Abstract
Mutations in the VHL gene are associated with highly vascular tumors of kidney, brain, retina, and adrenal gland. The ability of the VHL protein to destabilize HIF-1 plays a crucial role in malignant angiogenesis. VHL is also associated with ECM assembly but the molecular mechanisms of this activity remain unclear. We used expression arrays and cell lines with different VHL status to identify ECM-associated genes controlled by VHL. One of them, adhesion-associated TGFBI, was repressed by VHL and overexpressed in renal, gastrointestinal, brain, and other tumors. Analyzing the mechanism of TGFBI up-regulation in clear cell carcinoma, we identified a novel VHL target, a Kruppel-like transcriptional factor 10 (KLF10). The TGFBI promoter, which we isolated and studied in Luc-reporter assay, was induced by KLF10 but not hypoxia. These data provide the molecular basis for the observed VHL effect on TGFBI and stimulate further research into the KLF10 and TGFBI roles in cancer.
Keywords: VHL, TGFBI, KLF10, TGF-β, ECM, HIF-1
Introduction
The von Hippel-Lindau tumor suppressor (VHL) is the substrate recognition component of the E3 ligase that ubiquitinates HIF-1α (or HIF2-α) and plays a pivotal role in the control of hypoxia response [1, 2]. We previously identified several HIF-dependent VHL targets that helped in understanding the VHL function(s) during malignant growth [3–5]. VHL is also involved in HIF-independent regulation of cell-cell interaction, matrix signaling, and adhesion [6–9]. In this study, we asked if novel VHL targets related to ECM deposition can be identified.
We previously reported STRA13 as VHL/HIF-1 target up-regulated in multiple tumors. STRA13 can be also up-regulated by TGF-β [10]. Here we characterize two more targets common for both VHL and TGF-β, such as TGFBI, and KLF10 (TIEG1).
The TGFBI protein (Beta-ig, big-h3, keratoepithelin) is a 68 kDa ECM protein with four evolutionary conserved fasciclin-1 (FAS1) domains and a carboxy-terminal Arg-Gly-Asp (RGD) sequence [11, 12]. The protein is secreted into extracellular space and may bind to fibronectin and collagen [13] as well as integrins [14, 15]. TGFBI was discovered and associated with cancer as a gene induced in the lung adenocarcinoma cell line A549 by TGF-β [12]. The TGFBI function(s) in normal tissues is not fully understood. Mutations in this protein have been implicated in congenital corneal dystrophies [16]. TGFBI was also reported to stimulate adhesion, spreading, migration and proliferation in renal proximal tubular epithelial cells [17]. The RGD peptide of TGFBI can be released from the protein and induce apoptosis [18]. Although TGFBI was associated with cancers in various studies [19–26], the molecular mechanisms of its transcriptional regulation remained unknown.
KLF10, a Kruppel-like transcriptional factor induced by TGF-β, BMP-2, and EGF, mimics the effect of TGF-β in many cells [27, 28]. Although over-expression of KLF10 was recently reported in clear cell carcinomas, glioblastomas, and head-and-neck carcinomas (www.oncomine.org), its function in tumorigenesis remains unclear. In this study, we show that both TGFBI and KLF10 are down-regulated by VHL in 786-0 cells, and that KLF10 may serve as a transactivator of the TGFBI promoter.
Materials and Methods
Cell lines and hypoxic exposure
Human HEK293T cells were purchased from ATCC; U-87, U-251 (astrocytomas) and LN-229 (glioma) were kindly provided by Dr D. Zagzag, and MRC-5 cells (normal untransformed human fetal lung fibroblasts) were from Coriell Cell Repositories, Camden, NJ, USA. 1HAEo(−) cells were described elsewhere [29]. MEF (HIF1+/+ and HIF-1−/−) were obtained from Dr. R. Johnson (University of California San Diego). To produce hypoxia cells were exposed to 0.5% O2, 5% CO2, and 94.5% N2 at 37ºC or incubated with 0.5 mM NiSO4 for 20 h.
Gene expression analysis
RNA samples were isolated, 32P-labeled cDNA probes generated and hybridized with GEArrays HS-010 and HS-023 as recommended by the manufacturer (Superarray, Frederick, MD). Human MTN blots and cancer arrays were purchased from BD Biosciences (Palo Alto, CA). 786-0 clear cell RCC cells stably transfected with VHL transgenes were described previously [3]. Primers for RT-PCR were generated using GeneFisher server (http://bibiserv.techfak.uni-bielefeld.de/genefisher).
TGFBI Promoter and KLF10 construct
The TGFBI promoter was identified using the Genomatix software (http://www.genomatix.de) and isolated via PCR on human DNA with primers 5′-ggtaccTGTGTCTCCCCAGGGCTAG-3′, and 5′-aagcttTGCAGCACCAGCTGGTAG-3′. The promoter was cloned into KpnI/HindIII-digested pGL3-Basic vector and verified by sequencing. The KLF10/TIEG1 expressing construct in pCDNA4/TO was described earlier [28]. Luc-reporter assay was used as described previously [30].
