Abstract
The filamentous ascomycete Aspergillus niger is well known for its ability to produce a large variety of enzymes for the degradation of plant polysaccharide material. A major carbon and energy source for this soil fungus is starch, which can be degraded by the concerted action of α-amylase, glucoamylase and α-glucosidase enzymes, members of the glycoside hydrolase (GH) families 13, 15 and 31, respectively. In this study we have combined analysis of the genome sequence of A. niger CBS 513.88 with microarray experiments to identify novel enzymes from these families and to predict their physiological functions. We have identified 17 previously unknown family GH13, 15 and 31 enzymes in the A. niger genome, all of which have orthologues in other aspergilli. Only two of the newly identified enzymes, a putative α-glucosidase (AgdB) and an α-amylase (AmyC), were predicted to play a role in starch degradation. The expression of the majority of the genes identified was not induced by maltose as carbon source, and not dependent on the presence of AmyR, the transcriptional regulator for starch degrading enzymes. The possible physiological functions of the other predicted family GH13, GH15 and GH31 enzymes, including intracellular enzymes and cell wall associated proteins, in alternative α-glucan modifying processes are discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-008-0332-7) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus niger, Alpha-amylase, Alpha-glucosidase, Glucoamylase, AmyR, Starch, Maltose, Alpha-glucan, Cell wall, Starch-binding domain
Introduction
Aspergillus niger is a saprophytic fungus well known for its production and secretion of a variety of hydrolytic enzymes contributing to its ability to degrade plant polysaccharides such as cellulose, hemicellulose, pectin, starch and inulin (De Vries and Visser 2001; Tsukagoshi et al. 2001; Yuan et al. 2006). Starch is the most abundant storage carbohydrate in the plant kingdom and is present in tubers, seeds and roots of a variety of crop plants including cereals, potatoes and manioc (Peters 2006). Starch is composed of two different molecules: (1) amylose, an unbranched, single chain of α-(1,4)-linked glucose residues and (2) amylopectin, consisting of a α-(1,4)-linked glucose chain with α-(1,6)-branches on every 12–25 glucose residues along the α-(1,4)-linked backbone (Robyt 1998). The degradation of starch is performed by a variety of enzymes, which are divided over three Glycoside Hydrolase (GH) families based on their sequence similarity (http://www.cazy.org) (Coutinho and Henrissat 1999). The first step in starch degradation is the endo-hydrolysis of the long polysaccharide chains into shorter maltooligosaccharides and α-limit dextrins by α-amylases (EC 3.2.1.1). α-Amylases belong to family GH13, a large family containing various hydrolysing and transglycosylating enzymes, mostly acting on α-(1,4) or α-(1,6)-glycosidic bonds. Members of family GH13 have a (β/α)8 barrel structure and can be recognized by four highly conserved amino acid regions containing three catalytic residues (MacGregor et al. 2001; Nakajima et al. 1986). After endo-hydrolysis, subsequent steps in starch degradation involve exo-acting enzymes releasing glucose. This reaction is performed by glucoamylase type enzymes of family GH15 (EC 3.2.1.3), a relatively confined family with regard to enzyme specificity, as all its studied members hydrolyse either α-(1,4) or α-(1,6)-bonds to release β-glucose from the non-reducing end of maltooligosaccharides. Most GH15 enzymes described thus far possess a starch-binding domain (SBD) (Sauer et al. 2000), a discrete C-terminal region of the protein that binds to starch and facilitates hydrolysis (Southall et al. 1999). Additionally, α-(1,4)-glucosidases of family GH31 may release α-glucose from the non-reducing end of starch (EC 3.2.1.20). This family also harbours other enzyme specificities such as α-xylosidase activity.
