Abstract
Activation-induced cytidine deaminase (AID) initiates all postrearrangement processes that diversify the immunoglobulin repertoire by specific deamination of cytidines at the immunoglobulin (Ig) locus. As uncontrolled expression of AID is potentially mutagenic, different types of regulation, particularly nucleocytoplasmic shuttling, restrict the likelihood of AID–deoxyribonucleic acid encounters. We studied additional mechanisms of regulation affecting the stability of the AID protein. No modulation of protein accumulation according to the cell cycle was observed in a Burkitt's lymphoma cell line. In contrast, the half-life of AID was markedly reduced in the nucleus, and this destabilization was accompanied by a polyubiquitination that was revealed in the presence of proteasome inhibitors. The same compartment-specific degradation was observed in activated mouse B cells, and also in a non–B cell line. No specific lysine residues could be linked to this degradation, so it remains unclear whether polyubiquitination proceeds through several alternatives sites or through the protein N terminus. The nuclear-restricted form of AID displayed enhanced mutagenicity at both Ig and non-Ig loci, most notably at TP53, suggesting that modulation of nuclear AID content through proteasomal degradation may represent another level of control of AID activity.
B lymphocytes undergo two successive and regulated genomic modifications to generate the repertoire of specificities that allows them to cope with an almost infinite number of antigens (for review see [1, 2]). Early in development, the B cell receptor is assembled by V(D)J recombination, a site-specific recombination initiated by the lymphocyte-specific proteins RAG1 and RAG2, thus allowing the formation of the primary B cell repertoire. B cells reshape this repertoire in response to antigenic stimulation through two processes, class switch recombination (CSR) and somatic hypermutation (SHM), to generate secreted antibodies and memory B cells with improved antigen-binding capacities. SHM allows the introduction of point mutations, mainly in DNA encoding the V (variable) region of both rearranged heavy and light chain Ig genes, whereas CSR replaces the μ heavy chain constant (C) region by downstream C sequences (γ, ε, or α), for the expression of IgG, IgE, or IgA, respectively. Both processes, SHM and CSR, are initiated by a B cell–specific enzyme, activation-induced cytidine deaminase (AID). AID was first discovered and identified by Muramatsu et al. as a gene differentially expressed in cells induced to undergo isotype switching, and in centroblasts of mouse germinal centers (3). Its inactivation in mice abolishes both SHM and CSR in activated B cells (4), and a similar phenotype is observed in humans, the type 2 hyper-IgM syndrome (HIGM2) corresponding to the clinical manifestation of the inactivation of AID (5).
AID is closely related to APOBEC-1, which is an RNA-editing deaminase that targets the apolipoprotein B transcript and induces the formation of a shorter polypeptide by generating a premature stop codon (6). Genetic and biochemical analyses have, however, provided many lines of evidence that AID directly modifies DNA at the Ig locus by carrying out deamination of cytidine into uracil in exposed regions of single-stranded DNA (for review see [7]). The induced lesion triggers an error-prone repair cascade leading to the introduction of mutations in the Ig variable region or to double-strand breaks in switch regions allowing the rejoining of distantly located sequences. To modify bases on DNA, AID must be localized in the nucleus; however, data from several studies using either cell fractionation, microscopic examination of cells expressing AID fused to EGFP or immunohistological staining of lymphoid tissues, showed that AID localizes predominantly in the cytoplasm (8–12). This localization was shown to result, as for APOBEC1, from a constant nucleocytoplasmic shuttling mediated by the CRM1 pathway, in which nuclear export dominates (9–11). Active export has been clearly assigned to a nuclear export signal (NES) located in the C-terminal part of AID. The role of an N-terminal bipartite nuclear localization signal (NLS) remains in debate, but whether directly or indirectly, point mutations in this domain can clearly abolish the nuclear translocation of AID (13).
In a transgenic mouse model, constitutive expression of AID has been associated with the onset of tumors, particularly tumors of thymic origin (14). More recent data have clearly demonstrated the implication of AID in the induction of translocations between the c-myc and the heavy chain locus, similar to those observed in Burkitt's lymphoma (15–17). Therefore, the identification of AID as a potential tumor inducer has brought to light the importance of its regulation during the germinal center reaction. In this study, we report that the stability of AID is related to its subcellular localization, with the AID protein being destabilized in the nucleus through proteasomal-mediated degradation. Nuclear polyubiquitination of AID thus appears as another level of regulation that participates in controlling the access of this enzyme to its DNA target. The possibility that polyubiquitination may compete or interact with other posttranslational modifications modulating AID function is further discussed.
RESULTS
No cell-cycle regulation of AID expression at either the transcriptional or the posttranscriptional level
To facilitate the analysis of regulatory signals affecting the expression level of the AID protein, knock-in (KI) of the EGFP coding sequence at the AICDA locus was performed by homologous recombination in the BL2 cell line. EGFP was fused in frame with exon 5 at the 3′ end of the AID coding sequence, with the only other modification being the insertion of a loxP site in the intronic sequence between exons 2 and 3 (Fig. 1 A) after Cre-mediated excision of the hygromycin resistance marker. The expression of the AID protein from both KI and WT alleles was then compared and appeared to be similar. Their respective half-lives were assessed by [35S]methionine/cysteine pulse-chase labeling and immunoprecipitation with anti-AID monoclonal antibodies (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20070950/DC1; see the characterization of the anti-AID monoclonal antibodies used in this study in Fig. S2). Quantification of radiolabeled AID indicated that both forms of the proteins, whether they were linked to EGFP or not, had a comparable half-life (Fig. S1 B). IL-4 was added to the culture medium in this experiment to facilitate analysis, as it induced a two- to threefold increase in protein expression, an increase that was shown by Western blot analysis to affect both alleles comparably (unpublished data). In the presence of leptomycin B (LMB), which is an inhibitor of CRM1-mediated nuclear export, accumulation of AID-EGFP in the nucleus took place in a similar manner to the WT protein, suggesting that the shuttling of AID-EGFP between the nucleus and cytoplasm is not altered either (Fig. S1 C). Collectively, these data validate the use of the AID-EGFP knocked-in protein to investigate the regulation of the endogenous AID, and indicate that fusion with EGFP does not affect the stability or the trafficking of AID within the cell.
Figure 1.
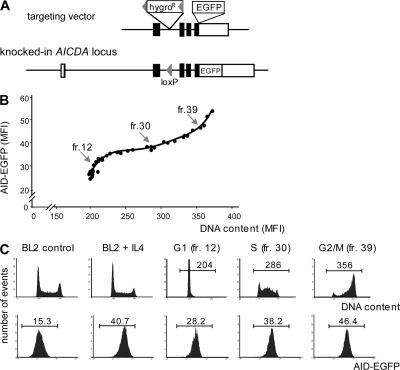
Variation of AID expression during the cell cycle. (A) EGFP KI at the AICDA locus in BL2 cells. The AID–EGFP KI construct includes the EGFP sequence inserted in-frame in exon 5 at the 3′ end of the AID coding region and a hygromycin resistance (hygroR) gene flanked by loxP sites. Configuration of the targeted AICDA locus is depicted after Cre-mediated excision of the hygroR gene. (B) Expression of AID-EGFP throughout the cell cycle. 48 h after IL-4 addition (10 ng/ml), AID-EGFP KI BL2 cells were fractionated according to their cell cycle status using counterflow elutriation. Collected fractions were stained with propidium iodide and analyzed for both DNA content and AID-EGFP MFI. Data for fractions 12, 30, and 39 are shown in C. (C) Cell cycle analysis and AID-EGFP expression level of the BL2 KI cell line with and without IL-4, and of representative elutriated fractions, corresponding to the different phases of cell cycle: G1 (fr.12), S (fr. 30), and G2/M (fr. 39).
