Abstract
The lysosomal storage disorder mucolipidosis type IV (MLIV) is caused by mutations in the transient receptor potential–mucolipin-1 (TRP-ML1) ion channel. The “biogenesis” model for MLIV pathogenesis suggests that TRP-ML1 modulates postendocytic delivery to lysosomes by regulating interactions between late endosomes and lysosomes. This model is based on observed lipid trafficking delays in MLIV patient fibroblasts. Because membrane traffic aberrations may be secondary to lipid buildup in chronically TRP-ML1–deficient cells, we depleted TRP-ML1 in HeLa cells using small interfering RNA and examined the effects on cell morphology and postendocytic traffic. TRP-ML1 knockdown induced gradual accumulation of membranous inclusions and, thus, represents a good model in which to examine the direct effects of acute TRP-ML1 deficiency on membrane traffic. Ratiometric imaging revealed decreased lysosomal pH in TRP-ML1–deficient cells, suggesting a disruption in lysosomal function. Nevertheless, we found no effect of TRP-ML1 knockdown on the kinetics of protein or lipid delivery to lysosomes. In contrast, by comparing degradation kinetics of low density lipoprotein constituents, we confirmed a selective defect in cholesterol but not apolipoprotein B hydrolysis in MLIV fibroblasts. We hypothesize that the effects of TRP-ML1 loss on hydrolytic activity have a cumulative effect on lysosome function, resulting in a lag between TRP-ML1 loss and full manifestation of MLIV.
The gene MCOLN1, coding for transient receptor potential–mucolipin-1 (TRP-ML1), is mutated in the rare lysosomal storage disorder mucolipidosis type IV (MLIV) (1–4), which is clinically characterized by severe developmental delays and psychomotor retardation, constitutive achlorohydria, and retinal degeneration and corneal opacities (5–8). As in all lysosomal storage disorders, MLIV is characterized at the cellular level by the buildup of membranous and electron-dense organelles containing undigested lipid products (9, 10). However, the stored lipid products are more heterogeneous than in most storage disorders and include gangliosides, sphingolipids, phospholipids, acidic mucopolysaccharides, and cholesterol (11–14). Lipid accumulations in other lysosomal storage disorders are a result of the abnormal targeting and/or activity of individual hydrolases involved in lipid processing. Although which lipid products accumulate in MLIV have been identified, there is no clear consensus as to which enzyme activities are compromised. A previous report suggested that there is deficient ganglioside sialidase activity associated with MLIV (15), whereas others argued against this enzyme deficiency (12, 16). Although some reports demonstrate that both phospholipase (17) and acid lipase (18) activities are normal in MLIV patient cells, other data suggest deficits in activities of these enzymes in MLIV cells (13). It is possible that such differences reflect variations between the primary fibroblasts used in these studies or compensatory changes in enzyme activity at the gene expression level.
Both immunofluorescence and subcellular fractionation studies demonstrate that TRP-ML1 is localized to lysosomes (19–22), and it is thought that lipid accumulations associated with MLIV are a result of imbalanced ion homeostasis along the endocytic pathway resulting from TRP-ML1 dysfunction (8, 12, 23). A consensus on TRP-ML1 permeability characteristics is only beginning to emerge, whereas native TRP-ML1 activity has not been studied. Recombinant TRP-ML1 was characterized in the plasma membrane, where it is targeted under overexpression conditions (19, 24), and in artificial lipid bilayers using TRP-ML1 purified from overexpressing cells or synthesized in a cell-free system (25, 26). These experimental systems yielded outwardly rectifying monovalent cation-permeable channels. On the other hand, “activating” mutations in TRP-ML1 resulted in an inwardly rectifying current (27). Such mutants were permeable to Ca2+, which is similar to some previously published data on wild-type TRP-ML1 (24, 28), whereas other studies have demonstrated a Ca2+ block of TRP-ML1 (13, 26).
The “biogenesis” model for MLIV progression suggests that TRP-ML1 regulates lipid trafficking by mediating specific fission and/or fusion events between late endosomes and lysosomes that occur during the process of lysosome biogenesis, a Ca2+-dependent process (29–34). In the absence of functional TRP-ML1, endocytosed material destined for degradation accumulates because of impaired access to the hydrolases necessary for catabolism. Moreover, this model predicts that there will be a global defect in the postendocytic delivery of both lipids and proteins to lysosomes. The notion that TRP-ML1 directly regulates membrane traffic is based on the observations that TRP-ML1–deficient cells display aberrant mixing of lysosomal and endosomal content (28, 33) and that trafficking of a fluorescent conjugate of BODIPY-C5-lactosylceramide (LacCer) along the endocytic pathway is delayed in these cells (13, 21, 23). This model is also supported by studies performed in Caenorhabditis elegans in which the functional TRP-ML1 orthologue CUP-5 has been identified (35, 36). Knockout of the cup-5 gene has been associated with defects in lysosome biogenesis. There is increased colocalization of late endosomal and lysosomal markers in cup-5 mutants, and loss of this gene results in the abnormal accumulation of vacuolar structures that are interpreted to represent hybrid late endosomal–lysosomal structures (33). Again, the observed endocytic abnormalities observed in cup-5 mutants were alleviated by exogenous expression of functional human TRP-ML1.
The “metabolic” model suggests that, similar to the ClC channels, TRP-ML1 regulates lysosomal ion homeostasis and thus directly affects the activity of lysosomal digestive enzymes (37). It was hypothesized that TRP-ML1 functions as a H+ leak pathway to prevent the overacidification of the lysosomal lumen, and that the activity of lysosomal lipases are disrupted as a consequence of the ionic imbalance in TRP-ML1–deficient lysosomes (13).
An additional complexity that must be clarified to properly describe MLIV pathogenesis and TRP-ML1 function is whether any defects in membrane traffic or lipid metabolism are the primary cause of MLIV, or are instead secondary effects caused by the chronic accumulation of undigested lipids in these cells. The membrane trafficking studies discussed earlier in this section were performed in chronically TRP-ML1–deficient fibroblasts. It is possible that the buildup of lipids and other undigested materials in these cells eventually impedes the entry of trafficking markers into lysosomes and manifests as delays in membrane traffic. Indeed, exactly the same lipid traffic delays were reported in several lysosomal storage disorders, whose main causes are entirely metabolic and are not directly related to membrane traffic (GM1 and GM2 gangliosidoses, Fabry's disease, and Niemann-Pick types A or B) (8, 38). To circumvent this issue, we used an siRNA approach to examine the consequences of acute down-regulation of TRP-ML1 function on postendocytic delivery to lysosomes.
Understanding whether TRP-ML1 regulates membrane traffic or lipolysis is a key step in determining whether enzyme replacement therapies will be effective as treatment for MLIV. A finding that TRP-ML1 directly regulates membrane traffic will make it unlikely that enzyme replacement therapies for MLIV will succeed. If, however, TRP-ML1 regulates lysosomal ion homeostasis, then replacement therapies, perhaps based on enzymes modified to work in an MLIV-specific lysosomal environment, are likely to be useful.
RESULTS
siRNA-mediated TRP-ML1 knockdown
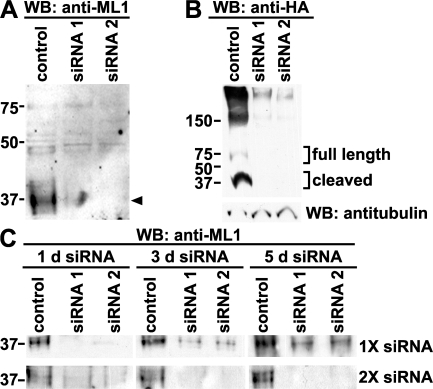
We identified two siRNA oligonucleotides specific for TRP-ML1 and tested their ability to knock down endogenous as well as heterologously expressed hemagglutinin (HA) epitope–tagged TRP-ML1 in HeLa cells. Cells were transfected with control (nonsilencing) or with one of two TRP-ML1–specific siRNA oligonucleotides. Cells were harvested 24 h after transfection and subjected to Western blot analysis. We previously demonstrated that the 65-kD full-length TRP-ML1 is intracellularly cleaved into two fragments of roughly equal molecular mass (40 and 37 kD) (19, 20). Because it contains only a single transmembrane domain, the N-terminal fragment is less prone to aggregation than the full-length protein or C-terminal half, and is thus easier to detect upon SDS-PAGE. The arrowhead in Fig. 1 A marks the migration of the N-terminal fragment of TRP-ML1. Transfection with either of the TRP-ML1–specific siRNA duplexes resulted in virtually complete knockout of native TRP-ML1. Similarly, knockdown of exogenously expressed TRP-ML1 was observed when cells transfected with siRNA were later retransfected with an HA-tagged version of TRP-ML1 (Fig. 1 B). Under these overexpression conditions, we could detect both the full-length protein and the HA-tagged cleaved fragments on Western blots of cells transfected with control but not with TRP-ML1–specific siRNA duplexes (Fig. 1 B).