Several A. niger enzymes involved in starch degradation, and their corresponding genes, have been characterized and isolated. A. niger glucoamylase GlaA (family GH15) is an important enzyme for the modification of starch in the food industry (Boel et al. 1984; van Dijck et al. 2003). Additionally, one GH31 α-glucosidase (AglA, renamed AgdA) (Nakamura et al. 1997) has been characterized previously, as well as three family GH13 α-amylases: acid amylase AamA, and the almost identical AmyA and AmyB (Boel et al. 1990; Korman et al. 1990). The transcriptional regulation of the genes encoding starch-degrading enzymes has been studied in several aspergilli (Nakamura et al. 2006). In general, their expression is high on starch and induced by the presence of (iso)maltose (Tsukagoshi et al. 2001; Kato et al. 2002a). The presence of the inducer activates the Zn(II)2Cys6 transcription factor AmyR which binds to CGGN8(C/A)GG sequences in the promoter regions of AmyR target genes thereby activating their transcription (Petersen et al. 1999; Gomi et al. 2000; Tani et al. 2001; Ito et al. 2004).
Recent studies have indicated that some GH13 enzymes in fungi may be involved in the synthesis or modification of α-glucan in the fungal cell wall, rather than in starch degradation. The cell wall of aspergilli contains four major classes of polysaccharides: chitin, α-glucan, β-(1,3)-glucan and galactomannan (Fontaine et al. 2000; Beauvais and Latgé 2001). The α-glucan fraction identified in A. niger consists of two types of molecules: a linear polymer with alternating α-(1,3)/(1,4)-glycosidic bonds called nigeran (Barker and Carrington 1953) and pseudonigeran, a linear α-(1,3)-glucan molecule with some (3–10%) α-(1,4)-linkages (Johnston 1965; Horisberger et al. 1972). Synthesis of α-glucan is thought to be carried out by α-glucan synthase enzymes encoded by ags genes. The first putative α-d-glucan synthase encoding gene (ags1) was identified in Schizosaccharomyces pombe (Hochstenbach et al. 1998). The ags1 gene encodes a large, three-domain protein. In addition to the multi-pass transmembrane domain in the C-terminal part of the protein, two predicted catalytic domains are present. The middle domain shows strong similarity to glycogen and starch synthases in Glycosyltransferase family (GT) 5 and is predicted to be involved in the synthesis of α-glucan. The N-terminal part of the protein is similar to α-amylases and belongs to family GH13. This part of the protein is predicted to be localized extracellularly and might be involved in connecting two α-(1,3)-glucan chains (Grün et al. 2005). Apart from the α-glucan synthases, two types of family GH13 enzymes were recently identified in fungi to play a role in fungal cell wall biosynthesis. Marion et al. (2006) provided evidence for the involvement of a putative α-amylase (Amy1p) in the formation of α-(1,3)-glucan in the cell wall of Histoplasma capsulatum. An AMY1 knockout was unable to produce α-(1,3) glucan and showed reduced virulence. In dimorphic fungi like H. capsulatum, cell wall α-glucan is a known virulence factor (Rappleye et al. 2004; Rappleye and Goldman 2006). The second α-amylase-like enzyme Aah3p was first studied in S. pombe (Morita et al. 2006). Disruption of aah3 encoding a GPI-anchored protein resulted in hypersensitivity towards cell wall-degrading enzymes and an aberrant cell shape, indicating that normal cell wall biosynthesis was affected. Disruption of a homologous gene (agtA) in A. niger also affected cell wall stability (van der Kaaij et al. 2007a).
We previously surveyed the A. niger genome sequence to identify all GH13, GH15 and GH31 family members present in this important industrial source for amylolytic enzymes (Pel et al. 2007). This resulted in identification of a surprisingly large number of previously unknown enzymes. In this study, we have analysed their phylogeny, the presence of specific protein features and synteny with other Aspergillus species, which allowed the division of the members of each GH family into several groups. Additionally, we studied the transcriptional regulation of the genes encoding these proteins in a wild type A. niger strain, and in a derived amyR deletion strain, during their growth on xylose and maltose. Only few of the identified proteins were induced by maltose. Expression of many of the identified groups of enzymes, including the homologues of both S. pombe Aah3p and H. capsulatum Amy1p, was not induced by maltose and was not dependent on the presence of AmyR. The possible involvement of these enzymes in cell wall α-glucan synthesis and modification is discussed.