We therefore used this KI clone to ask whether the AID protein is expressed in a specific phase of the cell cycle. A BL2 clone with one knock-out and one KI allele was selected because it displayed a brighter EGFP fluorescence, which facilitates such a study. Exponentially growing cells in the presence of IL-4 were submitted to elutriation, a selection procedure based on the size and density of the cell, which allows the collection of cell fractions according to their position in the cell cycle. Each collected fraction was analyzed for both AID-EGFP fluorescence and DNA content by flow cytometry, thus allowing the correlation of the relative abundance of the protein to the various phases of the cell cycle. Fig. 1 (B and C) shows that AID-EGFP increases steadily as the cell gains in size and progresses from G1 to G2/M, thus indicating that AID is continually expressed throughout the cell cycle. As EGFP-fluorescence varied within a factor of two, the expression level of AID appears roughly correlated with its gene copy number within the cell.
Differential accumulation of AID mutant proteins according to their subcellular localization
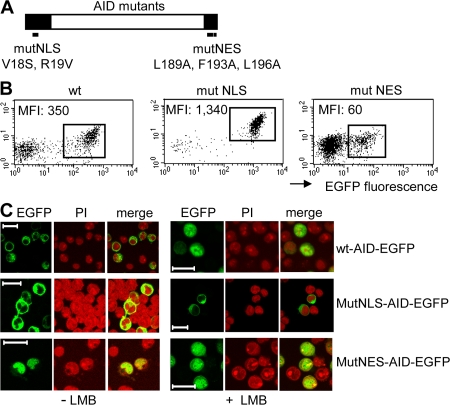
To obtain BL2 subclones expressing AID-EGFP with various subcellular localizations, we generated tetracycline-inducible expression vectors containing WT human AID tagged with EGFP (WT-AID-EGFP), or AID-EGFP containing mutations in either the NLS (MutNLS-AID-EGFP) or in the NES (MutNES-AID-EGFP), as indicated in Fig. 2 A. The different constructs were transfected in the BL2 cell line and the protein localization was confirmed by fluorescence microscopy, in the presence or absence of LMB (Fig. 2 C). Mutations of three hydrophobic residues within the NES blocked the exit of the fusion protein from the nucleus, which is in agreement with previous studies (9). Mutations in the NLS, performed as described by Shinkura et al. (13), completely prevented its entry into the nucleus as evidenced upon LMB treatment (Fig. 2 C), thus confirming the requirement for precise residues within the putative NLS to drive AID either directly or indirectly into the nucleus. The fluorescence intensity of the expressed AID-EGFP protein was monitored by analyzing three different clones for each mutant or WT construct, a representative example being shown in Fig. 2 B for each of them. We noticed that expression levels varied considerably according to the subcellular localization of the AID-EGFP protein, while being similar between different clones harboring the same construct. Fig. 2 B shows that, compared with WT-AID (mean fluorescence intensity [MFI] = 350), the cytoplasmic AID (Mut-NLS) exhibits a stronger expression (MFI = 1,340), whereas expression of the nuclear AID (Mut-NES) was by far the lowest (MFI = 60). Because all proteins have been expressed using the same expression vector driven by the same promoter, the level of expression of the fusion protein is likely to reflect its accumulation, and consequently its stability, within the cell. Our data therefore suggest that AID displays a different stability depending on the specific cell compartment in which it is located. The percentage of induced cells varied between the constructs as well, always being lower for mutNES-AID clones, a counterselection probably related to the higher mutagenicity of nuclear AID (see section The NES-AID-GFP protein is mutagenic in the BL2 cell line).
Figure 2.
Expression level of EGFP-tagged AID mutant proteins correlates with the specific cell compartment in which they reside. (A) Schematic representation of mutations affecting either the NLS (mutNLS, V18S, R19V, according to Shinkura et al. [13], with numbers indicating amino acid positions within the AID protein) or the NES (mutNES, L189A, F193A, and L196A), with both domains being represented as black boxes. (B) FACS analysis of the tetracycline-inducible clones, with values of the MFI of the EGFP-positive population. By comparison with endogenous AID expression levels in BL2 (corresponding to a MFI value of 15 in the KI clone [Fig. 1 C]), AID overexpression in EGFP-positive cells can be estimated at ∼4 for mutNES-AID, 24 for WT-AID, and 90 for mutNLS-AID constructs. (C) Confocal images of EGFP signals from inducible clones, with or without 2-h LMB treatment, are shown together with nuclear staining with propidium iodide (PI) of the same field. Clones with a lower percentage of inducible expression are shown, as indicated by the presence of EGFP-negative cells in the field, a shut-off of expression regularly observed in clones kept in culture in induced conditions. Bars, 20 μm.
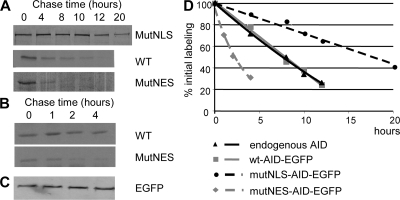
To investigate further the relationship between the cellular localization of the AID protein and its stability, we performed [35S]methionine/cysteine pulse-chase labeling of BL2 cells expressing either WT, nuclear, or cytoplasmic AID-EGFP. Protein levels were quantified, after immunoprecipitation with anti-EGFP antibodies, by autoradiography of PAGE-fractionated immunoprecipitated products (Fig. 3, A–C). The half-life of the nuclear AID-EGFP protein was estimated as ∼2.5 h, compared with ∼8 h for the WT and 18–20 h for the cytoplasmic protein (Fig. 3 D). To confirm this observation, the different BL2 subclones were incubated with cycloheximide to block protein synthesis, and the decay of the protein was followed by the decrease of its fluorescence intensity. Similar to pulse-chase data, AID localized in the nucleus displayed the fastest decay after cycloheximide treatment, with similar values of 2.5 and 8 h estimated for the half-lives of the MutNES and WT constructs, respectively (unpublished data).
Figure 3.
The half-life of AID differs according to its subcellular localization. (A and B) BL2 clones expressing WT-AID-EGFP, MutNES-AID-EGFP, or MutNLS-AID-EGFP were metabolically labeled for 1 h with [35S]-labeled cysteine and methionine and chased for the indicated length of time. AID-EGFP was immunoprecipitated from cell lysates with anti-EGFP antibody (or anti-AID for the endogenous protein) and subjected to SDS-PAGE and autoradiography. (C) Immunoprecipitated MutNES-AID-EGFP was transferred into nitrocellulose and probed with a monoclonal HRP-conjugated anti-EGFP antibody to control for immunoprecipitation efficiency. (D) Densitometry analysis of autoradiographs of pulse-chase immunoprecipitated products, expressed as percentage of initial labeling. The data shown are the mean of the results from two experiments.
AID is polyubiquitinated in the nucleus
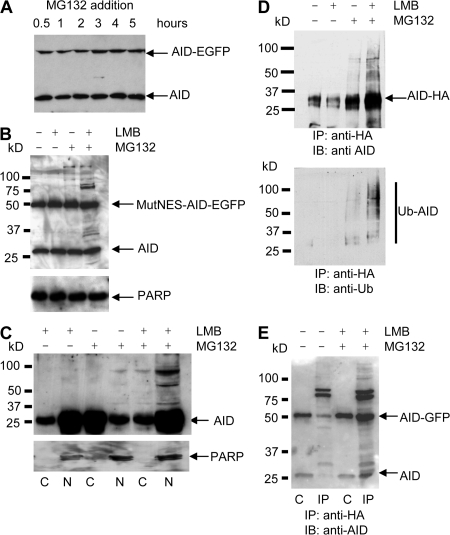
To determine whether proteasomal activity was involved in the nuclear AID degradation pathway in the BL2 cell line, we examined the effect of the 26S proteasome inhibitor MG132 on the turnover of AID. BL2 KI cells were incubated with MG132, total protein extracts were prepared at different time points, and the amount of AID and AID-EGFP proteins was assessed by Western blot using anti-AID antibodies. Fig. 4 A shows that incubation with MG132 has no obvious effect on either form of AID. However, when the cells containing MutNES-AID-EGFP were incubated with MG132, slower migrating protein species were detected (Fig. 4 B, third lane; see also Fig. 5 C), suggesting that the nuclear fraction of AID is sensitive to inhibition of the proteasome. When the modifications of the endogenous protein were studied in the same conditions, i.e., by first incubating BL2 cells with LMB for 1 h to block AID in the nucleus before MG132 addition, modifications were observed for the endogenous AID protein as well (Fig. 4 B, last lane). As incubation with proteasome inhibitors results in a stabilization that is not easily quantifiable by Western blot, the combined effect of MG132 and LMB was monitored for a WT-AID-EGFP–expressing clone through the evolution of its fluorescence intensity over time; the specific fluorescence decay observed upon nuclear sequestering of AID was prevented by the inhibition of proteasomal activity, whereas the fluorescence of a MutNLS-AID-EGFP clone treated in the same conditions was not affected (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070950/DC1). These results thus suggest that polyubiquitination as a prelude to proteasomal degradation may specifically take place in the nucleus.