Figure 1.
siRNA-mediated knockdown of TRP-ML1. (A) HeLa cells were transfected with either nonsilencing control or TRP-ML1–specific siRNA oligonucleotides. Cells were harvested for Western blot analysis after 24 h. Equal amounts of total protein were loaded for SDS-PAGE, as determined by protein assay. Samples were transferred to nitrocellulose and immunoblotted to detect endogenous levels of TRP-ML1 using an antibody directed against the first extracellular loop of the protein. The arrowhead denotes the migration of the cleaved form of TRP-ML1. (B) HeLa cells were transiently transfected with cDNA encoding HA epitope–tagged TRP-ML1 24 h after initial transfection with either control or TRP-ML1–specific siRNA duplexes. After an additional 24-h incubation, cells were solubilized and equal amounts of total protein were immunoprecipitated using anti-HA antibodies. After SDS-PAGE, proteins were transferred to nitrocellulose and probed using horseradish peroxidase–conjugated anti-HA antibody. 10% of the cell lysate was saved before immunoprecipitation and immunoblotted using antitubulin antibody as an additional loading control (bottom). (C) HeLa cells were transfected with either nonsilencing control or TRP-ML1–specific siRNA oligonucleotides and were harvested for Western blot analysis 1, 3, or 5 d after transfection (top), or were retransfected with siRNA duplexes after 2 d and were harvested for Western analysis at 3 or 5 d (bottom). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted to detect endogenous TRP-ML1. The migration of molecular mass markers (in kD) is noted on the left of the blots.
We next optimized conditions to knock down TRP-ML1 over longer time periods. To determine the duration of TRP-ML1 knockdown, cells were harvested for Western blot analysis at 1, 3, or 5 d after transfection with either a nonsilencing control siRNA duplex or with TRP-ML1–specific oligonucleotides. As before, knockdown was virtually complete within 1 d of transfection; however, native TRP-ML1 reappeared within 3 d after the initial siRNA duplex transfection (Fig. 1 C, top). To overcome this, we performed a similar experiment except that the 3- and 5-d samples were retransfected with TRP-ML1–specific or control siRNA duplexes 48 h after the initial transfection, and were subsequently harvested for Western blot analysis at 3 or 5 d. As shown in Fig. 1 C (bottom), using these conditions, we were able to knock down TRP-ML1 almost completely during this time course.
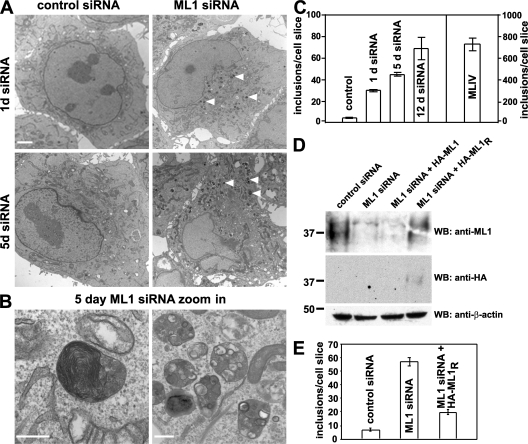
With specific TRP-ML1 knockdown conditions in place, we next sought to determine the onset of MLIV-like lipid inclusion formation over a 5-d knockdown period in HeLa cells. Electron microscopic analysis of cells transfected with either control or TRP-ML1–specific siRNA revealed the progressive accumulation of pleomorphic inclusions scattered throughout the cytoplasm of cells transfected with TRP-ML1 siRNA, whereas very few such inclusions were observed in control cells (Fig. 2 A). These inclusions are reminiscent of those observed in patient-derived MLIV fibroblasts, some of which contain multilamellar membranes, whereas others are filled with electron-dense, gray material (Fig. 2 B). Quantitation of the accumulation of storage bodies in both control and TRP-ML1 siRNA–transfected cells is shown in Fig. 2 C. In TRP-ML1 siRNA–treated cells, inclusions begin to accumulate as early as 1 d after siRNA transfection and gradually increased over the 5-d period. It should be noted that even after 12 d of siRNA treatment, the mean number of inclusions observed in HeLa cells (69 inclusions per cell slice; Fig. 2 C) was much lower than that previously determined for MLIV fibroblasts using the same method (∼720 inclusions per cell slice; Fig. 2 C).
Figure 2.
Time course of lipid inclusion formation in TRP-ML–deficient cells. (A and B) Electron micrographs of HeLa cells after either 1 or 5 d of TRP-ML1–specific or control siRNA oligonucleotide transfection. Bars: (A) 2 μm; (B, left) 200 nm; (B, right) 500 nm. (C) Quantitation of the effect of TRP-ML1 knockdown on the formation of storage inclusions. The number of inclusions was calculated using the automated particle counting function of ImageJ. Data are expressed as the number of inclusions per cell slice ± SEM (D) Western blot analysis of siRNA-treated cells expressing siRNA-resistant HA-ML1R. HeLa cells were transfected with either control or TRP-ML1–specific siRNA, as indicated. 24 h after siRNA treatment, cells were cotransfected with plasmids encoding GFP and either HA-ML1 (non-siRNA resistant) or HA-ML1R (siRNA resistant). The next day, cells were sorted by FACS analysis to identify GFP-positive (transfected) cells and were subsequently retransfected with the appropriate siRNA and returned to culture for 48 h. Cells were harvested, and 30 μg of total protein was loaded for SDS-PAGE. Samples were transferred to nitrocellulose and immunoblotted to detect endogenous levels of TRP-ML1 (top), HA-ML1 or HA-ML1R (middle), or β-actin (bottom) as a loading control. (E) Quantitation of the effect of HA-ML1R expression on formation of storage inclusions in TRP-ML1 siRNA–treated cells. The number of inclusions was calculated as described in C. The migration of molecular mass markers (in kD) is noted on the left of the blots.
To confirm that the accumulation of storage bodies in TRP-ML1 siRNA–treated cells is a direct consequence of TRP-ML1 loss, we performed knock-in experiments in which the TRP-ML1 siRNA–treated cells were transfected with an HA-tagged siRNA-resistant TRP-ML1 construct (HA-ML1R). Because transfection with cDNA was only ∼30% efficient in these cells, we cotransfected them with GFP-expressing plasmid and, the following day, enriched for GFP-expressing cells using FACS. The cells were then transfected with TRP-ML1–specific siRNA oligonucleotides and processed for electron microscopic analysis 48 h later. Knockdown of endogenous TRP-ML1 knockdown and expression of the siRNA-resistant HA-ML1R construct were confirmed by Western blotting of duplicate samples (Fig. 2 D). As shown in Fig. 2 E, siRNA-treated cells expressing HA-ML1R displayed a marked decrease in the number of inclusions compared with TRP-ML1 knockdown cells that were not transfected with cDNA, suggesting that HA-ML1R expression rescues or prevents inclusion formation in TRP-ML1–deficient cells. These studies confirm that inclusion formation is TRP-ML1 specific. Thus, this experimental system represents an appropriate model in which to study the early stages of MLIV progression. As a result, this approach will enable us to assess the function of TRP-ML1 under conditions that are unbiased by the chronic accumulation of undigested lipid material that may itself have adverse effects on specific lipid and protein trafficking events.
Lipid inclusions in TRP-ML1–deficient cells are lamp-1 positive and receive endocytosed cargo
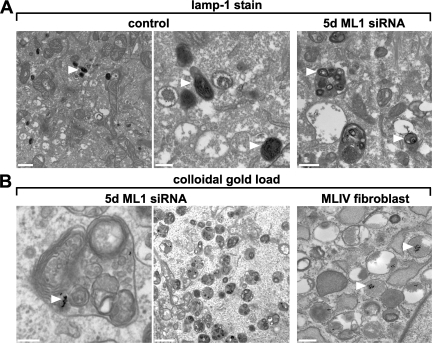
To examine whether the inclusions that accumulate in TRP-ML1–deficient cells represent active components of the endocytic pathway, control and 5-d TRP-ML1 siRNA–treated cells were labeled en bloc with antibodies directed against the lysosomal membrane protein lamp-1, and were subsequently labeled with biotinylated secondary antibodies. Immunoreactivity was detected using avidin–biotin peroxidase complex and 3,3′-diaminobenzidine. Fig. 3 A (right) shows that inclusions from TRP-ML1–deficient cells are positive for lamp-1 (dark stain identified by arrowheads). In contrast, lamp-1–positive compartments in cells transfected with a control siRNA duplex were smaller and more uniformly stained (Fig. 3 A, middle and left). This staining pattern is consistent with the idea that the lipid inclusions in TRP-ML1–deficient cells are of late endosomal or lysosomal origin.
Figure 3.
Inclusion bodies are lamp-1 positive and remain accessible to internalized cargo in TRP-ML–deficient cells. (A) Lamp-1 immunoreactivity in control and TRP-ML–deficient cells is indicated by arrowheads. Control or TRP-ML1 siRNA–treated HeLa cells (5 d) were labeled with antibodies directed against the lysosomal membrane protein lamp-1, as described in Materials and methods. Lysosomes from both control and TRP-ML1 siRNA–treated cells are positive for lamp-1 immunoreactivity. Arrowheads indicate lamp-1–positive labeled compartments. Bars: (left) 2 μm; (middle) 500 nm; (right) 500 nm. (B) Colloidal gold inside storage inclusions in TRP-ML–deficient cells. Control or TRP-ML1 siRNA–treated HeLa cells (5 d) were loaded with 10-nm colloidal gold particles and processed for electron microscopy as described in Materials and methods. A large fraction of the inclusions was gold positive in 5-d siRNA cells, indicating that the inclusions were recently formed. Gold was chased for 16 h. Arrowheads indicate colloidal gold–labeled compartments. Bars: (left) 200 nm; (middle) 2 μm; (right) 500 nm.