Material and methods
Database mining of A. niger genome and analysis of predicted proteins
The full genome sequence of A. niger strain CBS 513.88 has been deposited at the EMBL database with accession numbers AM270980–AM270998 (Pel et al. 2007) and was used for database mining. The nucleotide accession numbers of A. niger genes, as listed in Tables 1 and 2, refer to this database. Hidden Markov Model (HMM) profiles were built with the HMMER package (Durbin and Eddy 1998) (http://hmmer.wustl.edu/) based on the amino acid sequences of known members of GH13, GH15 and GH31. Proteins belonging to these families, originating from the different kingdoms of life, were retrieved from the CAZy website at http://www.cazy.org (Coutinho and Henrissat 1999), and the protein sequences were extracted from the GenBank/GenPept database at http://www.ncbi.nlm.nih.gov/entrez/ and Swiss-Prot database at http://www.expasy.org/sprot/. The A. niger genome was searched with the HMM profiles using the WISE2 package (Birney et al. 2004) (http://www.ebi.ac.uk/Wise2/).
Table 1.
All members of family GH13, GH15 and GH31 identified in the genome sequence of A. niger CBS 513.88 using HMM profiles
| Accession no. | Gene | Family | Enzyme activity | Featuresa | AmyR binding motifb | Proposed biological function | Refc |
|---|---|---|---|---|---|---|---|
| An11g03340 | aamA | GH13 | Acid α-amylase | SS | Starch degradation | 1 | |
| An12g06930 | amyA | GH13 | α-Amylase | SS | +970; +252 | Starch degradation | 2 |
| An05g02100 | amyB | GH13 | α-Amylase | SS | +252 | Starch degradation | 2 |
| An04g06930 | amyC | GH13 | α-Amylase | SS | +787; +664; −531 | Starch degradation | 3 |
| An09g03100 | agtA | GH13 | α-Glucanotransferase | SS, GPI | Cell wall α-glucan synthesis | 4 | |
| An12g02460 | agtB | GH13 | α-Glucanotransferase | SS, GPI | +810 | Cell wall α-glucan synthesis | 4 |
| An15g07800 | agtC | GH13 | Putative α-glucanotransferase | SS, GPI | Cell wall α-glucan synthesis | 4 | |
| An02g13240 | agdC | GH13 | Putative α-glucosidase | +368 | Unknown | ||
| An13g03710 | agdD | GH13 | Putative α-glucosidase | Unknown | |||
| An01g13610 | amyD | GH13 | Putative α-amylase | +504; −32 | Cell wall α-glucan synthesis | ||
| An09g03110 | amyE | GH13 | Putative α-amylase | −76 | Cell wall α-glucan synthesis | ||
| An01g06120 | gdbA | GH13 | Glycogen debranching enzyme | −487; +393 | Glycogen metabolism | ||
| An14g04190 | gbeA | GH13 | Glycogen branching enzyme | Glycogen metabolism | |||
| An04g09890 | agsA | GH13 | Putative α-glucan synthase | SS | Cell wall α-glucan synthesis | 5 | |
| An15g07810 | agsB | GH13 | Putative α-glucan synthase | SS | +287 | Cell wall α-glucan synthesis | 5 |
| An12g02450 | agsC | GH13 | Putative α-glucan synthase | SS | −973; +622, −185 | Cell wall α-glucan synthesis | 5 |
| An02g03260 | agsD | GH13 | Putative α-glucan synthase | SS | Cell wall α-glucan synthesis | 5 | |
| An09g03070 | agsE | GH13 | Putative α-glucan synthase | SS | Cell wall α-glucan synthesis | 5 | |
| An03g06550 | glaA | GH15 | Glucoamylase | SS, SBD | −792; −669; +423; −301 | Starch degradation | 6 |
| An12g03070 | glaB | GH15 | Putative glucoamylase | −878 | Unknown | ||
| An04g06920 | agdA | GH31 | α-Glucosidase | SS | +574; +191, | Starch degradation | 7 |
| An01g10930 | agdB | GH31 | Putative α-glucosidase | SS | +904; −334 | Starch degradation | |
| An09g05880 | agdE | GH31 | Putative α-glucosidase II | SS | Protein glycosylation | ||
| An18g05620 | agdF | GH31 | Unknown | Unknown | |||
| An07g00350 | agdG | GH31 | Unknown | SS | +402 | Unknown | |
| An09g03300 | axlA | GH31 | Putative α-xylosidase | SS | +126 | Xyloglucan degradation | |
| An01g04880 | axlB | GH31 | Putative α-xylosidase | +430; +138, | Xyloglucan degradation | ||
The newly identified proteins are indicated in bold
aSS predicted N-terminal Signal Sequence; GPI predicted Glycosylphosphatidylinositol anchor signal; SBD predicted starch-binding domain
bThe presence of consensus AmyR binding motif (CGGN8(A/C)GG) was analysed in the promoter region up to 1 kb upstream of the start codon
cReferences: 1 (Boel et al. 1990); 2 (Korman et al. 1990); 3 (R. M. Van der Kaaij and X. L. Yuan, unpublished). ; 4 (van der Kaaij et al. 2007); 5 (Damveld et al. 2005); 6 (Boel et al. 1984); 7 (Nakamura et al. 1997)
Table 2.