Figure 4.
Ubiquitination of AID and AID-EGFP requires both nuclear localization and proteasome inhibition. (A) AID-EGFP knocked-in BL2 cells, expressing both AID and AID-EGFP under its endogenous promoter, were incubated with MG132 to inhibit proteasome activity. Cell lysates from the indicated time points were analyzed by immunoblotting using anti-AID monoclonal antibodies. (B) BL2 cells expressing both endogenous AID and MutNES-AID-EGFP were incubated for 5 h with LMB and/or MG132, as indicated. AID status was analyzed as described in A. Poly(ADP-ribose) polymerase (PARP; 116 kD) was used as loading control. (C) BL2 cells were incubated with LMB, MG132, or both, as indicated. For each condition, cytoplasmic (C) and nuclear (N) protein extracts were prepared as described in Materials and methods and analyzed by immunoblotting, with monoclonal anti-AID antibodies. Anti-PARP antibody was used as a nuclear protein control. (D) A BL2 cell line expressing WT-AID-HA was incubated with LMB or MG132, or both, for 5 h. After treatment, cell lysates were denatured and immunoprecipitated with agarose-conjugated anti-HA antibodies before SDS-PAGE separation and probing with anti-AID (top) or antiubiquitin antibodies (bottom). MW markers and migration positions of AID-HA are indicated. (E) AID-EGFP knocked-in BL2 cells were transfected with an HA-tagged ubiquitin-expressing vector and either incubated or not with LMB and MG132 for 5 h. Immunoprecipitation was performed 16 h after transfection, as described in D. Western blot analysis was performed using anti-AID antibodies. Lanes: C, extract before immunoprecipitation (1/10th of total extract); IP, immunoprecipitated proteins.
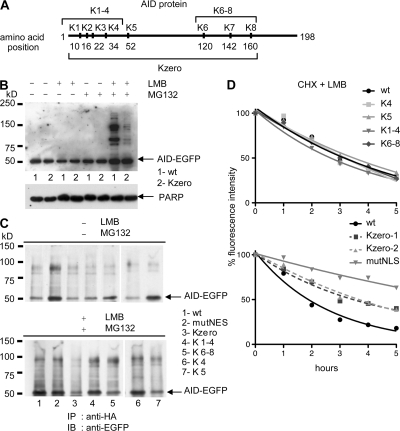
Figure 5.
Lysine mutants of AID are polyubiquitinated in BL2. (A) Schematic presentation of the human AID protein sequence along with the positions of the lysine residues mutated in the various constructs. (B) BL2 clones expressing either WT-AID-EGFP (sample 1, WT), or a lysineless mutant of AID (Kzero-AID-EGFP; sample 2, Kzero) were incubated with the indicated inhibitors for 5 h. Protein extracts were analyzed by immunoblotting with anti-AID. PARP (116 kD) was used as loading control. (C) BL2 cell clones stably expressing either WT-AID-EGFP (WT), mutNES-AID-EGFP (mutNES), a lysineless mutant of AID (Kzero), or the four other AID-EGFP lysine mutants depicted in A were transiently transfected with an HA-tagged ubiquitin-expressing vector and either left untreated (top) or incubated with both LMB and MG132 for 5 h (bottom). Denatured lysates were immunoprecipitated with agarose-conjugated anti-HA antibodies and analyzed by Western blotting using anti-EGFP antibody. Migration position of AID-EGFP is indicated. (D) Stable BL2 clones expressing the indicated AID-EGFP lysine mutants, WT-AID-EGFP, or mutNLS-AID-EGFP were incubated with cycloheximide (CHX, 50 μg/ml) and LMB (10 ng/ml). The decay of the protein was followed by the decrease in fluorescence intensity. The percentage of initial fluorescence is plotted at various time points after addition of the drugs. Kzero1 and 2 represent two independent clones. (top) Clones transfected with AID constructs cloned in the pIRES puro expression vector; (bottom) clones transfected with tetracycline-inducible pBI expression vectors.
To verify this hypothesis, nuclear and cytoplasmic fractions were extracted from normal BL2 cells after incubation with either MG132 or LMB, or both. High molecular weight (MW) species were already detectable in the presence of MG132 in the minor nuclear AID fraction, whereas they were absent in the major cytoplasmic fraction (Fig. 4 C). Addition of LMB alone did not lead to the appearance of such modified forms of AID, but their detection was markedly increased when LMB and MG132 were combined.
To demonstrate that these high MW species correspond to polyubiquitinylated forms of AID, BL2 cells stably transfected with an HA-tagged AID–expressing vector were treated with LMB or MG132, or with both inhibitors, and proteins were extracted after 5 h of incubation and denatured before immunoprecipitation with agarose-conjugated anti-HA antibodies. The immunoprecipitates were analyzed by Western blotting using either anti-AID or -ubiquitin antibodies (Fig. 4 D). The results showed that treatment with both LMB and MG132 led to the accumulation of multiple, higher MW species that are specifically recognized with an antiubiquitin antibody (Fig. 4 D, bottom). As an additional control for ubiquitin modification of AID, transfection of the BL2 KI cell line was performed with Ub-HA–expressing vectors, cells incubated with both MG132 and LMB, and denatured extracts immunoprecipitated with anti-HA antibodies. Western blot analysis using anti-AID antibodies revealed slower migrating bands for both forms of AID expressed from the endogenous loci, whether they were EGFP-tagged or not (Fig. 4 E). Modified forms of AID, although less abundant, were detectable in the absence of both inhibitors, a recurrent finding when ubiquitin-expressing vectors are transfected, suggesting that such massive expression of ubiquitin substrates alone compromises the proteasome machinery (Fig. 5 C and Fig. 6 C).
Figure 6.
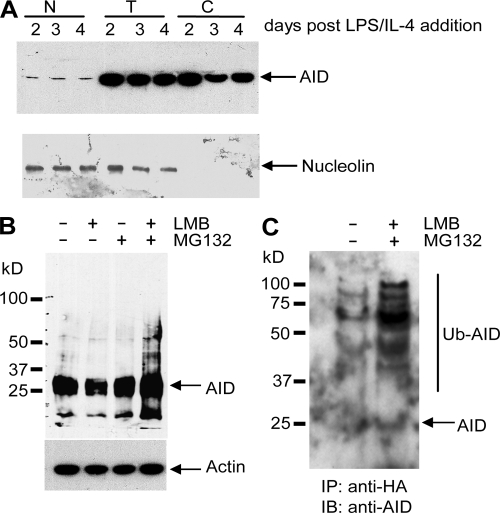
AID is ubiquitinated in the nucleus of activated mouse B splenocytes. (A) Purified B cells from mouse spleen were activated by incubation with LPS and IL-4. At the indicated days after activation, cells were collected and total (T), cytoplasmic (C), or nuclear (N) protein extracts were prepared as described in Materials and methods and analyzed by immunoblotting, with a monoclonal anti–mouse AID antibody. Antinucleolin antibody was used as a nuclear protein control. (B) At day 4, activated B cells were treated with LMB and/or MG132, as indicated. SDS-PAGE–fractionated extracts were probed with anti–mouse AID antibody. Actin was used as loading control. (C) At day 2 of IL-4/LPS activation, 2 × 107 cells were transfected with Ub-HA–expressing vector using Amaxa nucleofection, as indicated in Materials and methods, and either left untreated or incubated for 4 h with both MG132 and LMB, 16 h after transfection. After treatment, cell lysates were denatured and immunoprecipitated with agarose-conjugated anti-HA antibodies before Western blot analysis with anti-AID monoclonal antibodies. MW markers and migration positions of endogenous AID are indicated.