RESULTS
Identification of the adhesion-related VHL targets
To assess the VHL effect on adhesion-associated genes we took advantage of the renal clear cell carcinoma cell lines developed by us and successfully used for identification of novel VHL targets [3, 5, 30]. Original 786-0 cells express VHL devoid of functional domains. These cells were compared to 786-0 stably transfected with wtVHL (786-0/wt2) or mutVHL (786-0/mut2). We used mutVHL that lacks the elongin C-binding alpha domain as a negative control. The results of these experiments and subsequent validation of the VHL targets are shown in Fig. 1, Suppl. Fig. 1, and Table 1.
Fig. 1.
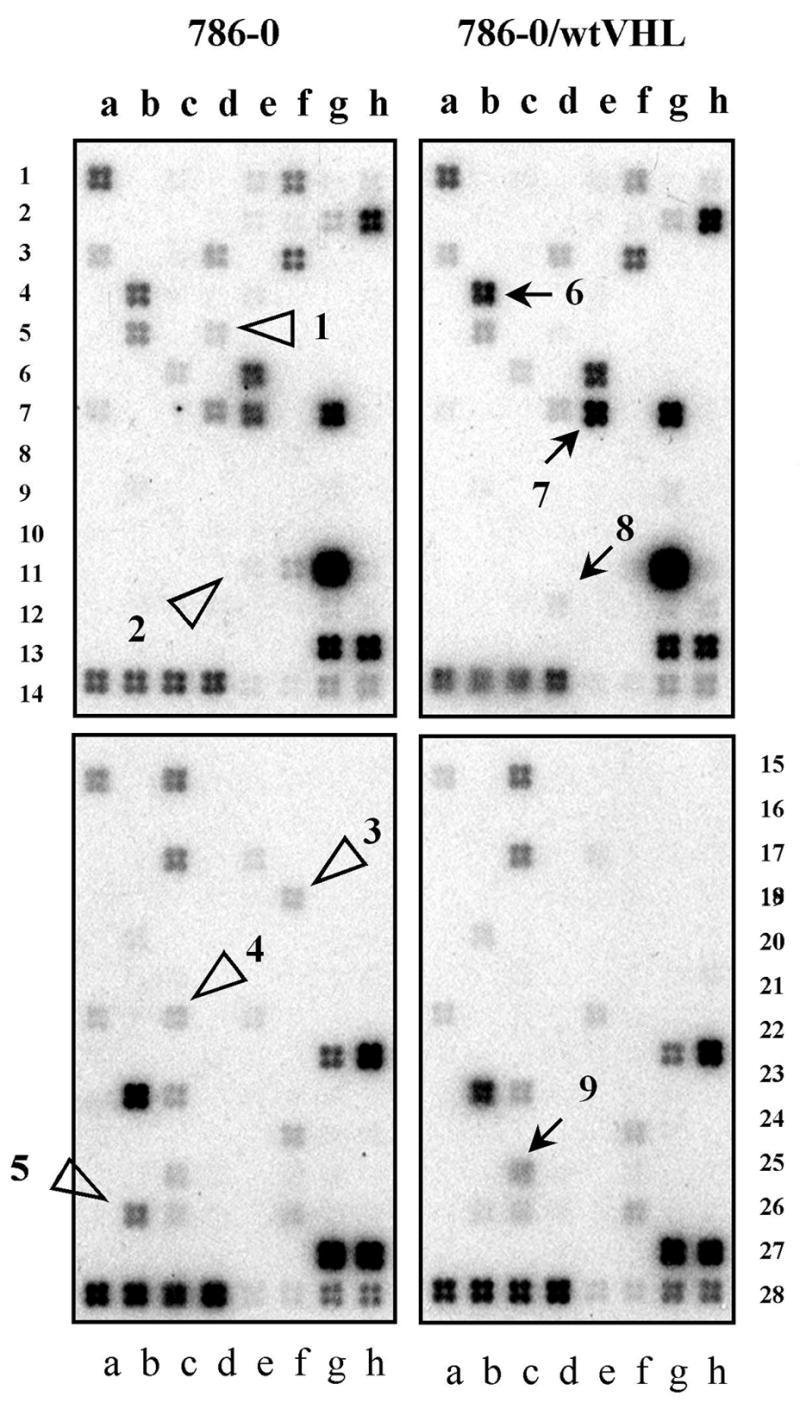
Identification of adhesion-associated TGF-β targets modulated by VHL. Extracellular Matrix and Adhesion (rows 1–14) and TGF-β/BMP (rows 15–28) arrays after hybridization with RNA samples extracted from the VHL-positive or VHL-negative cells. Open triangles indicate genes repressed by wtVHL while arrows show wtVHL-stimulated genes. Full gene charts can be obtained from http://www.superarray.com.
Table 1.