Functionally described family GH13 and GH31 members from other organisms, used for the multiple sequence alignments in Fig. 1
| Accession no. | Name | Family | Enzyme activity | Featuresa | Biological function | Organism | Refb |
|---|---|---|---|---|---|---|---|
| BAA78714 | AndGbe1 | GH13 | Glycogen branching enzyme | Glycogen metabolism | A. nidulans | 1 | |
| BAA34996 | ScGdb1 | GH13 | Glycogen debranching enzyme | Glycogen metabolism | S. cerevisiae | 2 | |
| P19571 | BsAmyA | GH13 | α-Amylase | SS | Starch degradation | Bacillus sp. | 3 |
| CAA54266 | BsAglA | GH13 | α-Glucosidase | Starch degradation | Bacillus sp. | 4 | |
| CAA21237 | SpAah1 | GH13 | Unknown | SS, GPI | α-Glucan biosynthesis | S. pombe | 5 |
| CAA91249 | SpAah2 | GH13 | Unknown | SS, GPI | α-Glucan biosynthesis | S. pombe | 5 |
| CAB40006 | SpAah3 | GH13 | Unknown | SS, GPI | α-Glucan biosynthesis | S. pombe | 5 |
| CAA16864 | SpAah4 | GH13 | Unknown | SS, GPI | α-Glucan biosynthesis | S. pombe | 5 |
| ABK62854 | HcAmy1 | GH13 | Unknown | α-Glucan biosynthesis | H. capsulatum | 6 | |
| ABF50883 | AN7345.2 | GH31 | α/β-Glucosidase | SS | Starch/cellulose degradation | A. nidulans | 7 |
| ABF50846 | AN7505.2 | GH31 | α-Xylosidase | Xylan degradation | A. nidulans | 7 | |
| BAB39856 | AndAgdB | GH31 | α-Glucosidase | SS | Starch degradation | A. nidulans | 8 |
| AAU87580 | TrAguII | GH31 | α-Glucosidase II | SS | Protein glycosylation | T. reesei | 9 |
| A45249 | CAMAL2 | GH31 | Maltase | Maltose degradation | C. albicans | 10 | |
For each A. niger protein identified, a functionally or biochemically characterized protein with the highest similarity was used in the phylogenetic analysis
aSS predicted N-terminal Signal Sequence; GPI predicted Glycosylphosphatidylinositol anchor signal
bReferences: 1 (Sasangka et al. 2002); 2 (Teste et al. 2000); 3 (Tsukamoto et al. 1988); 4 (Nakao et al. 1994); 5 (Morita et al. 2006); 6 (Marion et al. 2006); 7 (Bauer et al. 2006); 8 (Kato et al. 2002b); 9 (Geysens et al. 2005); 10 (Geber et al. 1992)
The presence of signal peptidase cleavage sites, glycosylphosphatidylinositol (GPI-) attachment sites and SBD in the obtained sequences were predicted by Web-based tools at URL: http://www.cbs.dtu.dk/services/SignalP/ (Bendtsen et al. 2004), URL: http://mendel.imp.ac.at/sat/gpi/gpi_server (Eisenhaber et al. 2004), and URL: http://www.ncbi.nlm.nih.gov/BLAST/ (Marchler-Bauer and Bryant 2004), respectively.