Although the formation of nonspecific aggregates cannot be ruled out even in the denaturing conditions used, the convergent results obtained using either Ub-HA– or AID-HA–tagged proteins and either antiubiquitin or anti-AID antibodies strongly argues against the fact that the detection of high MW species of AID could be an artifact. Collectively, these observations are thus consistent with an essential role of the ubiquitin–proteasomal pathway in the nuclear degradation of AID.
No specific lysine residue is involved in AID ubiquitination
Polyubiquitination proceeds most frequently through the conjugation of the first ubiquitin molecule to the ε-NH2 moiety of a lysine residue within the substrate protein. Five AID mutants, in which one or several lysines were mutated to arginine, including a lysineless mutant of AID (“Kzero-AID-EGFP”; depicted in Fig. 5 A), were transfected in the BL2 cell line, and stable clones were isolated. EGFP fluorescence decay in presence of both LMB and cycloheximide was followed for all these mutants, in parallel with WT-AID and mutNLS-AID–expressing clones. In the four clones with one to four lysines mutated, no alteration in the protein decay was observed (Fig. 5 D, top). The lysineless mutant behaved differently, being less destabilized by this treatment (Fig. 5 D, bottom). Moreover, incubation with either inhibitor indicated that the Kzero-AID-EGFP mutant is not efficiently destabilized in the presence of LMB, in spite of its nuclear localization (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070950/DC1), thus indirectly showing that nuclear destabilization is not an artifactual consequence of LMB addition. Western blot analysis after incubation with LMB, MG132, or both revealed that although higher MW species were observed for the lysineless mutant of AID after inhibition of both nuclear export and proteasome degradation, this occurred at a much lower level than for the WT-AID, thus correlating with the very different impact of LMB addition on the stability of this protein (Fig. 5 B). Accordingly, transfection of Ub-HA, followed by anti-HA immunoprecipitation of the different lysine mutants, confirmed a much lower level of modification of the Kzero-AID-EGFP protein compared with WT-AID, whereas all partial lysine modifications had no quantitative impact (Fig. 5 C). In the control immunoprecipitation performed in the absence of both inhibitors, the much higher level of modifications of the mutNES-AID-EGFP protein should be noted (Fig. 5 C, lane 2), reflecting, as discussed in the previous section, the proteasome saturation effect of massive ubiquitin expression, and which appears specifically pronounced for the sole AID mutant that resides in the nucleus without LMB addition.
No specific lysine was thus shown to be involved in AID destabilization, whereas the total absence of lysines had a stabilization effect that was intermediate between the WT and the cytoplasmic-only AID. The residual ubiquitination observed could explain this partial effect. Two alternative explanations are thus possible: either AID is, indeed, polyubiquitinated at its N terminus and not at internal lysines, with the low efficiency observed resulting from a conformational alteration of the protein induced by the eight mutations introduced. Alternatively, polyubiquitination of AID may proceed through several alternative lysine positions, with artifactual and inefficient conjugation taking place at its N terminus in the absence of internal acceptor sites.
Nuclear degradation of AID occurs in normal mouse B cells and in a human nonlymphoid cell line
We have shown that, in the BL2 Burkitt's lymphoma cell line, AID is polyubiquitinated in the nucleus, which leads to its rapid degradation via the 26S proteasome. To validate this observation in vivo, we studied the turnover and the ubiquitination of AID in mouse splenic B cells stimulated to undergo CSR. The culture of mouse spleen B cells in the presence of LPS and IL-4 drives cell proliferation and triggers the induction of AID gene expression, together with an increased accessibility and targeting to the Sγ1 switch region, allowing the cell to undergo CSR in culture from IgM to IgG1 (18). Cells were collected at different time points after LPS/IL-4 addition, and total or nuclear extracts were prepared as described in Materials and methods. As previously reported, AID expression was induced 48 h after LPS/IL-4 addition, with a small fraction of the protein being detectable in the nucleus (Fig. 6 A)(19, 20). 4 d after stimulation, cells were treated with LMB, MG132, or both, followed by total protein extraction. The direct Western blot analysis using anti–mouse AID antibody showed that addition of LMB, together with MG132, resulted in the accumulation of AID with the appearance of slow migrating forms of the protein (Fig. 6 B). Transient transfection of a Ub-HA–expressing vector was performed in LPS + IL-4–activated spleen cells 2 d after stimulation, and MG132 and LMB were added for 5 h, 16 h after transfection. Western blot analysis with an anti-AID antibody, after anti-HA immunoprecipitation performed as described in the previous section, revealed similarly high MW species, thus confirming that polyubiquitination of AID takes place in the nuclei of activated B cells (Fig. 6 C). As observed for the BL2 cell line, modifications were already observed in the control sample, but were quantitatively less important.
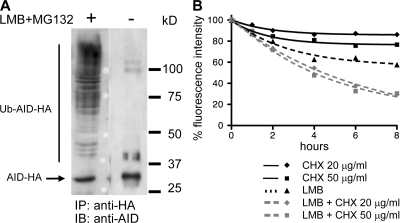
We next studied whether nuclear destabilization was restricted to the cells naturally expressing AID, i.e., to activated B lymphocytes, or whether it constitutes a general regulation of AID turnover. To this end, we transiently transfected the human embryonic kidney 293T cell line with the WT-AID-HA construct, and studied its ubiquitination in presence of MG132 and LMB, as described for the BL2 cell line. After anti-HA immunoprecipitation and analysis by Western blot, accumulation of high MW species of AID was clearly detectable in 293T cells (Fig. 7 A). 293T cells stably transfected with WT-AID-EGFP were treated with cycloheximide, LMB, or both, and the decay in fluorescence intensity was followed by FACS analysis as described for AID lysine mutants (Fig. 7 B). Addition of LMB alone was sufficient to induce a marked reduction in AID protein content, a degradation that was further increased by addition of cycloheximide. Thus, nuclear destabilization of the AID protein does not appear to be restricted to lymphoid B cells. However, it should be noted that although the relative difference in stability between the shuttling and the nuclear form of the protein appears similar in the 293T and the BL2 cell lines, the overall degradation of AID seems to occur faster in BL2 (Fig. S4).
Figure 7.
Nuclear destabilization of AID is not restricted to B cells. (A) Human 293T cells transfected with WT-AID-HA–expressing vector were incubated for 6 h with both LMB and MG132, or left untreated, as indicated. Immunoprecipitation was performed using agarose-conjugated anti-HA antibodies, and SDS-PAGE–fractionated protein extracts were analyzed by Western blot with anti-AID antibodies. Migration position of AID-HA is indicated. (B) 293T cells stably transfected with a WT-AID-EGFP–expressing vector were incubated with cycloheximide (CHX; 20 or 50 μg/ml) and LMB, either separately or in combination, and the decay of the protein was followed by the decrease in fluorescence intensity. The percentage of initial fluorescence is plotted at various time points after addition of the inhibitors of protein synthesis and/or protein nuclear export.
The NES-AID-EGFP protein is mutagenic in the BL2 cell line
Inducible MutNES-AID-EGFP and WT-AID-EGFP constructs were transfected in the previously described AID-deficient BL2 cell line (21). Individual clones were isolated, expression of AID was induced by tetracycline removal, and EGFP-positive cells were isolated by cell sorting 14 d after induction. Mutations in the functional V4-39 gene were analyzed for three mutNES-AID-EGFP and three WT-AID-EGFP clones. The average mutation frequency for the three clones analyzed was 2.3 times higher for the mutant compared with WT ones, with larger individual variations being observed between the mutant clones (Table S1, available at http://www.jem.org/cgi/content/full/jem.20070950/DC1). The mutNES-16 clone displayed the highest mutation frequency, 3.2–4.5 times higher than any of the WT-AID–expressing clones analyzed. Mutations were almost exclusively targeted at G/C positions. Moreover, G-to-A and C-to-T transitions predominate in the mutant clones (81%), consistent with a high number of C to U deaminations being carried over to replication. The proportion of transversions, essentially G-to-C or C-to-G, was higher in the WT-AID–expressing cell lines (45%; unpublished data), suggesting that saturation (or inhibition) of repair by uracil glycosylase occurs when AID is retained in the nucleus.