VHL targets involved in regulation of cell adhesion
| Pos. on Fig. 1 | pVHL effect | Gene | GeneBank #, Ref. | RT-PCR evaluation, Fold |
|---|---|---|---|---|
| 1 | ⇓ | ITGA5 | NM_002205 [31, 32] | 2.0 |
| 2 | ⇓ | SERPINE1 | NM_000602 [33, 34] | 10.0 |
| 3 | ⇓ | COL3A1 | NM_000090 | Not supported |
| 4 | ⇓ | IGFBP3 | NM_000598 [7] | 2.0 |
| 5 | ⇓ | TGFBi | NM_000358 | 3.5 |
| 6 | ⇑ | FN1 | NM_002026 [7] | Not done |
| 7 | ⇑ | LAMB1 | NM_002291 | 1.7 |
| 8 | ⇑ | TIMP2 | NM_003255 [35] | Not done |
| 9 | ⇑ | STAT1 | NM_007315 [30] | 2.5 |
Five out of nine adhesion-related genes identified by us as VHL-dependent, ITGA5, SERPINE1, IGFBP3, FN1, and TIMP2, turned out to be already associated with VHL targets (see refs in Table 1). STAT1 (number 9 in Fig. 1 and Table 1) that showed up-regulation by wtVHL in this study, was recently characterized by us as a VHL target that is controlled via STRA13 [30]. Identification of all these previously known VHL targets validated our approach as a reliable tool for finding genes transcriptionally modulated by VHL. We then focused on TGFBI (BIGH3, betaig-h3, keratoepithelin) that encodes a secreted matrix protein with apoptotic and adhesion-related growth activities. This gene, as we show by RT-PCR (Suppl Fig. 1A) and Northern analysis (Suppl Fig. 1B), was down-regulated by wtVHL but not by mutant in 786-0 cells.
Classical TGF-β components are not involved in the VHL-dependent effects
Expression of TGF-β1 (location f25), -β2 (h25), and -β3 (a26) was barely detectable (Fig. 1 and Suppl. Table 1), and no VHL effect was seen on these genes at a longer exposure (data not shown). The type II activin A receptor (ACVR2), c15; type I activin A receptors ALK-1, ALK-5, endoglin, Smad1–7, and Smad9 were also not affected. In a separate experiment on the same cells, no VHL-dependent changes in the amounts of phosphorylated SMAD1/5/8 or SMAD2 were detected (data not shown).
KLF10 is a VHL target that regulates the TGFBI promoter
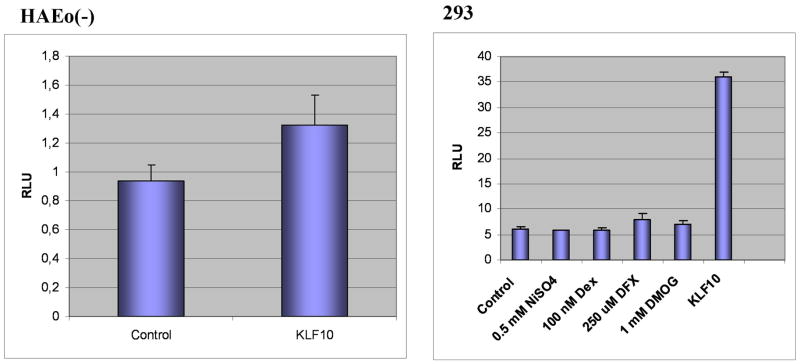
Seeking novel possible mediators of the VHL effect on TGFBI we focused on recently characterized transcriptional regulators KLF10 and KLF11 controlled by TGF-β [36, 37]. We found that KLF10 but not KLF11 was repressed in 786-0 cells by wtVHL but not mutVHL (Suppl Fig. 1B and data not shown) suggesting that it may also serve as a VHL target. We then identified the TGFBI promoter, isolated it via PCR, and assessed if KLF10 can transactivate it in a Luc-reporter assay. In this promoter, we identified a potential KLF10 binding site, which is also recognized as an Sp1-binding site and is very similar to the one found within the CD11d promoter [38]. This site in the TGFBI promoter is localized 89-80 nucleotides upstream from the transcription initiation site (Suppl. Fig. 2). No HRE motif typical for most of hypoxia-stimulated VHL targets was found in the promoter arguing against its direct activation by HIF-1. Co-expression of TGFBI-Luc reporter plasmid with KLF10 produced ~1.4–7-fold stimulation in 2 different cell lines (Fig. 2) suggesting that KLF10 may transactivate the TGFBI promoter in a cell type-specific manner. In agreement with the lack of the HRE site, no induction was observed by hypoxic mimetics and atmospheric hypoxia.
Fig. 2.
Stimulation of the TGFBI promoter by KLF10 in HAEo(−) and 293 cells.