We next asked whether the lipid inclusions present in TRP-ML1–deficient cells are accessible to internalized cargo. For these studies, cells transfected with TRP-ML1–specific siRNA for 5 d were loaded for 6 h with albumin-conjugated 10-nm colloidal gold particles, chased for an additional 12 h to allow accumulation in lysosomes, and processed for electron microscopy. Fig. 4 B (left and middle) shows extensive delivery of gold particles to lipid inclusions in TRP-ML1–deficient cells. Similar results were also obtained using MLIV fibroblasts (Fig. 4 B, right). These results demonstrate that the observed lipid inclusions are not “dead-end” structures but are able to receive material internalized along the endocytic pathway.
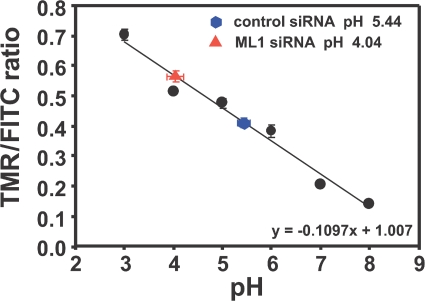
Figure 4.
Lysosomal pH is lower in TRP-ML1 siRNA–treated cells. Control or TRP-ML1 siRNA–treated (5 d) HeLa cells were loaded with 3 mg/ml of FITC- and TMR-conjugated dextrans for 12 h. Lysosomal pH was determined by calculating the ratio of TMR/FITC fluorescence. Images were acquired as described in Materials and methods. Ratiometric data were converted to absolute values of pH using TMR/FITC ratios determined from permeabilized cells equilibrated with calibration solutions. Data from 20 random fields of cells were quantified, and the pH determined is presented as mean pH ± SEM. Similar results were obtained in four independent experiments.
Lysosomal pH is reduced upon siRNA-mediated knockdown of TRP-ML1
Previous studies examining the measurement of lysosomal pH in TRP-ML1–deficient cells have produced conflicting results. Early studies performed using ratiometric imaging approaches determined that the lysosomal pH in MLIV cells was elevated almost one full pH unit higher when compared with control cells or to cells isolated from patients with other lysosomal storage diseases (39). In other studies, lysosomal pH in MLIV cells has been reported to be either more acidic (13) or unchanged (21, 40) compared with control cells. One unifying feature in all these studies is that they were performed using isolated patient fibroblasts. These cells are not necessarily appropriate for such experiments because of the long-term accumulation of undigested lipids that induce autofluorescence (41), which may interfere with dye loading and/or retention, and which may induce compensatory changes in these cells at the level of protein expression. Therefore, we examined lysosomal pH in cells treated with TRP-ML1–specific siRNA for 5 d, allowing us to measure pH in TRP-ML1–deficient cells without any potential effects caused by long-term lipid accumulation. To quantitatively determine lysosomal pH in cells treated with control or TRP-ML1–specific siRNA, cells were loaded for 12 h simultaneously with FITC- and tetramethylrhodamine (TMR)-conjugated 10-kD dextrans (3 mg/ml) to allow for accumulation in lysosomes. In separate experiments, we confirmed that both dextrans trafficked together and localized in lysosomes after the specified chase period (unpublished data). The FITC-conjugated dextran was used as a pH indicator, whereas the TMR-conjugated dextran provided a pH-insensitive readout for the ratiometric pH measurements. After dextran loading, measurements of lysosomal pH were obtained by calculating the ratio of TMR/FITC fluorescence and fitting these to a standard curve constructed as described in Materials and methods (42). As shown in a representative experiment in Fig. 4, TRP-ML1 knockdown was associated with lysosomal acidification. In this experiment, the mean lysosomal pH in cells treated with control siRNA was 5.44 ± 0.06, whereas in cells treated with TRP-ML1–specific siRNA, the mean pH was estimated to be 4.04 ± 0.09. Over the course of four experiments using identical imaging conditions, the mean difference in pH between control and TRP-ML1 siRNA–treated cells was 1.12 ± 0.16 pH units. These results indicated that loss of functional TRP-ML1 in cells results in a reduction of lysosomal pH even after a short period of TRP-ML1 knockdown, and that even lysosomes that have not yet accumulated significant amounts of undigested material are functionally affected by loss of TRP-ML1.
Lysosomal delivery of LacCer is unimpaired in TRP-ML1–deficient cells
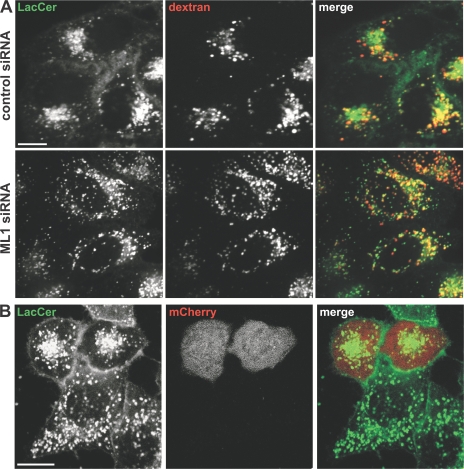
Numerous studies have reported that trafficking of the fluorescent lipid analogue LacCer is impaired in MLIV fibroblasts as well as in primary fibroblasts from several other lysosomal storage diseases (13, 21, 23, 38). We thus examined whether TRP-ML1 knockdown cells also display this phenotype. HeLa cells preloaded with fluorescent dextran were treated with control or TRP-ML1 siRNA were incubated with LacCer for 60 min and then chased for 5 h at 37°C. As shown in Fig. 5 A, LacCer staining in cells transfected with a control siRNA oligonucleotide was tightly clustered around the nucleus, consistent with efficient delivery to the Golgi complex (top). In contrast, the LacCer staining pattern in cells treated with TRP-ML1–specific siRNA was dispersed throughout the cytoplasm and colocalized partly with preinternalized dextran. These results are consistent with observations in MLIV fibroblasts (unpublished data) (13, 21, 23) and with another recent study examining TRP-ML1 knockdown in murine macrophages (43).
Figure 5.
Postlysosomal lipid traffic is impaired in TRP-ML–deficient cells. (A) Control or TRP-ML1 siRNA–treated (5 d) HeLa cells were preloaded for 12 h with Alexa Fluor 647–conjugated dextran. The next day, cells were labeled with LacCer, as described in Materials and methods, and chased for 5 h. Fluorescence images of both LacCer (green) and Alexa Fluor 647–dextran (red) were obtained for both control (top) and TRP-ML1 siRNA–treated (bottom) cells. Bar, 10 μm. (B) LacCer Golgi localization can be restored by expression of HA-ML1R in TRP-ML1 siRNA–treated cells. TRP-ML1 siRNA–treated cells (2 d) were cotransfected with plasmids encoding the siRNA-resistant construct HA-MLR cDNA and the fluorescent protein mCherry (to identify DNA-transfected cells) 24 h before siRNA treatment. Cells were loaded and chased with LacCer as in A. The juxtanuclear LacCer staining pattern reminiscent of normal cells is observed in mCherry-positive cells but not in neighboring cells that presumably do not express HA-ML1R. Bar, 10 μM.
We examined whether expression of the siRNA-resistant construct HA-ML1R in knockdown cells could rescue the LacCer sorting defect. Cells were cotransfected with a vector encoding the fluorescent protein mCherry (to identify transfected cells) and HA-MLR. The following day, these cells were transfected with TRP-ML1 siRNA and cultured for 48 h before labeling with LacCer as described. Fig. 5 B shows juxtanuclear localization of LacCer in mCherry-positive (presumably HA-MLR–expressing) cells, consistent with the staining pattern of control cells. Therefore, siRNA-mediated TRP-ML1 knockdown effectively recapitulates the cellular phenotype of MLIV cells.
A crucial question in MLIV pathogenesis is whether the delay in LacCer trafficking occurs at the step of endosome–lysosome interaction, as predicted by the biogenesis model. Although our previously published data did not support this idea (13), a detailed analysis of the membrane traffic in MLIV fibroblasts performed by Pryor et al. (21) suggested a delay in transfer of the endocytosed material from late endosomes to lysosomes. However, because these studies were performed in MLIV fibroblasts, this effect could be a secondary result of undigested lipid buildup. Therefore, we performed a series of experiments to quantitate the rate of protein and lipid delivery to lysosomes in control and TRP-ML1 knockdown HeLa cells. The premise of these experiments was that if TRP-ML1 directly regulates the delivery of endocytosed material to lysosomes, then acute TRP-ML1 down-regulation would result in prelysosomal buildup of endocytosed material, whose entry into lysosomes will be significantly delayed.