Multiple sequence alignments of GH13, 15 and 31 family members were performed using DNAMAN version 4.0 (Lynnon BioSoft, Canada). The alignments were based on the full length of the predicted proteins, except in case of predicted α-glucan synthases for which only the N-terminal part, encoding the family GH13 domain, was used for the alignment. The phylogenetic relationship was calculated by using Optimal Alignment (Thompson et al. 1994) with gap opening penalty and gap extension penalty of 10 and 0.05, respectively. A bootstrapped test of phylogeny was performed by the Neighbour-Joining method using 1,000 replicates. Wherever possible, one protein with described activity was included for each of the groups identified based on phylogenetic analysis.
Strains and transformations
The wild type A. niger strain used in this study is N402, a cpsA1 derivative of A. niger van Tieghem (CBS 120.49, ATCC 9029) (Bos et al. 1988). Strain AB4.1 is a pyrG negative derivative of N402 (van Hartingsveldt et al. 1987) and was used to construct the amyR disruption strain. A. niger strains were grown in Aspergillus minimal medium (MM) (Bennett and Lasure 1991), or Aspergillus complete medium (CM) consisting of MM with the addition of 0.5% (w/v) yeast extract and 0.1% (w/v) casamino acids. Growth medium was supplemented with 10 mM uridine (Serva, Germany) when required. Transformation of A. niger AB4.1 was performed as described earlier (Punt and van den Hondel 1992) using lysing enzymes (L1412, Sigma, USA) for protoplastation. The bacterial strain used for transformation and amplification of recombinant DNA was Escherichia coli XL1-Blue (Stratagene, USA). Transformation of XL1-Blue was performed according to the heat shock protocol (Inoue et al. 1990).
Disruption of the maltose utilization activator amyR in A. niger
Plasmid pJG01 containing the A. nigeramyR gene as a 4.3 kb NsiI fragment in pGEM11 was kindly provided by P. van Kuyk (Wageningen University, the Netherlands) and was used to disrupt the amyR gene. The construction of the amyR deletion cassette was performed as follows. The BamHI–EcoRI fragment and NsiI–SalI fragment flanking the amyR ORF at the 5′ and 3′ region, respectively, were isolated from pJG01. The isolated NsiI–SalI fragment was cloned into pUC19 to obtain plasmid pAmyRF3. Subsequently, a BamHI–SalI fragment carrying the A. oryzaepyrG gene, obtained from plasmid pAO4-13 (de Ruiter-Jacobs et al. 1989) was inserted into pAmyRF3 which resulted in plasmid pAmyRF3-pyrG. The BamHI–EcoRI fragment isolated from pJG10 was ligated into pAmyRF3-pyrG resulting in the amyR deletion plasmid (pΔamyR). Prior to transformation to AB4.1, pΔamyR was linearized with EcoRI. Uridine prototrophic transformants were selected by their ability to grow on MM without uridine. After two rounds of purification, transformants were tested for their ability to grow on starch. Approximately 10% of the pyrG+ transformants showed defective growth on MM agar plates containing starch as sole carbon source. Six independent putative amyR deletion strains (YvdM1.1-1.6) with identical phenotypes were obtained. Southern blot analysis confirmed proper deletion and a single integration of the amyR disruption cassette at the amyR locus. Strain YvdM1.1 was used for further analysis and we will refer to this strain as the ΔamyR strain in the remaining of this paper.
Culture conditions, RNA preparation, microarray experiments and data analysis
RNA extracted from the A. niger ΔamyR strain and its parental strain (N402) grown on different carbon sources were used for microarray experiments using custom-made ‘dsmM_ANIGERa_coll’ Affymetrix GeneChip® Microarrays kindly provided by DSM Food Specialties (Delft, The Netherlands). All experiments for each growth condition (culturing the mycelia, RNA extractions and microarray hybridizations) were performed twice as independent biological experiments.