TGFBI is commonly over-expressed in cancers
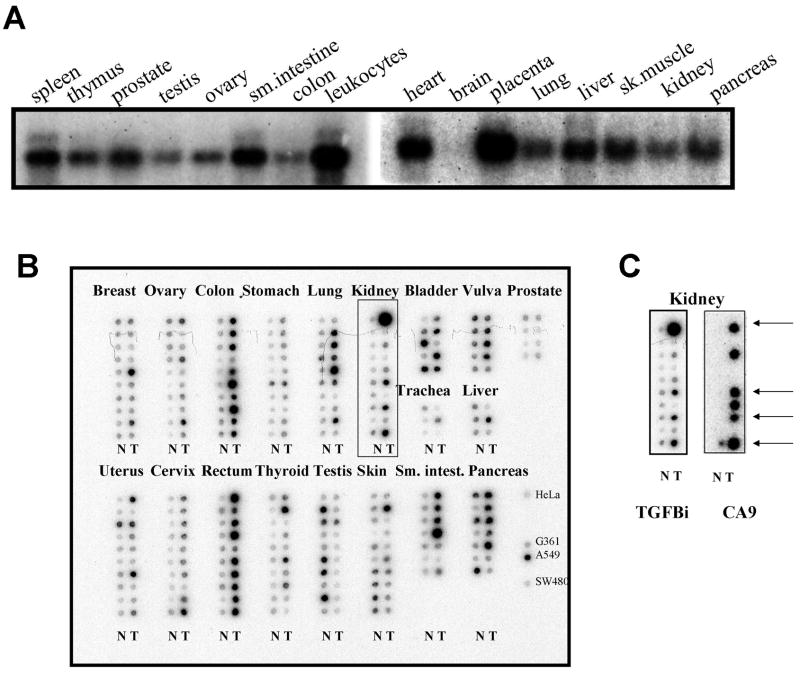
We found that TGFBI is broadly expressed in normal tissues with highest expression levels detected in placenta, leukocytes, and heart. TGFBI expression is moderate in kidney where it is localized predominantly to the epithelial cells of collecting ducts and distal as well as proximal tubules (Suppl. Fig. 3). TGFBI expression in brain was practically undetectable (Fig. 3A). Studying TGFBI expression in cancer we compared between matched tumor/normal RNA samples. As a result, 4 out of 10 renal, 5/7 pancreatic, 4/10 lung, 9/10 colon, 9/10 rectal, and 4/7 small intestine cancers showed TGFBI over-expression (Fig. 3BC). Our immunohistochemical study showed that in normal kidney TGFBI is expressed in distal and proximal tubular epithelium and in collecting duct epithelium. We also detected TGFBI expression in clinical specimens of renal clear cell carcinoma and glioblastoma (Suppl. Fig. 3). Analysis of the public cancer expression database Oncomine (http://www.oncomine.org) supported these observations and helped to identify more tumors that overexpress TGFBI, such as head-and-neck squamous cell carcinoma and seminoma (Suppl. Table 2).
Fig. 3.
Expression of TGFBI in normal and cancer tissues. A. Northern analysis with MTN blots. B. Expression analysis of 19 tumor types represented by paired tumor (T) and matched normal (N) RNA samples arranged in columns. Right bottom column shows TGFBI-expressing cancer cell lines. C. Comparison of TGFBI and CA9 expression in the set samples framed in B. Arrows show samples where both TGFBI and CA9 are induced.
TGFBI and KLF10 expression in cells with different HIF status
After showing that the TGFBI promoter does not possess HRE we asked if endogenous TGFBI can be induced by hypoxia and if KLF10 may mediate this effect. Endogenous TGFBI expression levels were compared by RT-PCR with the VHL/HIF targets CA9, CA12, and STRA13 characterized by us previously [3, 5] (Suppl. Fig. 4). While STRA13 showed consistent but moderate hypoxic up-regulation in each cell line studied, CA12 was induced in 2/5, and CA9 in 4/5. TGFBI showed a limited response to hypoxia in HAEo(−) cells only. KLF10 expression was not affected by hypoxic conditions.
To directly assess HIF involvement in regulation of endogenous TGFBI and KLF10, we compared their expression in Hif-1α-deficient and proficient MEFs (Fig. 4). Tgfbi expression in these cells was slightly elevated by hypoxia in a Hif-1α-dependent manner, while KLF10 was not affected by hypoxia. Altogether, these results suggest that the KLF-10 effect on TGFBI is hypoxia-independent, and that TGFBI shows a moderate response to hypoxia in a limited set of cell lines.
Fig. 4.
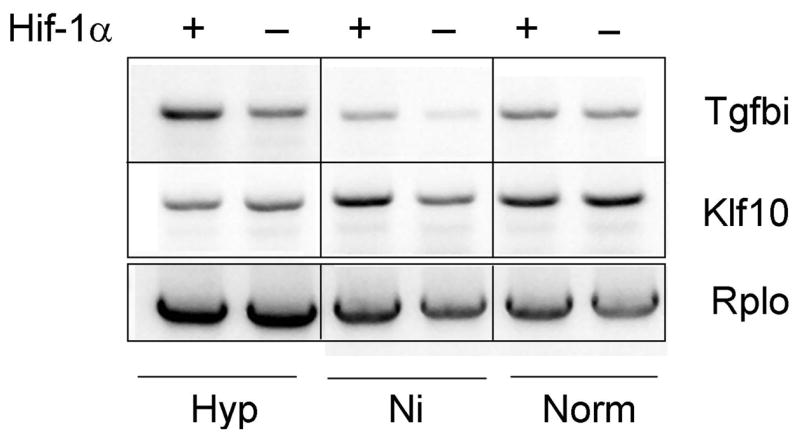
Hif-1α effect on endogenous Tgfbi and Klf10 expression in mouse fibroblasts.