Mutations induced by AID overexpression were also analyzed at the Cμ locus, as well as in several non-Ig genes, including the AID cDNA sequence of the expression construct, in one WT (WT-24) and one mutant (mutNES-16) clone. Among non-Ig genes, we selected BCL6 which is targeted by AID and mutated in normal centroblasts (22, 23), PAX5, which is mutated in some B cell lymphomas (particularly diffuse large B cell lymphomas, DLBCL) and has been recently shown to be targeted by AID, but subjected to error-free repair in normal B cells (24), and TP53, which has not been described as mutable in either DLBCL lymphomas or germinal center B cells (24, 25). A low mutation frequency was observed for Cμ, BCL6, and PAX5 genes in the WT clone, while being at background level for TP53 (Table I). Mutagenesis was increased for the BCL6 gene in the mutNES-16 clone, but, more surprisingly, also at the TP53 locus. The highest mutation frequency is observed for the AID transgene, in both WT and mutant clones, indicating that the strength of the transcriptional activation mediated by the tetracycline-regulated CMV promoter can possibly allow the targeting of AID at a level comparable to, if not higher than, the one occurring at the endogenous VH sequence. However, considerable variations in mutagenesis of the AID transgene were observed when all six clones were analyzed (unpublished data), confirming, as previously reported, that the insertion site can influence the accessibility to AID-induced mutagenesis of transgenes displaying equivalent transcriptional activity (26, 27).
Table I.
Mutation frequency at various Ig and non-Ig loci in BL2 clones transfected with WT-AID-EGFP or mutNES-AID-EGFP inducible expression vectors
|
|
WT-24 clone
|
mutNES-16 clone
|
|---|---|---|
| number of mutations/total number of sequences (mutation frequency) |
||
| Gene locus (domain sequenced) |
||
| V4-39 (390 bp) | 13/65 (0.51 × 10−3) |
69/74 (2.39 × 10−3) |
| Cmu (403 bp) | 5/74 (0.17 × 10−3) |
6/73 (0.20 × 10−3) |
| BCL6 (640 bp) | 3/47 (0.10 × 10−3) |
12/51 (0.37 × 10−3) |
| TP53 (1,064 bp) | 1/39 (0.024 × 10−3) |
14/43 (0.31 × 10−3) |
| PAX5 (885 bp) | 6/47 (0.14 × 10−3) |
8/46 (0.20 × 10−3) |
| AID cDNA (594 bp) | 14/22 (1.07 × 10−3) |
39/22 (2.98 × 10−3) |
DISCUSSION
In this work, we studied additional modes of regulation of AID that can impact on the availability of this protein in lymphoid cells, and may thus contribute to restrict access to its DNA target. The first question asked was whether expression and/or accumulation of AID were regulated according to the cell cycle. We chose the BL2 Burkitt's lymphoma cell line for this study. BL2 expresses AID, but it only undergoes minimal hypermutation of its immunoglobulin genes constitutively (21). It is nevertheless likely that this cell line constitutes a valid model for studying the basal regulation of AID because it represents the transformed counterpart of a germinal center B cell.
KI of EGFP at the AICDA locus was therefore performed in BL2, by fusing EGFP at the end of the last coding exon, and various measures showed that AID expression level and protein accumulation from both endogenous and KI alleles were similar. Fractionation of BL2 by counterflow elutriation and monitoring of AID-EGFP fluorescence in elutriated cells showed that AID expression varied approximately within a factor of 2 between the G1 and the G2/M phases, i.e., in proportion to its gene copy number. AID expression is thus not restricted to a specific phase of the cell cycle. Expression of AID and cytidine deamination throughout the cell cycle fit with current models of hypermutation, and we have proposed that mutations at A/T versus G/C bases might result from a differential handling of the deaminated base, according to its occurrence before or during DNA replication (28, 29).
AID has been shown to undergo constant nucleocytoplasmic shuttling, with a net balance toward export, thus resulting in a main cytoplasmic localization (8–12). In effect, incubation of AID-expressing cells with LMB, a specific inhibitor of CRM1, which is the protein responsible for this nuclear export, results in a rapid sequestering of AID in the cell nucleus (9). Alteration of a nuclear export sequence located at the 3′ end of the AID molecule, as well as of a putative NLS, has been shown to result in a protein with exclusive nuclear or cytoplasmic localization, respectively, a localization that is not further altered by addition of LMB (9, 10, 13). We generated such mutants, fused to the EGFP reporter gene, and obtained stable tetracycline inducible clones in BL2. We noticed that, although these mutants were all expressed from the same CMV promoter-driven inducible vectors, they displayed markedly different protein expression levels that consistently correlated with their specific subcellular localization, suggesting that such a differential accumulation might mirror their stability within the cell. The protein half-life was determined by either [35S] pulse-chase protein labeling or by incubation with cycloheximide and follow up of the radioactivity or fluorescence decay of the AID protein. When compared with the normal shuttling protein (WT-AID-EGFP), the protein sequestered in the nucleus (mutNES-AID-EGFP) showed a 3 time shorter half-life (t1/2 = 2.5 vs. 8 h), whereas the protein retained in the cytoplasm (mutNLS-AID-EGFP) was 3 times more stable (t1/2 = ∼20 h), resulting in a 10-fold difference in stability between the 2 cell compartment–restricted mutants of AID. AID nuclear localization thus appears to be linked with increased degradation.
Degradation of most cellular proteins requires tagging of protein substrates by polyubiquitin chains and proceeds via an energy-dependent process involving the 26S proteasome that constitutes the major neutral proteolytic activity in both the cytosol and the nucleus (for review see [30]). Incubation with proteasome inhibitors was shown to result in the appearance of high MW species of AID that were shown to be polyubiquitinylated. However, such modifications were essentially observed when AID was retained in the nucleus, either because of the addition of LMB or the mutation in the nuclear export sequence. We did not observe any ubiquitination of the cytoplasmic mutant, which displays the longest half-life (unpublished data). This could be caused by a modification rate that would be below the detection level of our assay, or possibly to a degradation that could be proteasome independent. Alternatively, AID could be complexed in the cytoplasm with proteins that would protect it from ubiquitin cross-linking and degradation. Specific interaction between cytoplasmic AID and protein factors has indeed been described in the Ramos cell line (31).
Nuclear-specific degradation of endogenous mouse AID was similarly observed in mouse splenic B cells, after its induction by LPS and IL-4 stimulation, indicating that this regulation is an intrinsic part of its physiological turnover. This process is nevertheless not restricted to B cells, as it is similarly observed, albeit with some quantitative differences, upon transfection of the human embryonic kidney 293T cell line.
In proteins destined for degradation by the proteasome, polyubiquitin chains are commonly anchored to the ε-amine group of one or several internal lysine residues. To identify the putative lysine target within the eight candidate amino acids present in the human AID protein, various mutants were generated by replacement of lysines by arginines, including a lysineless form of AID. No single lysine residue could be identified as the internal site of ubiquitin conjugation, as no single or multiple (up to four) mutations were able to stabilize the nuclear form of the protein. In contrast, the lysineless AID was still polyubiquitinated upon addition of proteasome inhibitors and sequestration in the nucleus, but at a much lower level, and displayed accordingly a nuclear stability intermediate between the normal and cytoplasmic-only protein. Thus, polyubiquitination of AID may proceed through several alternative internal sites, as has been shown for the IκBα protein (32), the low level of modification still observed in the lysineless mutant representing an N-terminal modification that would be artificially forced. Or, alternatively, N-terminal ubiquitination, observed for a few proteins such as the ARF/INK4a tumor suppressor protein or the MyoD and Id2 transcriptional regulators (33–36), could be the physiological process, its relative inefficiency being indirectly generated by the eight mutations that would alter the targeting of the ubiquitin ligase involved. Stepwise reversion of mutant lysines may allow us to settle this issue.