Aspergillus niger spores (2 × 106 spores ml−1) were inoculated in 250 ml MM supplemented with 2% (w/v) xylose (Sigma) and 0.1% (w/v) casamino acids and grown for 18 h at 30°C on a rotary shaker at 300 rpm. The mycelium was harvested by suction over a nylon membrane and washed with MM without carbon source. Aliquots of 1.6 g wet weight of mycelium were transferred to 300 ml Erlenmeyer flasks containing 70 ml MM supplemented with 1% (w/v) carbon source [maltose (Sigma) or xylose] and incubated at 30°C for a further 2 or 8 h. The pH of all cultures grown for 2 h was equal to the pH at the time of transfer (pH 6.2). Cultures grown for 8 h were buffered at pH 4 by the addition of 100 mM of citric acid/sodium citrate to allow comparison between the N402 and the ΔamyR strain at this time point. The mycelium was harvested over Miracloth filter, frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from mycelia using TRIzol reagent (Invitrogen) and RNA quality was verified by analyzing aliquots with glyoxal/DMSO gel electrophoresis and Agilent Bioanalyzer “Lab on chip” system (Agilent Technologies, USA). Processing, labeling and hybridization of cRNA to A. niger Affymetrix GeneChips were performed according to the corresponding Affymetrix protocols for “Eukaryotic Target Preparation” and “Eukaryotic Target hybridization”. For probe array washing and staining, the protocol “Antibody Amplification for Eukaryotic Targets” was followed. Hybridized probe array slides were scanned with Agilent technologies G2500A Gene Array Scanner at a 3 μm resolution and a wavelength of 570 nm. Affymetrix Microarray Suite software MAS5.0 was used to calculate the signal and P-values and to set the algorithm’s absolute call flag, which indicates the reliability of the data points according to P (present), M (marginal) and A (absent). The data on each chip were globally scaled to an arbitrary target gene intensity of 500. The complete microarray data were deposited into ArrayExpress with an accession E-TABM-324 at http://www.ebi.ac.uk/miamexpress.
The pre-scaled data from each hybridization experiment was then normalized for statistical analysis using Genespring 7.0 software (Silicon Genetics, USA). The per chip normalization was performed to ensure that the overall characteristic of the expression distribution such as median should be the same for all the chips. For the genome-wide analysis, we focused on maltose-induced genes and therefore a pre-filtering of data was performed to select for genes whose detection calls are present in both maltose duplicate samples in the wild-type strain (N402). The selected dataset was used to perform a one-way ANOVA analysis under the test type of “parametric test, don’t assume variances equal”. Fold changes in expression between two different conditions were then computed for genes with P < 0.08 based on one-way ANOVA analysis.
Results
Identification of glycoside hydrolase family 13, 15 and 31 genes in the A. niger CBS 513.88 genome sequence
α-Amylases, glucoamylases and α-glucosidases, members of families GH13, 15 and 31, respectively, are the three main types of enzymes involved in breakdown of starch by aspergilli (Tsukagoshi et al. 2001). To identify all genes encoding enzymes that might play a role in starch utilization, or other α-glucan modifying processes in A. niger, the genome of A. niger CBS 513.88 was searched with HMM profiles based on known enzymes from families GH13, 15 and 31. This resulted in the retrieval of a total of 27 protein sequences including 17 previously unknown proteins (Pel et al. 2007), as listed in Table 1. The predicted proteins were annotated based on their similarity to known enzymes. Gene names were assigned based on this annotation and do not necessarily match the activity of the enzymes, as this is often unknown.
Two approaches were combined to predict putative functions in cellular processes for this surprisingly large number of newly identified proteins. First, phylogenetic trees were constructed using the GH13, GH15 and GH31 family members identified in the A. niger genome, as well as functionally characterized proteins from other organisms with similarity to the identified A. niger proteins (Fig. 1). Second, using DNA microarrays, the expression of all the A. niger genes encoding GH13, GH15 and GH31 enzymes was examined in both the A. niger wild type strain N402 and the ΔamyR strain derived, after growth on different carbon sources (Fig. 2; Table S1). Both the N402 and the ΔamyR strains were pregrown in xylose for 18 h, and mycelia were transferred to either xylose or maltose media and grown further for 2 or 8 h. Expression levels were determined based on geometric mean data of biological duplicate samples. We will discuss each enzyme family in detail and combine the findings in A. niger with the predicted proteins present in the genomes of A. fumigatus (Nierman et al. 2005), A. nidulans (Galagan et al. 2005) and A. oryzae (Machida et al. 2005).