Cell specificity of TGFBI response to hypoxia was further evaluated on 26 cancer cell lines that represented 13 different types of common tumors (Suppl. Fig. 5). Treatment with a hypoxia mimetic desferrioxamine induced TGFBI expression in 6 out of 26. This finding further substantiated high cell-specificity of the hypoxic effect on TGFBI.
To assess if HIF can be specifically involved in up-regulation of TGFBI in clinical renal carcinoma specimens, we stripped the membrane shown in Fig. 3B and re-hybridized it with CA9, a well-characterized hypoxia-inducible gene [4]. As seen from Fig. 4C, TGFBI was strongly stimulated in 1 and moderately induced in 3 out of 6 CA9-positive tumors. Taken together, all these results suggest that the up-regulation of TGFBI commonly observed in tumor specimens and cancer cell lines cannot be fully and reliably explained by hypoxic stimulation.
Expression of TGFBI in cancer cell lines in response to stress factors and chemotherapeutic agents
Since TGFBI was previously associated with apoptosis [18, 39] and apoptosis is linked with cell survival in general and with hypoxia in particular, we asked if TGFBI expression can be induced by stress-related stimuli. To this end, we studied TGFBI expression in 26 cancer cell lines exposed to 26 chemotherapeutic agents (Suppl. Fig. 5). Among all cancer types, breast (MDA-MB-435S, MCF-7), skin (SK-N-SH), and epidermal (A-431) cancer cells showed the highest amplitude of TGFBI induction in response to different treatment conditions. In 9/26 cell lines TGFBI expression was up-regulated by UV irradiation, heat shock, desferrioxamine, hydrogen peroxide, and gamma irradiation. TGFBI was also induced by common chemotherapeutic agents, e.g. a ribonucleotide reductase inhibitor hydroxyurea, a topoisomerase 2 inhibitor etoposide, a DNA synthesis inhibitors 5′-fluoracil, and cisplatin. Chemoresistant H460 non-small cell lung cancer cells turned out to be the most responsive showing TGFBI induction in 23 out of 26 conditions. Using the Gene Expression Omnibus repository at NCBI we found Tgfbi overexpression during ischemic/hypoxic brain and lung injury (accession ## GDS1017 and GDS247, respectively), lung hyperoxic and inflammatory injury (GDS247 and GDS1239), as well as allergic reactions in lung (GDS42 and GDS958). All these data relate TGFBI expression with stress response suggesting that hypoxia is one of multiple stress-related stimuli that may induce TGFBI expression.
DISCUSSION
The molecular mechanisms of VHL effect(s) on tumor progression are not fully defined and novel HIF-independent VHL targets identified recently, such as recently characterized secreted tumor marker clusterin [40] or PL6 (our study [41]) provide more insight into the VHL tumor suppressor function. Research into the molecular mechanisms of the VHL effect on the extracellular matrix may provide novel therapeutic approaches against stroma-supported tumor growth and invasion [42]. In this study, using our assay for identification of novel VHL targets, we characterized ITGA5, SERPINE(PAI-1), IGFBP3, TGFBI, FN1, LAMB, and TIMP2 as common VHL and TGF-β transcriptional targets. Our research was then focused on TGFBI, an important extracellular matrix protein with as yet unknown function in cancer. Our data linked TGFBI over-expression with mutations in the von Hippel-Lindau gene and VHL-associated cancers, such as clear cell carcinoma and hemangioblastoma. We demonstrated that KLF10, a yet another novel VHL target that we identified, may potently transactivate the TGFBI promoter independently of hypoxia. This finding suggests that the KLF10 may mediate up-regulation of TGFBI in VHL-deficient tumors and other cancers.
Addressing clinical significance of our findings we found TGFBI up-regulation in 40% of kidney, 90% of the colorectal and ~60% of intestinal cancers. TGFBI was also highly expressed in brain tumors but not in normal brain tissue suggesting that it may serve as a diagnostic marker. Since similar observations were reported by other investigators [43, 44], we suggest that it is now imperative to see if TGFBI protein can be secreted by malignant brain cells into the cerebrospinal fluid and serve for early detection of brain tumors. What role(s) may TGFBI play in tumorigenesis? Our finding that TGFBI is expressed in epithelial cells of normal kidney is in agreement with the role suggested for TGFBI in promoting adhesion, migration, and regeneration of these cells [17]. Likewise, up-regulation of TGFBI in cancer cells may stimulate their survival via ECM-dependent signaling. Indeed, a TGFBI ortholog periostin is also linked with tumorigenesis where it may promote angiogenesis and metastasizing [45, 46]. In this regard, our study provides the first evidence that TGFBI expression in tumors may be controlled by stress-associated pathways.
Supplementary Material
A.Verification of the VHL targets by RT-PCR. Five genes were confirmed to be under the VHL control by RT-PCR. Four of them were down-regulated by wtVHL, while LAMB1 was stimulated. As a loading control, we used RT-PCR on beta-2-microglobulin. B. Verification of the VHL targets by Northern analysis. TGFBI and KLF10 are down-regulated by VHL. Membrane was stained with methylene blue and 23S rRNA was used as a loading control.