The fraction of ubiquitinated AID observed appears to always be low in BL2, which may be linked to the intrinsic difficulty of observing such modifications outside the classical readouts that 293T cells or fibroblasts constitute. We cannot exclude on the other hand that an ubiquitin-independent pathway may contribute to AID turnover, as has been recently described for the p21 cell cycle regulator, which is channeled for proteasomal degradation by the REGγ complex in the absence of ubiquitin cross-linking (37, 38). In addition to the proteasome-mediated nuclear degradation that we have described, we do not exclude either that there may be other steps at which AID turnover might be regulated, which would impact its interaction with other protein partners or its assembly as a dimeric or multimeric complex, for example.
The natural target of AID-induced mutagenesis in normal germinal center B cells is the Ig locus, as well as several genes like BCL6, which nevertheless harbor a much lower mutation frequency (22, 23, 39). Moreover, numerous genes, including oncogenes like PAX5, PIM1, MYC, and RhoH/TTF, which undergo a low level of mutagenesis in some B cell lymphomas constitutively expressing AID (25, 40), appear to be targeted by AID in normal B cells, but are spared from mutations caused by error-free repair (24). Deliberate overexpression of AID has been shown to increase mutation levels of several transcribed genes in both prokaryotic and mammalian cells (41–45). However, although the nuclear-restricted AID protein induces enhanced mutagenesis both in Escherichia coli and on a transfected substrate in fibroblasts, its mutagenicity in B lymphocytes is not clearly established (9, 10). We obtained inducible clones overexpressing the WT or mutant form of AID in the AICDA−/− BL2 cell line, and confirmed that, although different clones show variable mutation frequencies of their endogenous rearranged VH gene, higher mutagenicity can be achieved by the nuclear restricted AID compared with the WT one. As nuclear AID is expressed at a lower level than the normal, shuttling protein, the mutagenicity related to the amount of AID protein present in a cell is thus 6–10-fold higher for a protein with an exclusive nuclear residence. When several non-Ig loci were studied in the mutNES-AID-EGFP clone showing the highest mutation frequency, mutagenesis at the BCL6 locus was also found to be increased, as well as, and more surprisingly, at TP53. However, it should be mentioned that, whereas p53 expression is repressed by Bcl-6 in Burkitt's lymphoma, as well as in germinal center B cells, and is thus logically spared from hypermutation (24, 46), its expression has been shown to be inducible in specific stress conditions in BL2 (47). The TP53 gene could thus become an AID target in cells that failed to repress its expression. Such an AID-induced mutagenesis of the p53 coding sequence has been recently described in human gastric epithelial cells (48), even though in this case mutations were focused in the core p53 coding region and not in the region studied here, which is located downstream from the promoter. Abnormal AID expression in nonlymphoid cells could thus unmask additional AID targets that would become mutated depending on the specific gene expression profile of a given cell type, and would thus increase the number of genes recently described as susceptible to AID deamination (24).
HIGM2 patients with heterozygous or homozygous mutations in the nuclear export sequence of AID do not show markedly different mutation frequencies of their endogenous Ig genes, suggesting that transcriptional regulation and nuclear destabilization during germinal center B cell differentiation are sufficient to keep AID activity under control (49, 50). However, in transformed B cells displaying ongoing AID expression (25, 40, 51), or in conditions of chronic inflammation leading to AID activation (48, 52), factors that would modify the nuclear turnover of AID could aggravate its oncogenic potential and thus contribute actively to aberrant mutagenesis.
In conclusion, our study shows that ubiquitin-mediated nuclear degradation of AID is part of a multilayer control that contributes to the regulation of AID function during hypermutation and CSR. A fraction of AID has been detected in activated B cells as being linked to chromatin, and phosphorylated AID is found enriched in such a chromatin fraction (18, 53). Phosphorylation at serine 38 has also been shown to impact AID activity in vivo (31, 53, 54). It is therefore likely that specific posttranslational modifications, among them phosphorylation, may contribute to sequester AID from the nucleus. Such stabilization in chromatin domains would at the same time prevent AID from being constantly exported from the nucleus, while possibly allowing it to escape the nuclear-specific degradation described in this study. This competition between attachment to DNA, degradation and export could also be central in limiting the magnitude of AID activity, which has been proposed to be mainly nonprocessive (55, 56), a mode of action that would, after each enzymatic round, render this enzyme accessible to destabilization and nuclear exclusion. Whether such a competition between phosphorylation and polyubiquitination, be it direct or indirect, occurs, and whether additional posttranslational modifications, like for example sumoylation, orchestrate the assembly of AID into an active mutasome remain to be addressed.
MATERIALS AND METHODS
Construction and transfection of expression vectors for AID mutants.
Full-length cDNA encoding human AID without stop codon was fused in frame with the sequence coding for 3 repeats of YPYDVPDYA, corresponding to residues 99–107 of the influenza virus hemagglutinin (HA), cloned into pIRESpuro vector. Stable clones were obtained by electroporation in the Burkitt's lymphoma BL2 cell line. HA-tagged ubiquitin-expressing vector was a gift from M. Treier (European Molecular Biology Laboratory, Heidelberg, Germany). Transient transfection was performed using the Amaxa device (program C-009). AID-EGFP fusion sequences were cloned in the pIRESpuro vector or in the tetracycline-inducible pBI expression vector (Clontech Laboratories), in which a blasticidin resistance gene amplified from the pcDNA6/TR plasmid (Invitrogen) was inserted in its AatII site to allow for selection of transfected clones. The EGFP coding sequence was fused at the 3′ end of AID. Mutants were constructed by stepwise PCR mutagenesis, introducing the following mutations. MutNES: L189A, F193A, L196A; MutNLS: V18S, R19V (13); Kzero: all lysines (K10, 16, 22, 34, 52, 120, 142, 160) mutated to arginines (AAA/G to AGA/G); K1-4: first four lysines to arginines; K6-8: last three lysines to arginines; K4, K5: fourth and fifth lysines (positions 34 and 52), respectively, to arginines. Inducible vectors were electroporated in the BTZ cell line (a tetracycline-inducible subclone of BL2, obtained by transfection of the pTet-tTAK vector [Invitrogen] in which a zeocin resistance gene was inserted [vector and cell line provided by M. Muramatsu and T. Honjo, Kyoto University, Kyoto, Japan]). Tetracycline-inducible AID-deficient BL2 clones were obtained by electroporation of the same pTet-tTAK-zeo vector in the 60–129 AID-deficient cell line (21). Selection of transfected clones was performed in the following conditions: 350 ng/ml puromycin, 5 μg/ml blasticidin, 500 μg/ml zeocin, and 1 μg/ml tetracycline. Tetracycline-free serum (Clontech Laboratories) was used for the culture of inducible clones.
Inducible 293T cells were similarly obtained after transfection of the pTet-tTAK vector, and inducible WT-AID-EGFP–expressing cells were selected as a pool. Transient transfection in 293T was performed using Amaxa conditions (program Q-001).
Mouse B cell culture, stimulation, and transfection.
B cells were purified from mouse spleen using the mouse B cell isolation kit (Miltenyi Biotec) and cultured in complete RPMI 1640 medium with 20 ng/ml murine IL-4 (PeproTech) and 25 μg/ml LPS (Sigma-Aldrich). Cells were collected at different days after stimulation. 2 d after stimulation, 4 × 107 cells were collected and transfected with Ub-HA–expressing vector using the Test Nucleofector kit 104A1 (Amaxa), by sets of 5 × 106 cells and 5 μg of Ub-HA DNA per transfection. Increasing the transfection volume to 200 μl had a major incidence on cell viability. Transfection of 2.5 μg EGFP-expressing vector (pMaxGFP, Amaxa) in parallel resulted in 60–70% transfection efficiency and 50% cell viability estimated by 7-amino-actinomycin-D exclusion. Mouse experiments were performed according to the Institut National de la Santé et de la Recherche Médicale guidelines for laboratory animals and approved by the Scientific Committee of the Necker Animal Facility.
Immunofluorescence microscopy.
BL2 cells transfected with AID-EGFP fusion constructs were spun down on poly-lysine–coated glass coverslips for 5 min at 100 g. Cells were fixed in cold methanol for 20 min at −20°C. Coverslips were mounted on slide using Vectashield mounting medium containing propidium iodide (Vector Laboratories). EGFP fluorescence was visualized using an LSM 510 confocal microscope (Carl Zeiss, Inc.).