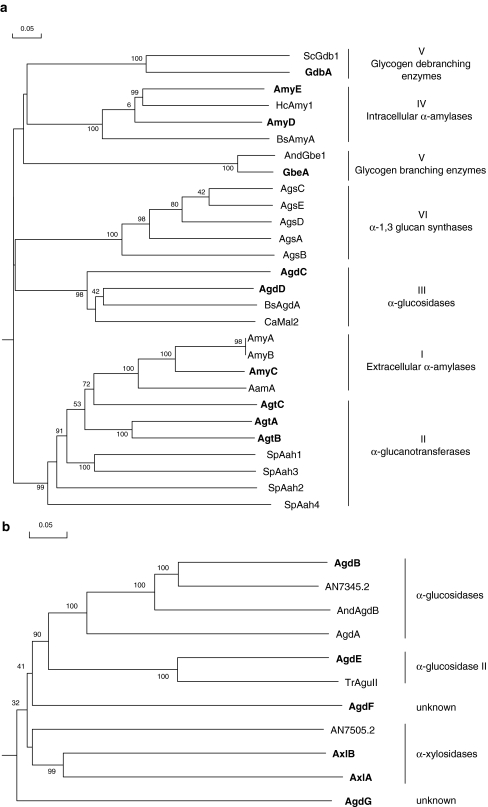
Fig. 1.
Bootstrapped phylogenetic tree of A. niger GH13 (a) and GH31 (b) enzymes and several closest homologues from other species. Newly identified proteins in the genome of A. niger are shown in bold. A description of each protein is listed in Tables 1 and 2. Bootstrap values are indicated on the node of each branch. The tree was created with DNAMAN 4.0 using gap and extension penalties of 10 and 0.5, respectively. The scale bar corresponds to a genetic distance of 0.05 substitution per position
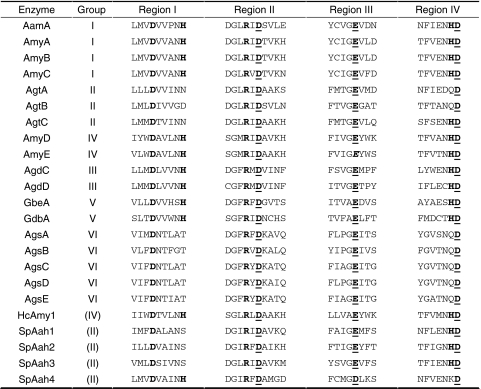
Fig. 2.
Expression profiles of A. niger family GH13, 15 and 31 enzymes. Accession numbers of the gene names are given in Tables 1 and 2. Strain and time points after transfer from the preculture are indicated on the right-hand side. The numeric values and Present/Absent calls from the expression data are provided as Supplementary Tables 1, 2
Identification and transcriptional regulation of GH13 family members
The HMMer search for family GH13 enzymes in the A. niger genome resulted in the identification of 18 protein sequences of which 10 had not been identified previously (Table 1). Table 3 displays the four conserved regions typical for family GH13 proteins as identified in these enzymes. A phylogenetic tree was produced combining the A. niger family GH13 enzymes with several functionally characterized GH13 family proteins from other organisms (Fig. 1). The combination of this phylogenetic analysis with a functional annotation of the proteins revealed six recognizable subgroups.
Table 3.