TGFBI promoter used for Luc-reporter studies. Putative protein-binding motifs are shown in bold and underlined. The 5′-end of the transcript is shown in italic and the protein-coding sequence marked in bold.
Immunohistochemical analysis of TGFBI expression in kidney and clear cell carcinoma. Normal human kidney (A, B) shows TGFBI expression in epithelial cells of distal tubules and collecting duct and, to a lesser extent, in proximal tubules. C, D – sections of clear cell carcinoma stained with TGFBI antibodies. E- sections of glioblastoma stained with TGFBI antibodies. Intense staining in necrotic/hypoxic areas is observed with a 1:200 delution of antibodies. F- specificity of the staining in E was verified with 10 nM antigenic peptide.
Responsiveness of TGFBI and KLF10 to hypoxic conditions as compared to known VHL targets.
C. TGFBI expression in 26 cancer cell lines treated with chemotherapeutic agents and stress stimuli. Rectangles show significant up-regulation of TGFBI expression as compared to controls, while circles indicate induction of TGFBI with desferrioxamine.
Acknowledgments
This research was supported by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research and by Contract No. NO1-CO-56000 to Basic Research Program, SAIC-Frederick, Inc. The content of the publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Authors thank Robert Nalewaik for technical help with experimentation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- 1.Kaelin WG., Jr Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2(9):673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov SV, Kuzmin I, Wei MH, Pack S, Geil L, Johnson BE, Stanbridge EJ, Lerman MI. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc Natl Acad Sci U S A. 1998;95(21):12596–12601. doi: 10.1073/pnas.95.21.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158(3):905–919. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanova AV, Ivanov SV, Danilkovitch-Miagkova A, Lerman MI. Regulation of STRA13 by the von Hippel-Lindau tumor suppressor protein, hypoxia, and the UBC9/ubiquitin proteasome degradation pathway. J Biol Chem. 2001;276(18):15306–15315. doi: 10.1074/jbc.M010516200. [DOI] [PubMed] [Google Scholar]
- 6.Kamada M, Suzuki K, Kato Y, Okuda H, Shuin T. von Hippel-Lindau protein promotes the assembly of actin and vinculin and inhibits cell motility. Cancer Res. 2001;61(10):4184–4189. [PubMed] [Google Scholar]
- 7.Bluyssen HA, Lolkema MP, van Beest M, Boone M, Snijckers CM, Los M, Gebbink MF, Braam B, Holstege FC, Giles RH, et al. Fibronectin is a hypoxia-independent target of the tumor suppressor VHL. FEBS Lett. 2004;556(1–3):137–142. doi: 10.1016/s0014-5793(03)01392-9. [DOI] [PubMed] [Google Scholar]
- 8.Davidowitz EJ, Schoenfeld AR, Burk RD. VHL induces renal cell differentiation and growth arrest through integration of cell-cell and cell-extracellular matrix signaling. Mol Cell Biol. 2001;21(3):865–874. doi: 10.1128/MCB.21.3.865-874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteban-Barragan MA, Avila P, Alvarez-Tejado M, Gutierrez MD, Garcia-Pardo A, Sanchez-Madrid F, Landazuri MO. Role of the von Hippel-Lindau tumor suppressor gene in the formation of beta1-integrin fibrillar adhesions. Cancer Res. 2002;62(10):2929–2936. [PubMed] [Google Scholar]
- 10.Zawel L, Yu J, Torrance CJ, Markowitz S, Kinzler KW, Vogelstein B, Zhou S. DEC1 is a downstream target of TGF-beta with sequence-specific transcriptional repressor activities. Proc Natl Acad Sci U S A. 2002;99(5):2848–2853. doi: 10.1073/pnas.261714999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamoto T, Noshiro M, Shen M, Nakamasu K, Hashimoto K, Kawashima-Ohya Y, Gotoh O, Kato Y. Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim Biophys Acta. 1998;1395(3):288–292. doi: 10.1016/s0167-4781(97)00172-3. [DOI] [PubMed] [Google Scholar]
- 12.Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol. 1992;11(7):511–522. doi: 10.1089/dna.1992.11.511. [DOI] [PubMed] [Google Scholar]
- 13.Billings PC, Whitbeck JC, Adams CS, Abrams WR, Cohen AJ, Engelsberg BN, Howard PS, Rosenbloom J. The transforming growth factor-beta-inducible matrix protein (beta)ig-h3 interacts with fibronectin. J Biol Chem. 2002;277(31):28003–28009. doi: 10.1074/jbc.M106837200. [DOI] [PubMed] [Google Scholar]