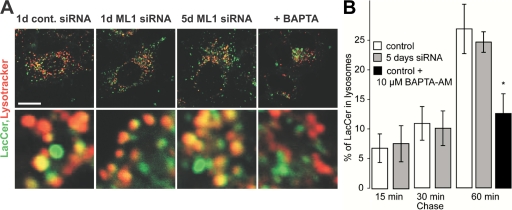
Fig. 6 shows the results of our experiments. Control and siRNA-transfected cells were loaded with 2 μg/ml LacCer for 15 min at 37°C to incorporate LacCer into the plasma membrane. The cells were washed and chased for brief periods (up to 60 min) in the presence of 5 mg/ml BSA. After the chase, cells were loaded with LysoTracker Red to identify lysosomes, and confocal images of cells were analyzed for overlap between LacCer and LysoTracker Red to quantify lysosomal delivery of LacCer. As shown in Fig. 6, we found no difference in kinetics of lysosomal delivery of LacCer in control versus TRP-ML1–deficient cells. As a positive control, we pharmacologically suppressed membrane fusion and confirmed that this induces an observable delay in lipid traffic. To do this, we loaded untransfected HeLa cells with 1 μM of the cell-permeable Ca2+ chelator BAPTA-AM. Because vesicular fusion in the endocytic pathway requires Ca2+, chelation with BAPTA should suppress membrane fusion and, thus, replicate the conditions predicted by the biogenesis model associated with the loss of TRP-ML1 Ca2+ conductance. Fig. 6 shows that, unlike TRP-ML1 down-regulation, BAPTA-AM inhibited delivery of LacCer to lysosomal compartments. Therefore, the delivery of LacCer to lysosomes is unimpaired in TRP-ML1–deficient cells.
Figure 6.
Lysosomal delivery of LacCer is normal in TRP-ML1 siRNA–treated cells. (A) Control and TRP-ML1 siRNA–treated cells were loaded with 2 μg/ml LacCer for 15 min at 37°C and LysoTracker Red to label lysosomes, as described in Materials and methods. Bar, 10 μm. (B) Where indicated, cells were incubated with 10 μM BAPTA-AM for 1 h. Delivery of LacCer to lysosomes was measured by quantifying the percent overlap between LacCer and LysoTracker. Data are mean ± SEM. *, P = 0.013 (obtained in six separate measurements).
Low density lipoprotein (LDL) trafficking and degradation in TRP-ML1–deficient cells
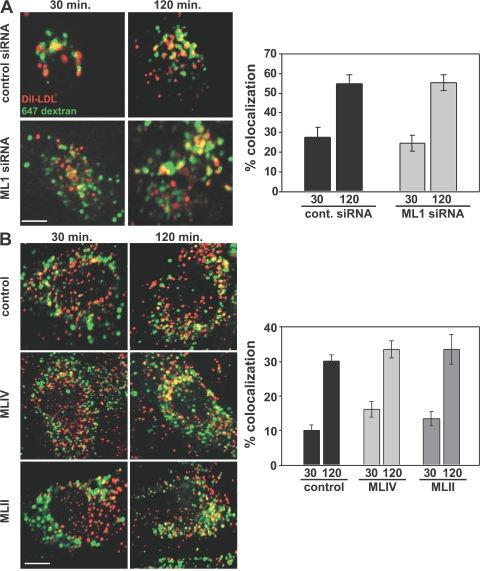
To more quantitatively assess lysosomal delivery and function in TRP-ML1 knockdown cells, we examined the kinetics of lysosomal delivery, apolipoprotein B (apoB) proteolysis, and cholesterol deesterification of LDL. Previous studies in MLIV fibroblasts have specifically probed lipid metabolism along the LDL receptor–mediated pathway of lysosomal degradation using radiolabeled forms of both sphingomyelin and cholesteryl oleate (CO) (18). These studies found that MLIV fibroblasts metabolize both lipids more slowly compared with control cells. Therefore, our first approach was to examine whether delivery of LDL to lysosomes in TRP-ML1–deficient cells was impaired. To determine this, we used fluorescence microscopy to examine and quantitate the delivery of DiI-labeled LDL to lysosomes of HeLa cells transfected with TRP-ML1–specific siRNA. In these studies, cells were treated with either control or TRP-ML1 siRNA duplexes for 5 d. Alexa Fluor 647–conjugated dextran was preaccumulated in lysosomes during a 12-h preincubation before the start of the experiment. The following day, cells were incubated with 20 μg/ml DiI-LDL on ice for 1 h and were subsequently chased in fresh medium for 30 or 120 min at 37°C. At either time point, cells were fixed and coverslips were mounted for analysis. Delivery of DiI-LDL to lysosomes was measured by quantifying the percentage of overlap between DiI-LDL and the preloaded Alexa Fluor 647–dextran expressed as the amount of Di-LDL present in dextran-positive compartments. As shown in Fig. 7 A, ∼60% of the internalized DiI-LDL colocalized with the preloaded Alexa Fluor 647–dextran by 120 min of chase in both control and TRP-ML1 siRNA–treated cells, suggesting that delivery of LDL in TRP-ML1–deficient cells is unimpaired. We also did not observe a delay in parallel studies quantifying DiI-LDL delivery in MLIV and MLII fibroblasts relative to control cells, suggesting that chronic lipid buildup in these cells does not affect LDL delivery to lysosomes (Fig. 7 B). Collectively, these results suggest that delivery of LDL to lysosomes is unaffected by both acute and chronic loss of TRP-ML1 function.
Figure 7.
LDL delivery to lysosomes is normal in TRP-ML–deficient cells. Control or TRP-ML1 siRNA–treated (5 d) HeLa cells (A) or fibroblasts (B) were preloaded for 12 h with Alexa Fluor 647–conjugated dextran. Cells were incubated with DiI-LDL on ice for 60 min and were subsequently chased in prewarmed media at 37°C for an additional 30 or 120 min. At the indicated time points, cells were fixed and processed for immunofluorescence. Delivery of DiI-LDL to lysosomes in cells was measured by quantifying the percent overlap between DiI-LDL and the preloaded Alexa Fluor 647–dextran. Graphical representations of the quantifications are shown (A and B, right) and are expressed as the percent overlap ± SEM for cells under each condition (n = 20). Bars, 10 μm.
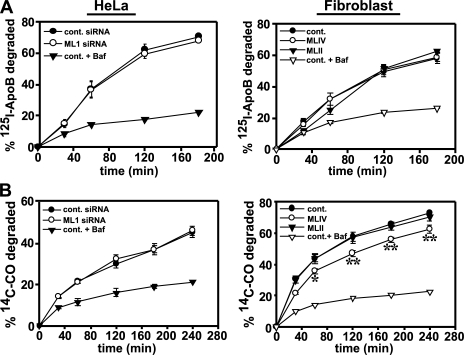
Our second approach was to examine the delivery and lysosomal degradation of both the lipid and protein components of LDL in both control and MLIV fibroblasts as well as in control and in TRP-ML1 siRNA–treated cells. If TRP-ML1 is involved in the interactions between late endosomes and lysosomes, then there should be a defect in the degradation of both the protein and lipid components of LDL along this pathway. To monitor the degradation of the protein component of LDL, cells were incubated with 125I-apoB-LDL for 2 h on ice. After ligand binding, cells were extensively washed and incubated with prewarmed medium for various times. At the indicated times, the medium was removed and replaced, and degradation kinetics were determined by trichloroacetic acid (TCA) precipitation of the medium and of cell lysates solubilized at the end of the time course. The rate of apoB degradation was determined by calculating the cumulative release of TCA-soluble counts into the medium. As shown in Fig. 8 A, the kinetics of degradation of apoB were identical in both control and TRP-ML1–deficient cells, as well as in control and MLIV fibroblasts. Similarly, we found no difference in the kinetics of 125I-transferrin recycling in MLIV and control fibroblasts, or in the degradation of 125I–epidermal growth factor in both siRNA-treated cells and MLIV fibroblasts (unpublished data). These results suggest that there is no defect in postendocytic recycling or lysosomal delivery in TRP-ML1–deficient cells. These results are consistent with the observations of Chen et al., who (using a similar 125I-transferrin recycling assay) also found no defects in the endocytosis or recycling of transferrin in MLIV fibroblasts (23).
Figure 8.
Degradation of LDL cholesterol is impaired in MLIV fibroblasts. (A) siRNA-transfected HeLa (5 d) or patient fibroblasts were incubated on ice for 2 h with 125I-apoB-LDL complexes. For control cells, 0.5 μM bafilomycin was added 30 min before LDL labeling and maintained in the culture medium throughout the experiment. After ligand binding, cells were washed and incubated in prewarmed medium. At the indicated time points, the medium was collected and replaced, and cells were solubilized after the final time point. The rate of apoB degradation was determined by calculating the cumulative release of TCA-soluble counts into the medium, as described in Materials and methods. (B) siRNA-transfected HeLa (5 d) or patient fibroblasts were labeled with medium containing 10% LPDS and [14C]CO–LDL complexes (50 μg/ml LDL protein) for 4 h at 37°C, washed, and incubated for 30 min at 18°C in medium supplemented with 10% FBS. The time course was initiated by the addition of fresh culture medium supplemented with 10% LPDS at 37°C. At the indicated times, the medium was collected and replaced. After the final time point, cells were harvested, and radioactivity was measured in both the medium and cell pellet. The cumulative percentage of preinternalized [14C]CO released into the medium at each time point was calculated as described in Materials and methods. The mean ± SEM of three experiments (two for bafilomycin A1–treated samples) performed in triplicate is plotted in each panel. *, P = 0.032; and **, P < 0.008 by using the Student's t test.