Flow cytometry, cell sorting, and elutriation.
Transfected BL2 clones were analyzed with Cell Quest software on a FACScan apparatus (BD Biosciences). For AID stability estimates, EGFP fluorescence was measured at various time points after cycloheximide, LMB, or MG132 addition. For the determination of mutation frequencies, EGFP-positive cells were sorted on a FACS Aria cell sorter (Becton Dickinson). Fractionation of BL2 cells according to their cell cycle status was performed by counterflow elutriation, and collected fractions were stained and analyzed for DNA content, as previously described (21), after determination of their EGFP fluorescence intensity.
Pulse-chase assay.
Exponentially growing BL2 cells were washed twice with methionine and cysteine-free DME medium, and then incubated at 106 cell/ml in the same medium containing 5% of dialyzed FCS. After 1 h at 37°C, 100 μCi/ml of [35S]-L-methionine/cysteine promix (GE Healthcare) was added and incubated for an additional 2 h. The medium was then replaced with chase medium (RPMI containing 10% FCS and 10-fold excess of cold methionine and cysteine). Protein extracts were prepared at various times after the chase in radioimmune precipitation assay (RIPA) buffer and the protein concentration of each extract was normalized and precleared with protein G–agarose before immunoprecipitation. Immunoprecipitation was performed using human monoclonal anti-AID or -GFP antibodies (Invitrogen), and protein G–agarose beads (Santa Cruz Biotechnology). Immunoprecipitated proteins were resolved by SDS-PAGE. Gels were fixed in 10% acetic acid/40% ethanol before being dried and autoradiographed. The half-life curves were calculated from densitometry readings of the autoradiograph.
Preparation of cytoplasmic and nuclear extracts.
Nuclear and cytoplasmic extracts were prepared either from BL2 or from activated mouse B cells, as follows: cells were washed twice with cold PBS and suspended in cold hypotonic buffer (HB; 10 mM Hepes, pH 7.9, 10 mM KCl, 0.2 mM EDTA, 1 mM DTT, 1x protease inhibitor mixture [Roche], and 0.25 mM PMSF). After incubation on ice for 15 min, NP-40 was added to a final concentration of 0.5%. The mixtures were vortexed at high speed for 10 s. The nuclei were recovered by centrifugation at 13,000 rpm for 30 s. The supernatants were collected as cytoplasmic extracts. The pellet was washed once with buffer HB, and proteins were extracted by adding high-salt (HS) buffer (20 mM Hepes, pH 7.9, 420 mM NaCl, 2 mM EDTA, 1 mM DTT, 1x protease inhibitor mixture, and 0.25 mM PMSF), followed by incubation at 4°C for 30 min under gentle agitation. The mixtures were spun at 13,000 rpm for 15 min, and the supernatants were collected as nuclear extracts. In ubiquitination experiments, 10 μg/ml of N-ethylmaleimide (Sigma-Aldrich) was added to PBS during the cell washes and to both the HB and HS buffers.
Total extract preparation, immunoprecipitation, and immunoblotting.
The whole-cell protein lysates were washed twice with cold PBS, and incubated under agitation in the cold room for 20 min in RIPA (50 mM Hepes, pH 7.9, 150 mM NaCl, 1.25 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, 1 mM dithiothreitol, 1x protease inhibitor mixture, and 0.25 mM PMSF). Insoluble material was removed with 30-min centrifugation at 15,000 rpm at 4°C. The protein concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific). Protein samples were mixed with an equal volume of electrophoresis loading buffer containing 2% SDS and heated at 95°C for 5 min before SDS-PAGE separation and transfer to nitrocellulose membranes. The membrane was then incubated with antibodies, followed by chemiluminescence detection, as previously described (21).
Immunoprecipitation was performed after denaturation in immunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail) plus 1% SDS, and then heated twice for 5 min at 95°C. Supernatants were diluted 10-fold with immunoprecipitation buffer containing 5 mM N-ethylmaleimide and 0.5% NP-40 before antibody addition and immunopurification.
In addition to our monoclonal anti–human AID antibodies, the following antibodies were used: monoclonal anti–mouse AID (Cell Signaling Technology), monoclonal anti-nucleolin, agarose-conjugated anti-HA (Santa Cruz Biotechnology), rabbit polyclonal anti-actin (Abcam), monoclonal antiubiquitin (clone FK2; BioMol), HRP-conjugated monoclonal anti-EGFP (Miltenyi Biotec), HRP-coupled anti–mouse and –rabbit IgG (Bio-Rad Laboratories), monoclonal anti-PARP (Serotec), and 7-amino-actinomycin-D (BD Biosciences). MG132 and LMB (both from Calbiochem) were used at concentrations of 10 μM and 10 ng/ml, respectively, and cycloheximide (Sigma-Aldrich) was used at either 20 or 50 μg/ml.
Online supplemental material.
Supplemental materials and methods describe the production of the anti–human AID monoclonal antibodies used in this study, the generation of the BL2 cell line with EGFP knocked-in at the AICDA locus, and the conditions for the analysis of AID-induced mutations at various loci. Fig. S1 compares the protein turnover of AID and AID-EGFP expressed from the endogenous loci. Fig. S2 shows the specificity of the anti–human AID monoclonal antibodies used. Fig. S3 shows the effect of the MG132 proteasome inhibitor on AID stabilization. Fig. S4 shows the lack of nuclear destabilization of the lysineless AID mutant. Table S1 describes the mutation frequency at the VH locus of various AID-expressing BL2 clones. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20070950/DC1.
Supplementary Material
Acknowledgments
We thank Damiana Lecoeuche for excellent technical assistance, Meriem Garfa for assistance with confocal microscopy, and Jérôme Mégret for performing cell sorting. We thank T. Honjo and M. Muramatsu for the gift of the pTet-tTAK-zeo plasmid and the BTZ cell line, and Novartis Pharma (Basel, Switzerland) for graciously providing recombinant human IL-4.
This work was supported by the Ligue Nationale contre le Cancer (Equipe labellisée) and the European Commission Sixth Framework Program (HYBLIB project, NEST pathfinder program in Synthetic Biology).
The authors declare no competing financial interest.
Abbreviations used: AID, activation-induced cytidine deaminase; CSR, class switch recombination; HA, hemagglutinin; KI, knock-in; LMB, leptomycin B; MFI, mean fluorescence intensity; MW, molecular weight; NES, nuclear export signal; NLS, nuclear localization signal; SHM, somatic hypermutation.