- 14.Kim JE, Jeong HW, Nam JO, Lee BH, Choi JY, Park RW, Park JY, Kim IS. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein betaig-h3 that interact with the alphavbeta5 integrin. J Biol Chem. 2002;277(48):46159–46165. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- 15.Kim MO, Yun SJ, Kim IS, Sohn S, Lee EH. Transforming growth factor-beta-inducible gene-h3 (beta(ig)-h3) promotes cell adhesion of human astrocytoma cells in vitro: implication of alpha6beta4 integrin. Neurosci Lett. 2003;336(2):93–96. doi: 10.1016/s0304-3940(02)01260-0. [DOI] [PubMed] [Google Scholar]
- 16.Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mutat. 2006;27(7):615–625. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- 17.Park SW, Bae JS, Kim KS, Park SH, Lee BH, Choi JY, Park JY, Ha SW, Kim YL, Kwon TH, et al. Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with alpha3beta1 integrin. Exp Mol Med. 2004;36(3):211–219. doi: 10.1038/emm.2004.29. [DOI] [PubMed] [Google Scholar]
- 18.Kim JE, Kim SJ, Jeong HW, Lee BH, Choi JY, Park RW, Park JY, Kim IS. RGD peptides released from beta ig-h3, a TGF-beta-induced cell-adhesive molecule, mediate apoptosis. Oncogene. 2003;22(13):2045–2053. doi: 10.1038/sj.onc.1206269. [DOI] [PubMed] [Google Scholar]
- 19.Arimoto T, Katagiri T, Oda K, Tsunoda T, Yasugi T, Osuga Y, Yoshikawa H, Nishii O, Yano T, Taketani Y, et al. Genome-wide cDNA microarray analysis of gene-expression profiles involved in ovarian endometriosis. Int J Oncol. 2003;22(3):551–560. [PubMed] [Google Scholar]
- 20.Sasaki H, Kobayashi Y, Nakashima Y, Moriyama S, Yukiue H, Kaji M, Kiriyama M, Fukai I, Yamakawa Y, Fujii Y. Beta IGH3, a TGF-beta inducible gene, is overexpressed in lung cancer. Jpn J Clin Oncol. 2002;32(3):85–89. doi: 10.1093/jjco/hyf021. [DOI] [PubMed] [Google Scholar]
- 21.Schneider D, Kleeff J, Berberat PO, Zhu Z, Korc M, Friess H, Buchler MW. Induction and expression of betaig-h3 in pancreatic cancer cells. Biochim Biophys Acta. 2002;1588(1):1–6. doi: 10.1016/s0925-4439(02)00052-2. [DOI] [PubMed] [Google Scholar]
- 22.Tang J, Zhou HW, Jiang JL, Yang XM, Li Y, Zhang HX, Chen ZN, Guo WP. BetaIg-h3 is involved in the HAb18G/CD147-mediated metastasis process in human hepatoma cells. Exp Biol Med (Maywood) 2007;232(3):344–352. [PubMed] [Google Scholar]
- 23.Becker J, Erdlenbruch B, Noskova I, Schramm A, Aumailley M, Schorderet DF, Schweigerer L. Keratoepithelin suppresses the progression of experimental human neuroblastomas. Cancer Res. 2006;66(10):5314–5321. doi: 10.1158/0008-5472.CAN-05-3049. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, El-Gabry M, Hei TK. Loss of Betaig-h3 protein is frequent in primary lung carcinoma and related to tumorigenic phenotype in lung cancer cells. Mol Carcinog. 2006;45(2):84–92. doi: 10.1002/mc.20167. [DOI] [PubMed] [Google Scholar]
- 25.Walker G, MacLeod K, Williams AR, Cameron DA, Smyth JF, Langdon SP. Estrogen-regulated gene expression predicts response to endocrine therapy in patients with ovarian cancer. Gynecol Oncol. 2007;106(3):461–468. doi: 10.1016/j.ygyno.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Buckhaults P, Rago C, St Croix B, Romans KE, Saha S, Zhang L, Vogelstein B, Kinzler KW. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61(19):6996–7001. [PubMed] [Google Scholar]
- 27.Subramaniam M, Hawse JR, Johnsen SA, Spelsberg TC. Role of TIEG1 in biological processes and disease states. J Cell Biochem. 2007;102(3):539–548. doi: 10.1002/jcb.21492. [DOI] [PubMed] [Google Scholar]
- 28.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21(37):5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 29.Salnikow K, Li X, Lippmann M. Effect of nickel and iron co-exposure on human lung cells. Toxicol Appl Pharmacol. 2004;196(2):258–265. doi: 10.1016/j.taap.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Ivanov SV, Salnikow K, Ivanova AV, Bai L, Lerman MI. Hypoxic repression of STAT1 and its downstream genes by a pVHL/HIF-1 target DEC1/STRA13. Oncogene. 2006 doi: 10.1038/sj.onc.1209842. [DOI] [PubMed] [Google Scholar]
- 31.Wykoff CC, Sotiriou C, Cockman ME, Ratcliffe PJ, Maxwell P, Liu E, Harris AL. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br J Cancer. 2004;90(6):1235–1243. doi: 10.1038/sj.bjc.6601657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koike T, Kimura N, Miyazaki K, Yabuta T, Kumamoto K, Takenoshita S, Chen J, Kobayashi M, Hosokawa M, Taniguchi A, et al. Hypoxia induces adhesion molecules on cancer cells: A missing link between Warburg effect and induction of selectin-ligand carbohydrates. Proc Natl Acad Sci U S A. 2004;101(21):8132–8137. doi: 10.1073/pnas.0402088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Los M, Zeamari S, Foekens JA, Gebbink MF, Voest EE. Regulation of the urokinase-type plasminogen activator system by the von Hippel-Lindau tumor suppressor gene. Cancer Res. 1999;59(17):4440–4445. [PubMed] [Google Scholar]