To examine the degradation of the lipid component of LDL, we used an experimental approach previously used by Groener et al. (44) to monitor the metabolism of radiolabeled CO. Cells were incubated with [14C]CO in complex with LDL for 4.5 h at 37°C. After labeling, cells were incubated for various times with prewarmed medium supplemented with lipoprotein-deficient serum (LPDS). At each time point, the medium was removed and replaced, and degradation kinetics were determined by quantitating the cumulative release of free fatty acids into the cell-culture medium compared with the amount that remained cell associated. As shown in Fig. 8 B, the kinetics of [14C]CO degradation were not different in cells treated with either control or TRP-ML1–specific siRNA for 5 d. Additionally, treatment with TRP-ML1–specific siRNA over a 12-d period also had no effect on cholesterol hydrolysis (unpublished data). However, the rate of [14C]CO degradation in MLIV fibroblasts was significantly slower when compared with the rates of degradation in either control or MLII fibroblasts.
DISCUSSION
The internalization and processing of cargo via the endocytic pathway is a complex process involving numerous steps that include cargo binding and entry, receptor–ligand dissociation, membrane fusion, degradation of internalized lipids, proteins, and carbohydrates, and excretion of digested products through the lysosomal membrane (45, 46). The efficiency of these processes depends on the maintenance of specific ionic environments that are established by the combination actions of ion channels and pumps residing in endocytic compartments (32, 47). New information about these ion transporters regularly emerges from studies on the genetic determinants of lysosomal storage diseases. MLIV is one of several examples of lysosomal storage diseases caused by a dysfunction in the ionic balance of the endocytic machinery. The severity of this disease is compounded by the fact that no unified model exists for MLIV pathogenesis, thus hindering the development of pharmacological interventions.
Because the identification that mutations in the MCOLN1 gene are responsible for the MLIV disease phenotype (1), it has become apparent that this lysosomal cation channel plays a key role in the maintenance and regulation of postendocytic events (5, 12, 18, 23). As described in the Introduction, two models exist to explain the primary defect in MLIV. The biogenesis model suggests that MLIV is caused by delayed membrane traffic to lysosomes as a result of impaired endosome–lysosome fusion or fission (21, 31, 33, 34). The metabolic model postulates that MLIV results from an ionic imbalance in lysosomes that precludes efficient processing of internalized lipids and other molecules (13).
In principle, a comprehensive model for the regulation of TRP-ML1 channel activity would enable the discrimination between these models. Fusion between various membrane-bound vesicular endocytic compartments is regulated by Ca2+ concentration in the vicinity of these compartments, whereas the trafficking and activity of various proteases and lipases depends on the acidic environment maintained within both endosomal and lysosomal compartments (32, 47). TRP-ML1 has been reported to be permeable to both Ca2+ and to H+; however, electrophysiological characterization of TRP-ML1 activity under physiological conditions has yet to be performed (13, 24–26, 28).
In the present studies, we sought to determine whether MLIV results from aberrant membrane traffic to lysosomes. An additional issue is whether any observed defects in membrane trafficking or lipid metabolism are a primary result of MLIV pathophysiology, or are instead secondary effects that result from the chronic accumulation of undigested lipids in TRP-ML1–deficient cells. Our goal was to design an experimental system in which we could examine these membrane-trafficking steps in cells that lacked TRP-ML1 but that had not yet chronically accumulated undigested lipids. Because MLIV has been unequivocally identified as a “single-gene disorder”, acute TRP-ML1 knockdown using siRNA is a valid model for the early stages of MLIV pathogenesis. Using this approach, we were able to down-regulate TRP-ML1 expression in cells over a period of several days. During this time, lysosomes from these cells gradually accumulated lipid inclusions, but at a level significantly less than previously described in MLIV fibroblasts (13). Formation of inclusions was efficiently rescued by transfection of an siRNA-resistant version of TRP-ML1. Lipids accumulated in lamp-1–positive organelles, suggesting that the inclusions are of lysosomal origin. Furthermore, colloidal gold internalized by fluid-phase endocytosis could access inclusions in both isolated patient fibroblasts and in siRNA-treated cells, demonstrating that these organelles remain active components of the endocytic pathway.
Several studies have reported that the trafficking of fluorescent conjugates of LacCer is hindered in MLIV fibroblasts as well as in other lysosomal storage diseases (13, 21, 23, 38). Consistent with these observations, we observed a similar defect in LacCer handling in cells treated with TRP-ML1–specific siRNA. However, we observed no difference in the delivery of this lipid to lysosomes in control and TRP-ML1 knockdown cells. Therefore, the altered trafficking of LacCer may represent an effect on postlysosomal membrane traffic and/or lipid metabolism in cells after prolonged accumulation of undigested materials. These results are consistent with a recent report by Thompson et al. (43) suggesting that the primary defect in MLIV lipid handling may be the exit of internalized lipids from lysosomes. We found no effect of acute TRP-ML1 knockdown on the rate of LDL delivery to lysosomes or on the degradation of both the lipid and protein components of this complex. Collectively, these data argue strongly against the biogenesis model for MLIV progression, which predicts that acute loss of TRP-ML1 function will result in a global defect in the delivery of both internalized lipids and proteins to lysosomes.
The role of TRP-ML1 in lysosomal ion homeostasis is currently disputed. Increased accumulation of acridine orange in MLIV fibroblasts has previously been observed (13), consistent with increased acidity of these organelles; however, quantitation of lysosomal pH in MLIV fibroblasts by other groups has yielded discrepant results (21, 39, 40). Similar to Pryor et al. (21), we were unable to document any difference in lysosomal pH between control and MLIV fibroblasts by fluorescence ratio imaging; however, our studies were compromised by the intense autofluorescence in MLIV lipid inclusions that has previously been reported (41). However, we reproducibly observed that lysosomal pH in HeLa cells lacking functional TRP-ML1 was more acidic than control (a decrease of ∼1.12 pH units). In principle, this finding is consistent with the metabolic model for MLIV pathogenesis, which postulates that the increased acidity of lysosomes disrupts lipid hydrolysis.
Previous groups have demonstrated defects in lipase handling in MLIV fibroblasts, including delayed deesterification of cholesterol esters and decreased lysosomal acid lipase activity (13, 18). Consistent with this, we found that the release of free fatty acids from [14C]CO-labeled LDL was slowed in MLIV fibroblasts. Surprisingly, however, we did not detect a deficit in cholesterol metabolism in siRNA-treated cells (after 1, 5, or 12 d).
The apparently normal hydrolysis of LDL cholesterol upon acute loss of TRP-ML1 function demonstrates that the increased lysosomal acidity observed in these cells does not critically impair acid lipase activity. Rather, there appears to be a gradual effect on lysosomal hydrolysis that manifests as a lag period between loss of TRP-ML1 function and the full elaboration of the MLIV disease phenotype. It is possible that a minor deficit in lipid hydrolysis that is undetectable early after TRP-ML1 loss has a cumulative effect whose consequences slowly develop as the disease progresses. Indeed, other lysosomal storage disease models such as Niemann-Pick type C (NPC) (48, 49) teach us that a defect in a single component of lysosomal machinery may sabotage processing of unrelated classes of lipids. NPC disease is caused by the mutation in either the late endosomal membrane protein NPC1 or the soluble lysosomal protein NPC2, and results in abnormal cholesterol transport along the late endocytic pathway. Similar to MLIV, NPC is caused by defective cholesterol trafficking rather than by a specific enzymatic abnormality. Although the primary defect is in cholesterol transport, the accumulation of other lipids, including sphingolipids, has also been demonstrated in NPC (48).
In summary, our results suggest that TRP-ML1 does not directly regulate membrane traffic; thus, enzyme replacement therapies remain a potentially viable treatment option for MLIV. Future studies are needed to determine whether TRP-ML1 plays an essential role in maintaining lysosomal ion homeostasis directly so that replacement enzymes may be designed to operate in the unique environment of the TRP-ML1–deficient lysosome.
MATERIALS AND METHODS
Cell lines and reagents.
HeLa SS6 cells were maintained in DME (Sigma-Aldrich) supplemented with 10% FBS (Atlanta Biologicals) and 100 μg/ml penicillin/streptomycin (Invitrogen). Skin fibroblasts from patients with MLIV (WG0909), a heterozygous relative (WG0987), and MLII (WG0229) were obtained from the Repository for Mutant Human Cell Strains. All skin fibroblast cell lines were maintained in DME supplemented with 10% FBS and 100 μg/ml penicillin/streptomycin. All reagents were obtained from Sigma-Aldrich unless indicated otherwise.
siRNA-mediated knockdown of TRP-ML1.