References
- 1.Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. [DOI] [PubMed] [Google Scholar]
- 2.Li, Z., C.J. Woo, M.D. Iglesias-Ussel, D. Ronai, and M.D. Scharff. 2004. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 18:1–11. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 5.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S.H., G. Habib, C.Y. Yang, Z.W. Gu, B.R. Lee, S.A. Weng, S.R. Silberman, S.J. Cai, J.P. Deslypere, M. Rosseneu, et al. 1987. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 238:363–366. [DOI] [PubMed] [Google Scholar]
- 7.Neuberger, M.S., R.S. Harris, J. Di Noia, and S.K. Petersen-Mahrt. 2003. Immunity through DNA deamination. Trends Biochem. Sci. 28:305–312. [DOI] [PubMed] [Google Scholar]
- 8.Rada, C., J.M. Jarvis, and C. Milstein. 2002. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc. Natl. Acad. Sci. USA. 99:7003–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito, S., H. Nagaoka, R. Shinkura, N. Begum, M. Muramatsu, M. Nakata, and T. Honjo. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 101:1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride, K.M., V. Barreto, A.R. Ramiro, P. Stavropoulos, and M.C. Nussenzweig. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brar, S.S., M. Watson, and M. Diaz. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279:26395–26401. [DOI] [PubMed] [Google Scholar]
- 12.Cattoretti, G., M. Buttner, R. Shaknovich, E. Kremmer, B. Alobeid, and G. Niedobitek. 2006. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 107:3967–3975. [DOI] [PubMed] [Google Scholar]
- 13.Shinkura, R., S. Ito, N.A. Begum, H. Nagaoka, M. Muramatsu, K. Kinoshita, Y. Sakakibara, H. Hijikata, and T. Honjo. 2004. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5:707–712. [DOI] [PubMed] [Google Scholar]
- 14.Okazaki, I.M., H. Hiai, N. Kakazu, S. Yamada, M. Muramatsu, K. Kinoshita, and T. Honjo. 2003. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 16.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotani, A., N. Kakazu, T. Tsuruyama, I.M. Okazaki, M. Muramatsu, K. Kinoshita, H. Nagaoka, D. Yabe, and T. Honjo. 2007. Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice. Proc. Natl. Acad. Sci. USA. 104:1616–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nambu, Y., M. Sugai, H. Gonda, C.G. Lee, T. Katakai, Y. Agata, Y. Yokota, and A. Shimizu. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 302:2137–2140. [DOI] [PubMed] [Google Scholar]
- 19.Dedeoglu, F., B. Horwitz, J. Chaudhuri, F.W. Alt, and R.S. Geha. 2004. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFkappaB. Int. Immunol. 16:395–404. [DOI] [PubMed] [Google Scholar]
- 20.Schrader, C.E., E.K. Linehan, S.N. Mochegova, R.T. Woodland, and J. Stavnezer. 2005. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J. Exp. Med. 202:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faili, A., S. Aoufouchi, Q. Gueranger, C. Zober, A. Leon, B. Bertocci, J.C. Weill, and C.A. Reynaud. 2002. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 3:815–821. [DOI] [PubMed] [Google Scholar]
- 22.Shen, H.M., A. Peters, B. Baron, X. Zhu, and U. Storb. 1998. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 280:1750–1752. [DOI] [PubMed] [Google Scholar]
- 23.Pasqualucci, L., A. Migliazza, N. Fracchiolla, C. William, A. Neri, L. Baldini, R.S. Chaganti, U. Klein, R. Kuppers, K. Rajewsky, and R. Dalla-Favera. 1998. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA. 95:11816–11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, M., J.L. Duke, D.J. Richter, C.G. Vinuesa, C.C. Goodnow, S.H. Kleinstein, and D.G. Schatz. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. [DOI] [PubMed] [Google Scholar]
- 25.Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R.S. Chaganti, R. Kuppers, and R. Dalla-Favera. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346. [DOI] [PubMed] [Google Scholar]
- 26.Wang, C.L., R.A. Harper, and M. Wabl. 2004. Genome-wide somatic hypermutation. Proc. Natl. Acad. Sci. USA. 101:7352–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, A., and M.D. Scharff. 2002. Somatic hypermutation of the AID transgene in B and non-B cells. Proc. Natl. Acad. Sci. USA. 99:12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delbos, F., S. Aoufouchi, A. Faili, J.C. Weill, and C.A. Reynaud. 2007. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 204:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weill, J.C., and C.A. Reynaud 2008. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. In press. [DOI] [PubMed]
- 30.Glickman, M.H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373–428. [DOI] [PubMed] [Google Scholar]
- 31.Pasqualucci, L., Y. Kitaura, H. Gu, and R. Dalla-Favera. 2006. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA. 103:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldi, L., K. Brown, G. Franzoso, and U. Siebenlist. 1996. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of I kappa B-alpha. J. Biol. Chem. 271:376–379. [DOI] [PubMed] [Google Scholar]
- 33.Fajerman, I., A.L. Schwartz, and A. Ciechanover. 2004. Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem. Biophys. Res. Commun. 314:505–512. [DOI] [PubMed] [Google Scholar]
- 34.Ben-Saadon, R., I. Fajerman, T. Ziv, U. Hellman, A.L. Schwartz, and A. Ciechanover. 2004. The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J. Biol. Chem. 279:41414–41421. [DOI] [PubMed] [Google Scholar]
- 35.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17:5964–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo, M.L., W. den Besten, and C.J. Sherr. 2004. N-terminal polyubiquitination of the ARF tumor suppressor, a natural lysine-less protein. Cell Cycle. 3:1367–1369. [DOI] [PubMed] [Google Scholar]
- 37.Chen, X., L.F. Barton, Y. Chi, B.E. Clurman, and J.M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell. 26:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, X., L. Amazit, W. Long, D.M. Lonard, J.J. Monaco, and B.W. O'Malley. 2007. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell. 26:831–842. [DOI] [PubMed] [Google Scholar]
- 39.Muschen, M., D. Re, B. Jungnickel, V. Diehl, K. Rajewsky, and R. Kuppers. 2000. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J. Exp. Med. 192:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaidano, G., L. Pasqualucci, D. Capello, E. Berra, C. Deambrogi, D. Rossi, L. Maria Larocca, A. Gloghini, A. Carbone, and R. Dalla-Favera. 2003. Aberrant somatic hypermutation in multiple subtypes of AIDS-associated non-Hodgkin lymphoma. Blood. 102:1833–1841. [DOI] [PubMed] [Google Scholar]
- 41.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 42.Martin, A., P.D. Bardwell, C.J. Woo, M. Fan, M.J. Shulman, and M.D. Scharff. 2002. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 415:802–806. [DOI] [PubMed] [Google Scholar]
- 43.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 44.Yoshikawa, K., I.M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 296:2033–2036. [DOI] [PubMed] [Google Scholar]
- 45.Bachl, J., C. Carlson, V. Gray-Schopfer, M. Dessing, and C. Olsson. 2001. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166:5051–5057. [DOI] [PubMed] [Google Scholar]
- 46.Phan, R.T., and R. Dalla-Favera. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 432:635–639. [DOI] [PubMed] [Google Scholar]
- 47.Gao, C., T. Nakajima, Y. Taya, and N. Tsuchida. 1999. Activation of p53 in MDM2-overexpressing cells through phosphorylation. Biochem. Biophys. Res. Commun. 264:860–864. [DOI] [PubMed] [Google Scholar]
- 48.Matsumoto, Y., H. Marusawa, K. Kinoshita, Y. Endo, T. Kou, T. Morisawa, T. Azuma, I.M. Okazaki, T. Honjo, and T. Chiba. 2007. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 13:470–476. [DOI] [PubMed] [Google Scholar]
- 49.Ta, V.T., H. Nagaoka, N. Catalan, A. Durandy, A. Fischer, K. Imai, S. Nonoyama, J. Tashiro, M. Ikegawa, S. Ito, et al. 2003. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 4:843–848. [DOI] [PubMed] [Google Scholar]
- 50.Imai, K., Y. Zhu, P. Revy, T. Morio, S. Mizutani, A. Fischer, S. Nonoyama, and A. Durandy. 2005. Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin. Immunol. 115:277–285. [DOI] [PubMed] [Google Scholar]
- 51.Feldhahn, N., N. Henke, K. Melchior, C. Duy, B.N. Soh, F. Klein, G. von Levetzow, B. Giebel, A. Li, W.K. Hofmann, H. Jumaa, and M. Muschen. 2007. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1–transformed acute lymphoblastic leukemia cells. J. Exp. Med. 204:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Endo, Y., H. Marusawa, K. Kinoshita, T. Morisawa, T. Sakurai, I.M. Okazaki, K. Watashi, K. Shimotohno, T. Honjo, and T. Chiba. 2007. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 26:5587–5595. [DOI] [PubMed] [Google Scholar]
- 53.McBride, K.M., A. Gazumyan, E.M. Woo, V.M. Barreto, D.F. Robbiani, B.T. Chait, and M.C. Nussenzweig. 2006. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA. 103:8798–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basu, U., J. Chaudhuri, C. Alpert, S. Dutt, S. Ranganath, G. Li, J.P. Schrum, J.P. Manis, and F.W. Alt. 2005. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 438:508–511. [DOI] [PubMed] [Google Scholar]
- 55.Xue, K., C. Rada, and M.S. Neuberger. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 203:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coker, H.A., and S.K. Petersen-Mahrt. 2007. The nuclear DNA deaminase AID functions distributively whereas cytoplasmic APOBEC3G has a processive mode of action. DNA Repair (Amst.). 6:235–243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.