- 34.Zatyka M, da Silva NF, Clifford SC, Morris MR, Wiesener MS, Eckardt KU, Houlston RS, Richards FM, Latif F, Maher ER. Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 2002;62(13):3803–3811. [PubMed] [Google Scholar]
- 35.Koochekpour S, Jeffers M, Wang PH, Gong C, Taylor GA, Roessler LM, Stearman R, Vasselli JR, Stetler-Stevenson WG, Kaelin WG, Jr, et al. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol Cell Biol. 1999;19(9):5902–5912. doi: 10.1128/mcb.19.9.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23(23):4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273(40):25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 38.Noti JD, Johnson AK, Dillon JD. The zinc finger transcription factor transforming growth factor beta-inducible early gene-1 confers myeloid-specific activation of the leukocyte integrin CD11d promoter. J Biol Chem. 2004;279(26):26948–26958. doi: 10.1074/jbc.M310634200. [DOI] [PubMed] [Google Scholar]
- 39.Morand S, Buchillier V, Maurer F, Bonny C, Arsenijevic Y, Munier FL, Schorderet DF. Induction of apoptosis in human corneal and HeLa cells by mutated BIGH3. Invest Ophthalmol Vis Sci. 2003;44(7):2973–2979. doi: 10.1167/iovs.02-0661. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura E, Abreu-e-Lima P, Awakura Y, Inoue T, Kamoto T, Ogawa O, Kotani H, Manabe T, Zhang GJ, Kondo K, et al. Clusterin is a secreted marker for a hypoxia-inducible factor-independent function of the von Hippel-Lindau tumor suppressor protein. Am J Pathol. 2006;168(2):574–584. doi: 10.2353/ajpath.2006.050867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanova A, Vortmeyer A, Ivanov S, Nickerson M, Maher E, Lerman M. Loss of PL6 protein expression in renal clear cell carcinomas and other VHL-deficient tumours. J Pathol. 2007 doi: 10.1002/path.2252. [DOI] [PubMed] [Google Scholar]
- 42.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15(1):97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golembieski WA, Rempel SA. cDNA array analysis of SPARC-modulated changes in glioma gene expression. J Neurooncol. 2002;60(3):213–226. doi: 10.1023/a:1021167211131. [DOI] [PubMed] [Google Scholar]
- 44.Colin C, Baeza N, Bartoli C, Fina F, Eudes N, Nanni I, Martin PM, Ouafik L, Figarella-Branger D. Identification of genes differentially expressed in glioblastoma versus pilocytic astrocytoma using Suppression Subtractive Hybridization. Oncogene. 2006;25(19):2818–2826. doi: 10.1038/sj.onc.1209305. [DOI] [PubMed] [Google Scholar]
- 45.Kudo Y, Ogawa I, Kitajima S, Kitagawa M, Kawai H, Gaffney PM, Miyauchi M, Takata T. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res. 2006;66(14):6928–6935. doi: 10.1158/0008-5472.CAN-05-4540. [DOI] [PubMed] [Google Scholar]
- 46.Puglisi F, Puppin C, Pegolo E, Andreetta C, Pascoletti G, D’Aurizio F, Pandolfi M, Fasola G, Piga A, Damante G, et al. Expression of periostin in human breast cancer. J Clin Pathol. 2007 doi: 10.1136/jcp.2007.052506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A.Verification of the VHL targets by RT-PCR. Five genes were confirmed to be under the VHL control by RT-PCR. Four of them were down-regulated by wtVHL, while LAMB1 was stimulated. As a loading control, we used RT-PCR on beta-2-microglobulin. B. Verification of the VHL targets by Northern analysis. TGFBI and KLF10 are down-regulated by VHL. Membrane was stained with methylene blue and 23S rRNA was used as a loading control.
TGFBI promoter used for Luc-reporter studies. Putative protein-binding motifs are shown in bold and underlined. The 5′-end of the transcript is shown in italic and the protein-coding sequence marked in bold.
Immunohistochemical analysis of TGFBI expression in kidney and clear cell carcinoma. Normal human kidney (A, B) shows TGFBI expression in epithelial cells of distal tubules and collecting duct and, to a lesser extent, in proximal tubules. C, D – sections of clear cell carcinoma stained with TGFBI antibodies. E- sections of glioblastoma stained with TGFBI antibodies. Intense staining in necrotic/hypoxic areas is observed with a 1:200 delution of antibodies. F- specificity of the staining in E was verified with 10 nM antigenic peptide.
Responsiveness of TGFBI and KLF10 to hypoxic conditions as compared to known VHL targets.
C. TGFBI expression in 26 cancer cell lines treated with chemotherapeutic agents and stress stimuli. Rectangles show significant up-regulation of TGFBI expression as compared to controls, while circles indicate induction of TGFBI with desferrioxamine.