Two double-stranded siRNAs targeting the human form of ML1 (TRP-ML1; available from GenBank/EMBL/DDBJ under accession no. BC005149) were designed and synthesized using Invitrogen's proprietary BLOCK-iT RNAi protocol. The first TRP-ML1–specific target sequence (ML1 siRNA no. 1) used was 5′-CCCACATCCAGGAGTGTAA-3′, corresponding to nucleotides 959–977 of the human TRP-ML1 mRNA. The second target sequence (ML1 siRNA no. 2) used was 5′-CCGCTACCTGACCTTCTT-3′, corresponding to nucleotides 1328–1345 of the human TRP-ML1 mRNA. All experiments, unless otherwise noted, were performed used TRP-ML1 siRNA no. 1. To generate the siRNA-resistant construct HA-ML1R, the codons encoding amino acids IQEC starting at position 281 of TRP-ML1 (targeted by siRNA no. 1) were mutated from CATCCAGGAGTG to TATTCAAGAATG to create siRNA resistance without affecting the corresponding amino acid sequence. For knockdown of endogenous TRP-ML1, HeLa SS6 cells were plated in 24-well dishes and allowed to grow to ∼50–60% confluence. Cells were transfected with a nonspecific control siRNA duplex (Thermo Fisher Scientific) or TRP-ML1–specific siRNA duplexes. For each individual well, 3 μl of 20 μM siRNA oligonucleotide and 4.5 μl TransIT-TKO oligonucleotide transfection reagent (Mirus) were used. Cells were harvested for protein concentration or Western blot analysis 1, 3, or 5 d after transfection. For cells transfected with siRNA twice over the 5-d knockdown period, 2 d after initial transfection, cells were replated onto 24-well plates, retransfected as described the same day, and harvested for analysis either 24 or 72 h later. For knockdown of exogenously expressed HA epitope–tagged ML1 (HA-ML1) (19, 20), cells were plated and transfected with siRNA as described. 24 h after siRNA transfection, cells were transiently transfected with cDNA encoding HA-ML1 using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol, and harvested for Western blot analysis 24 h later. Samples were immunoprecipitated and immunoblotted as previously described (19, 20).
Electron microscopy.
Cells grown on plastic dishes were fixed by a 30-min incubation with a solution containing 2.5% glutaraldehyde in 0.1M Na-cacodylate, washed with 0.1M Na-cacodylate, postfixed with a solution containing 1% OsO4, washed with PBS, and stained en bloc for 30 min with 2% uranyl acetate. After dehydration by immersion in 30–100% ethanol, the samples were embedded in resin by immersion in 30–100% resin/propylene oxide mixtures. Fixed samples were mounted on grids and analyzed with a transmission electron microscope (100CX; JEOL Ltd.). For immunostaining with lamp-1 antibodies, after fixation and freeze–thaw permeabilization of the membranes at –80°C in a cryoprotectant solution containing glycerol and sucrose, cells were blocked in BSA with goat serum, incubated overnight with monoclonal anti–lamp-1 antibodies (Santa Cruz Biotechnology, Inc.), and rinsed, and biotinylated secondary antibodies were added. After an extensive wash, the samples were incubated with avidin–peroxidase complexes for 30 min, followed by another wash. Next, 3,3′-diaminobenzidine and H2O2 were added for 4–10 min and, after another wash, the samples underwent secondary fixation with 1% OsO4 for 1 h. This procedure yields the dark stain associated with lamp-1 immunoreactivity.
For colloidal gold uptake experiments, 10-nm gold particles were incubated with gelatin and BSA (pH 5), to coat the particles. Next, the particles were washed and added to the cells for 1 h to load the endocytic pathway. After a 6–16-h chase, cells were fixed and processed for electron microscopy as described in the previous paragraph.
Measurement of lysosomal pH.
Determination of lysosomal pH was performed as previously described (50, 51). HeLa cells treated with siRNA for 5 d were plated onto 0.17-mm ΔT live-cell cover glass dishes (Biotechs). 12 h before the start of imaging, cells were loaded with 3 mg/ml each of FITC- and TMR-conjugated 10,000-kD dextrans (Invitrogen) to allow accumulation in lysosomes. To measure pH differences between cells, a standard curve was determined for each experiment. Before imaging for the standard curve, cells were rinsed once with MES buffer (115 mM KCl, 5 mM NaCl, 1.2 mM MgSO4, 25 mM MES) that had been calibrated using a pH meter (Accumet; Thermo Fisher Scientific) to 3, 4, 5, 6, 7, or 8. After rinsing, the cells were imaged in the same buffer supplemented with 10 μM nigericin and 10 μM monesin to equilibrate intracellular and extracellular pH. Experimental dishes were imaged in DME without sodium bicarbonate. Images were taken using a microscope (IX81; Olympus) with a 60× 1.4 NA PlanApo oil-immersion objective. Pairs of images were captured from random fields of cells using a spinning disc confocal system (Perkin Elmer). Images were acquired and analyzed with Metamorph software (MDS Analytical Technologies). For analysis, equivalent FITC and TMR images were subjected to a morphological filter (Tophat) to remove background fluorescence. A binary mask was applied so that only matched spots from each image were compared. After applying a threshold, the total gray value for each image was recorded for both channels, and the TMR/FITC ratio was determined. TMR/FITC ratios were plotted against pH values, and curves were fitted using linear regression analysis. At least 20 images were analyzed for each condition in four independent experiments. The Student's t test was used to determine statistical significance.
LacCer traffic.
To examine LacCer trafficking at long chase times, HeLa cells were labeled with 5 μM LacCer complexed to BSA in serum-free DME for 60 min at 37°C. Cells were gently washed three times with PBS and incubated with DME plus 10% FCS for 5 h at 37°C. After chase, cell-surface LacCer was back-extracted by washing cells for 30 min in DME without bicarbonate supplemented with 2% (wt/vol) fatty acid–free BSA at 4°C. Cells were washed briefly with PBS, and images were acquired in DME without bicarbonate at 20°C. Images were acquired using the Olympus IX81 equipped with a spinning disc confocal microscope system (UltraVIEW; PerkinElmer). Cells were imaged using a 60× objective, and the LacCer was excited using a 488-nm laser, whereas the dextran was excited using a 647-nm laser. These two excitation filters were used to account for the aggregation-dependent shift in red fluorescence that is characteristic of the BODIPY fluorophore. Under these experimental conditions, little red LacCer fluorescence was observed, so LacCer images in Figs. 5 and 6 were acquired using only the 488-nm laser.
For shorter experiments examining lysosomal delivery kinetics, HeLa cells were loaded with LacCer (Invitrogen) and analyzed as before (13). After loading and chase, cells were incubated with the lysosomal marker LysoTracker Red (Invitrogen) and confocal images were taken. The images were analyzed using the RGB colocalization add-in to ImageJ (available at http://rsb.info.nih.gov/ij/), as previously described (13, 52). In each image, the percentage of LacCer in lysosomes (%laccerLys) was estimated by dividing the number of pixels contained within the area of overlap between LacCer and LysoTracker stains (Nover) by the number of pixels covered by the LacCer staining pattern (Nlaccer): %laccerLys = 100 × Nover/Nlaccer. To estimate Nlaccer, the threshold settings of the LacCer image were adjusted to remove the signal from the cytoplasm and binarized by assigning 1 to each LacCer-positive pixel and 0 to LacCer-negative pixels. Nlaccer, the number of nonzero pixels, was calculated using the analysis function of ImageJ. Next, to calculate Nover, red (LysoTracker), green (LacCer), and blue (null) images of the same field of view were merged, and the resulting RGB image was subjected to the RGB colocalization algorithm, yielding a single binary image in which each pixel positive for both green (LacCer) and red (LysoTracker) signals has a value of 1 and each pixel negative for either green or red signals has a value of 0. Nover is the number of pixels with nonzero values.
DiI-LDL labeling.
48 h before imaging, cells were plated onto coverslips and preincubated in DME supplemented with 10% LPDS to up-regulate LDL receptor surface expression. Alexa Fluor 647–conjugated dextran (Invitrogen) was preaccumulated in lysosomes during a 12-h incubation before the start of the experiment. The next day, cells were incubated with 20 μg/ml DiI-LDL (Invitrogen) on ice for 60 min to allow LDL binding. Cells were subsequently washed in PBS and chased in fresh DME for either 30 or 120 min at 37°C. At the end of the time course, cells were fixed in 3.7% paraformaldehyde (Sigma-Aldrich) solution diluted in PBS for 10 min and mounted for image analysis. Confocal imaging was performed on an Olympus IX81 equipped with an UltraVIEW spinning disc confocal head, and an argon–ion, argon–krypton, and helium–cadmium laser combiner. Images were acquired with a 100× plan-apochromat objective (NA 1.4) and the appropriate filter combination. The extent of colocalization between Alexa Fluor 647–dextran and DiI-LDL–positive compartments was determined using image analysis software (Metamorph; MDS Analytical Technologies). The TIFF images were imported into Photoshop (Adobe) to adjust contrast and image size.
Preparation of [14C]CO–LDL complexes.
