: Genetics and Biometry Laboratory, University of Geneva.
- Slade R, Moritz C, Hoelzel A, Burton H. Molecular population genetics of the southern elephant seal Mirounga leonina. Genetics. 1998;149:1945–1957. doi: 10.1093/genetics/149.4.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutes in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Weir B.S. Sinauer Associates, Inc; Sunderland, MA: 1996. Genetic data analysis II: methods for discrete population genetic data. [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;6:111–178. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]