In vitro preparation of cholesteryl ester–radiolabeled lipoproteins was performed according to a modified version of the methods previously described by Terpstra et al. (53) and Brown et al. (54). 600 μg LDL (Biomedical Technologies, Inc.) and 120 mg LPDS (Biomedical Technologies, Inc.) were mixed in 0.15 M NaCl containing 0.3 mM thiomersol, 1 μg/ml aprotinin, and 0.65 mM glutathione at a final volume of ∼1.6 ml. 5 μCi [14C]CO (GE Healthcare) in denatured toluene solution was transferred to a 2.-ml conical tube and evaporated to dryness under a steady stream of nitrogen for ∼15 min, then resuspended in 50 μl of absolute ethanol. The resuspended [14C]CO was incubated for ∼30 min in a 37°C water bath with frequent vortexing to ensure complete resuspension. The LDL–LPDS was then added to the resuspended [14C]CO, and this mixture was incubated overnight at 37°C under nitrogen. The next day, the sample was transferred to a 0.5–3-ml Slide-A-Lyzer (10,000 kD) dialysis cassette (Thermo Fisher Scientific) and dialyzed for 8–12 h at 4°C against 4 liter of buffer containing 0.15 M NaCl and 0.3 mM EDTA (pH 7). After dialysis, the solution was centrifuged in a benchtop microcentrifuge at 12,000 rpm for 5 min. The supernatant solution was collected, supplemented with 0.5% human serum albumin, and stored for up to 3 wk at 4°C.
Degradation of [14C]CO–LDL.
[14C]CO–LDL degradation was performed essentially as previously described by Groener et al. (44), with slight modification. HeLa SS6 cells or human skin fibroblasts were cultured in DME supplemented with 10% FBS and 100 μg/ml penicillin/streptomycin. Cells were plated in 12-well plates at ∼50% confluence. 48h before the start of an experiment, cells were preincubated with DME supplemented with 10% LPDS to up-regulate cell-surface expression of the LDL receptor. Cells were then incubated with 0.4 ml DME containing 10% LPDS and [14C]CO–LDL (50 μg/ml LDL protein) for 4h at 37°C. The labeling medium was removed, and cells were washed with PBS and incubated for 30 min at 18°C in DME supplemented with 10% FBS. The experiment was initiated by the addition of fresh DME supplemented with 10% LPDS at 37°C to scavenge the released free fatty acids (44). Where indicated, 0.5 μM bafilomycin A1 (Sigma-Aldrich), was added to culture medium 30 min before labeling and maintained throughout the time course. At various times (0.5–4 h), the medium was collected and replaced. At the end of the time course, cells were harvested in buffer containing 50 mM Tris-HCl (pH 7.4) and 1% Tx-100. Radioactivity was measured in both the medium and the cell pellet by liquid scintillation counting. The percentage of [14C]CO–LDL degraded was calculated as the amount of radioactivity present in the medium divided by the total radioactivity present in the medium and cell pellet.
Degradation of 125I-apoB-LDL.
HeLa SS6 cells or human skin fibroblasts were cultured in DME supplemented with 10% FBS and 100 μg/ml penicillin/streptomycin. Cells were plated in six-well plates at ∼50% confluence. 48 h before the start of an experiment, cells were preincubated with DME supplemented with 10% LPDS to up-regulate cell-surface expression of the LDL receptor. Cells were incubated in DME supplemented with 125I-apoB-LDL (25 μg/ml LDL protein at 50 μCi/ml; Biomedical Technologies, Inc.) on ice for 2h (0.6 ml). Cells were extensively washed in DME containing BSA for three 10-mim periods. For control samples, 0.5 μM bafilomycin A1 was added 30 min before LDL labeling and maintained throughout the time course. At the start of the experiment, cells were incubated with 0.6 ml of prewarmed DME per well at 37°C. At various times (10–180 min), the medium was collected from cells and replaced. After the time course was completed, the cells were solubilized with buffer containing 50 mM Tris-HCl (pH 7.4) and 1% Tx-100 for 15 min. After solubilization, TCA was added to the medium collected over the time course and to the solubilized cells (final concentration = 10% [vol/vol]). The samples were incubated on ice for 20 min and centrifuged at maximum speed in a microcentrifuge at 4°C for 15 min. Radioactivity in the corresponding supernatants and pellets was counted using a γ counter (PerkinElmer). The rate of 125I-apoB-LDL degradation was determined by calculating the cumulative release of TCA-soluble counts into the medium over the experimental time course.
Acknowledgments
This work was supported by an ML4 Foundation grant (to K. Kiselyov), by a seed money award from the Pittsburgh Life Science Greenhouse (to K. Kiselyov), by National Institutes of Health grant DK54407 (to O.A. Weisz), by an American Heart Association Predoctoral Fellowship (to C.J. Guerriero), and by the Clinical and Translational Science Institute Multidisciplinary Predoctoral Fellowship program, awarded through the Clinical and Translational Science Institute and the Institute for Clinical Research Education at the University of Pittsburgh (grant 5TL1RR024155-02 to M.T. Miedel).
The authors have no conflicting financial interests.
Abbreviations used: apoB, apolipoprotein B; CO, cholesteryl oleate; HA, hemagglutinin; LacCer, BODIPY-C5-lactosylceramide; LDL, low density lipoprotein; LPDS, lipoprotein-deficient serum; MLII, mucolipidosis type II; MLIV, mucolipidosis type IV; NPC, Niemann-Pick type C; TCA, trichloroacetic acid; TMR, tetramethylrhodamine; TRP-ML1, transient receptor potential–mucolipin-1.
References
- 1.Bargal, R., N. Avidan, E. Ben-Asher, Z. Olender, M. Zeigler, A. Frumkin, A. Raas-Rothschild, G. Glusman, D. Lancet, and G. Bach. 2000. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 26:118–123. [DOI] [PubMed] [Google Scholar]
- 2.Bassi, M.T., M. Manzoni, E. Monti, M.T. Pizzo, A. Ballabio, and G. Borsani. 2000. Cloning of the gene encoding a novel integral membrane protein, mucolipidin–and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 67:1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slaugenhaupt, S.A., J.S. Acierno Jr., L.A. Helbling, C. Bove, E. Goldin, G. Bach, R. Schiffmann, and J.F. Gusella. 1999. Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am. J. Hum. Genet. 65:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun, M., E. Goldin, S. Stahl, J.L. Falardeau, J.C. Kennedy, J.S. Acierno Jr., C. Bove, C.R. Kaneski, J. Nagle, M.C. Bromley, et al. 2000. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9:2471–2478. [DOI] [PubMed] [Google Scholar]
- 5.Bach, G. 2001. Mucolipidosis type IV. Mol. Genet. Metab. 73:197–203. [DOI] [PubMed] [Google Scholar]
- 6.Schiffmann, R., N.K. Dwyer, I.A. Lubensky, M. Tsokos, V.E. Sutliff, J.S. Latimer, K.P. Frei, R.O. Brady, N.W. Barton, E.J. Blanchette-Mackie, and E. Goldin. 1998. Constitutive achlorhydria in mucolipidosis type IV. Proc. Natl. Acad. Sci. USA. 95:1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slaugenhaupt, S.A. 2002. The molecular basis of mucolipidosis type IV. Curr. Mol. Med. 2:445–450. [DOI] [PubMed] [Google Scholar]
- 8.Zeevi, D.A., A. Frumkin, and G. Bach. 2007. TRPML and lysosomal function. Biochim. Biophys. Acta. 1772:851–858. [DOI] [PubMed] [Google Scholar]
- 9.Bargal, R., H.H. Goebel, E. Latta, and G. Bach. 2002. Mucolipidosis IV: novel mutation and diverse ultrastructural spectrum in the skin. Neuropediatrics. 33:199–202. [DOI] [PubMed] [Google Scholar]
- 10.Lubensky, I.A., R. Schiffmann, E. Goldin, and M. Tsokos. 1999. Lysosomal inclusions in gastric parietal cells in mucolipidosis type IV: a novel cause of achlorhydria and hypergastrinemia. Am. J. Surg. Pathol. 23:1527–1531. [DOI] [PubMed] [Google Scholar]
- 11.Bargal, R., and G. Bach. 1989. Phosphatidylcholine storage in mucolipidosis IV. Clin. Chim. Acta. 181:167–174. [DOI] [PubMed] [Google Scholar]
- 12.Bargal, R., and G. Bach. 1997. Mucolipidosis type IV: abnormal transport of lipids to lysosomes. J. Inherit. Metab. Dis. 20:625–632. [DOI] [PubMed] [Google Scholar]
- 13.Soyombo, A.A., S. Tjon-Kon-Sang, Y. Rbaibi, E. Bashllari, J. Bisceglia, S. Muallem, and K. Kiselyov. 2006. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281:7294–7301. [DOI] [PubMed] [Google Scholar]
- 14.Raghavan, S., E. Leshinsky, and E.H. Kolodny. 1999. G(M2)-ganglioside metabolism in situ in mucolipidosis IV fibroblasts. Neurochem. Res. 24:475–479. [DOI] [PubMed] [Google Scholar]
- 15.Bach, G., M. Zeigler, T. Schaap, and G. Kohn. 1979. Mucolipidosis type IV: ganglioside sialidase deficiency. Biochem. Biophys. Res. Commun. 90:1341–1347. [DOI] [PubMed] [Google Scholar]
- 16.Lieser, M., E. Harms, H. Kern, G. Bach, and M. Cantz. 1989. Ganglioside GM3 sialidase activity in fibroblasts of normal individuals and of patients with sialidosis and mucolipidosis IV. Subcellular distribution and some properties. Biochem. J. 260:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bargal, R., and G. Bach. 1988. Phospholipids accumulation in mucolipidosis IV cultured fibroblasts. J. Inherit. Metab. Dis. 11:144–150. [DOI] [PubMed] [Google Scholar]
- 18.Jansen, S.M., J.E. Groener, W. Bax, and B.J. Poorthuis. 2001. Delayed lysosomal metabolism of lipids in mucolipidosis type IV fibroblasts after LDL-receptor-mediated endocytosis. J. Inherit. Metab. Dis. 24:577–586. [DOI] [PubMed] [Google Scholar]
- 19.Kiselyov, K., J. Chen, Y. Rbaibi, D. Oberdick, S. Tjon-Kon-Sang, N. Shcheynikov, S. Muallem, and A. Soyombo. 2005. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J. Biol. Chem. 280:43218–43223. [DOI] [PubMed] [Google Scholar]
- 20.Miedel, M.T., K.M. Weixel, J.R. Bruns, L.M. Traub, and O.A. Weisz. 2006. Posttranslational cleavage and adaptor protein complex-dependent trafficking of mucolipin-1. J. Biol. Chem. 281:12751–12759. [DOI] [PubMed] [Google Scholar]
- 21.Pryor, P.R., F. Reimann, F.M. Gribble, and J.P. Luzio. 2006. Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic. 7:1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vergarajauregui, S., and R. Puertollano. 2006. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 7:337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, C.S., G. Bach, and R.E. Pagano. 1998. Abnormal transport along the lysosomal pathway in mucolipidosis, type IV disease. Proc. Natl. Acad. Sci. USA. 95:6373–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaPlante, J.M., J. Falardeau, M. Sun, M. Kanazirska, E.M. Brown, S.A. Slaugenhaupt, and P.M. Vassilev. 2002. Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 532:183–187. [DOI] [PubMed] [Google Scholar]
- 25.Raychowdhury, M.K., S. Gonzalez-Perrett, N. Montalbetti, G.A. Timpanaro, B. Chasan, W.H. Goldmann, S. Stahl, A. Cooney, E. Goldin, and H.F. Cantiello. 2004. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum. Mol. Genet. 13:617–627. [DOI] [PubMed] [Google Scholar]
- 26.Cantiello, H.F., N. Montalbetti, W.H. Goldmann, M.K. Raychowdhury, S. Gonzalez-Perrett, G.A. Timpanaro, and B. Chasan. 2005. Cation channel activity of mucolipin-1: the effect of calcium. Pflugers Arch. 451:304–312. [DOI] [PubMed] [Google Scholar]
- 27.Xu, H., M. Delling, L. Li, X. Dong, and D.E. Clapham. 2007. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc. Natl. Acad. Sci. USA. 104:18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaPlante, J.M., C.P. Ye, S.J. Quinn, E. Goldin, E.M. Brown, S.A. Slaugenhaupt, and P.M. Vassilev. 2004. Functional links between mucolipin-1 and Ca2+-dependent membrane trafficking in mucolipidosis IV. Biochem. Biophys. Res. Commun. 322:1384–1391. [DOI] [PubMed] [Google Scholar]
- 29.Luzio, J.P., V. Poupon, M.R. Lindsay, B.M. Mullock, R.C. Piper, and P.R. Pryor. 2003. Membrane dynamics and the biogenesis of lysosomes. Mol. Membr. Biol. 20:141–154. [DOI] [PubMed] [Google Scholar]
- 30.Luzio, J.P., P.R. Pryor, and N.A. Bright. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. [DOI] [PubMed] [Google Scholar]
- 31.Piper, R.C., and J.P. Luzio. 2004. CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 14:471–473. [DOI] [PubMed] [Google Scholar]
- 32.Pryor, P.R., B.M. Mullock, N.A. Bright, S.R. Gray, and J.P. Luzio. 2000. The role of intraorganellar Ca2+ in late endosome–lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J. Cell Biol. 149:1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treusch, S., S. Knuth, S.A. Slaugenhaupt, E. Goldin, B.D. Grant, and H. Fares. 2004. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA. 101:4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaheen, L., H. Dang, and H. Fares. 2006. Basis of lethality in C. elegans lacking CUP-5, the Mucolipidosis Type IV orthologue. Dev. Biol. 293:382–391. [DOI] [PubMed] [Google Scholar]
- 35.Fares, H., and I. Greenwald. 2001. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 159:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fares, H., and I. Greenwald. 2001. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 28:64–68. [DOI] [PubMed] [Google Scholar]
- 37.Jentsch, T.J., M. Poet, J.C. Fuhrmann, and A.A. Zdebik. 2005. Physiological functions of CLC Cl− channels gleaned from human genetic disease and mouse models. Annu. Rev. Physiol. 67:779–807. [DOI] [PubMed] [Google Scholar]
- 38.Pagano, R.E. 2003. Endocytic trafficking of glycosphingolipids in sphingolipid storage diseases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358:885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bach, G., C.S. Chen, and R.E. Pagano. 1999. Elevated lysosomal pH in Mucolipidosis type IV cells. Clin. Chim. Acta. 280:173–179. [DOI] [PubMed] [Google Scholar]
- 40.Kopitz, J., C. Gerhard, P. Hofler, and M. Cantz. 1994. [14C]Methylamine accumulation in cultured human skin fibroblasts–a biochemical test for lysosomal storage and lysosomal diseases. Clin. Chim. Acta. 227:121–133. [DOI] [PubMed] [Google Scholar]
- 41.Goldin, E., E.J. Blanchette-Mackie, N.K. Dwyer, P.G. Pentchev, and R.O. Brady. 1995. Cultured skin fibroblasts derived from patients with mucolipidosis 4 are auto-fluorescent. Pediatr. Res. 37:687–692. [DOI] [PubMed] [Google Scholar]
- 42.Belhoussine, R., H. Morjani, S. Sharonov, D. Ploton, and M. Manfait. 1999. Characterization of intracellular pH gradients in human multidrug-resistant tumor cells by means of scanning microspectrofluorometry and dual-emission-ratio probes. Int. J. Cancer. 81:81–89. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, E.G., L. Schaheen, H. Dang, and H. Fares. 2007. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groener, J.E., W. Bax, and B.J. Poorthuis. 1996. Metabolic fate of oleic acid derived from lysosomal degradation of cholesteryl oleate in human fibroblasts. J. Lipid Res. 37:2271–2279. [PubMed] [Google Scholar]
- 45.Maxfield, F.R., and T.E. McGraw. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. [DOI] [PubMed] [Google Scholar]
- 46.Mukherjee, S., R.N. Ghosh, and F.R. Maxfield. 1997. Endocytosis. Physiol. Rev. 77:759–803. [DOI] [PubMed] [Google Scholar]
- 47.Weisz, O.A. 2003. Acidification and protein traffic. Int. Rev. Cytol. 226:259–319. [DOI] [PubMed] [Google Scholar]
- 48.Ikonen, E. 2006. Mechanisms for cellular cholesterol transport: defects and human disease. Physiol. Rev. 86:1237–1261. [DOI] [PubMed] [Google Scholar]
- 49.Maxfield, F.R., and I. Tabas. 2005. Role of cholesterol and lipid organization in disease. Nature. 438:612–621. [DOI] [PubMed] [Google Scholar]
- 50.Di, A., M.E. Brown, L.V. Deriy, C. Li, F.L. Szeto, Y. Chen, P. Huang, J. Tong, A.P. Naren, V. Bindokas, et al. 2006. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat. Cell Biol. 8:933–944. [DOI] [PubMed] [Google Scholar]
- 51.Holopainen, J.M., J. Saarikoski, P.K. Kinnunen, and I. Jarvela. 2001. Elevated lysosomal pH in neuronal ceroid lipofuscinoses (NCLs). Eur. J. Biochem. 268:5851–5856. [DOI] [PubMed] [Google Scholar]
- 52.Jennings, J.J., Jr., J.H. Zhu, Y. Rbaibi, X. Luo, C.T. Chu, and K. Kiselyov. 2006. Mitochondrial aberrations in mucolipidosis Type IV. J. Biol. Chem. 281:39041–39050. [DOI] [PubMed] [Google Scholar]
- 53.Terpstra, A.H., R.J. Nicolosi, and P.N. Herbert. 1989. In vitro incorporation of radiolabeled cholesteryl esters into high and low density lipoproteins. J. Lipid Res. 30:1663–1671. [PubMed] [Google Scholar]
- 54.Brown, M.S., S.E. Dana, and J.L. Goldstein. 1975. Receptor-dependent hydrolysis of cholesteryl esters contained in plasma low density lipoprotein. Proc. Natl. Acad. Sci. USA. 72:2925–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]