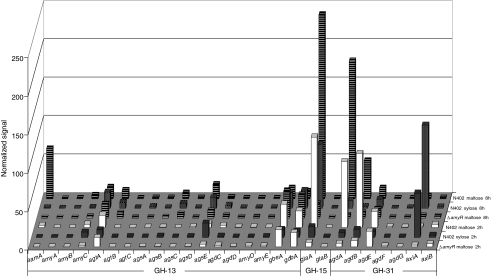
Alignment of the four conserved regions of all family GH13 enzymes identified in A. niger, as well as in four Aah proteins from S. pombe and Amy1p from H. capsulatum
The seven residues generally conserved in family GH13 are indicated in bold and the three catalytic residues are additionally underlined. The group to which the proteins are assigned, as described in this paper, is indicated
Group I consists of four extracellular α-amylases. Three of these are the previously characterized extracellular α-amylases acid-amylase (AamA) and α-amylases AmyA and AmyB (Korman et al. 1990). One new extracellular α-amylase was identified and named AmyC. This protein displays high similarity with the known A. niger α-amylases (74% identity to AmyA and AmyB, 65% identity to AamA). The amyC gene is located in a gene cluster also containing an α-glucosidase gene (agdA) and the amyR gene encoding the AmyR transcription factor. Expression of aamA in A. niger N402 was not detectable in xylose media, but was strongly induced in the presence of maltose (Fig. 2). The expression of aamA was reduced to a non-detectable level in the ΔamyR strain. Expression of amyA and amyB was not detected in A. niger N402 in any of the conditions tested (Fig. 2) (see Discussion). The expression of the newly identified amyC gene was relatively low compared to the aamA gene. At 2 h after transfer from the preculture, the expression level of amyC was independent of the carbon source, while after 8 h the expression level was reduced 3-fold on xylose compared to maltose. Additionally, the expression on maltose was reduced 2- to 3-fold in the ΔamyR strain (Table S1). The presence of three putative AmyR-binding elements in the promoter region of amyC further suggests that its expression is controlled by AmyR (Table 1).
The A. niger acid amylase (encoded by aamA) in the CBS 513.88 strain does not contain a predicted SBD, whereas its homologues in A. nidulans, A. fumigatus and other aspergilli contain a full-length SBD (Table S2). In addition, the presence of a functional SBD in the AamA protein of A. niger N402 was suggested because of the purification of AamA via its SBD from culture fluid of a N402 ΔglaA strain, and subsequent demonstration of the SBD with specific antibodies (M. F. Coeffet-Le Gal and D. Archer, personal communication, University of Nottingham, UK). We therefore PCR amplified the aamA gene and its 3′ flanking regions using N402 genomic DNA as a template, determined its DNA sequence and compared it to the aamA gene and its 3′ flanking region from CBS 513.88. The comparison revealed that the aamA gene of A. niger strain N402 does include an SBD and that the CBS513.88 strain harbours a deletion of 230 nucleotides right after the part encoding the (β/α)8 barrel, causing a frame shift and the introduction of a stop codon resulting in a truncated protein (Fig. S2). After the deletion, the DNA sequence continues by encoding part of a predicted SBD, but this part is not translated (Fig. S2). The SBD in AamA found in the A. niger N402 strain is also predicted to be present in A. niger strain ATCC 1015 (Baker 2006).
Group II contains three putative GPI-anchored enzymes, recently identified as α-glucanotransferases, and named AgtA, AgtB and AgtC, respectively (van der Kaaij et al. 2007a). This subgroup of proteins is characterized by the presence of two hydrophobic signal sequences. The N-terminal signal sequence is predicted to serve for translocation to the endoplasmic reticulum (ER), whereas the C-terminal sequence is predicted to be replaced by a preassembled glycosylphosphatidylinositol (GPI) anchor in the ER (Orlean 1997). The three enzymes cluster together with the α-amylases in the phylogenetic tree (Fig. 1a), but can be distinguished from the α-amylases by their catalytic domains which are clearly different from the consensus sequence for the α-amylase family. In all three proteins, one or two highly conserved histidines in conserved regions I and IV are replaced by other hydrophilic residues (Table 3). Family GH13 members without these histidine residues, which are part of the active site, are very rare (Uitdehaag et al. 1999). Mutation of these residues generally results in reduced activity or altered reaction specificity of the enzymes (Chang et al. 2003; Leemhuis et al. 2004). Interestingly, both the conserved His-residues in Regions I and IV are also missing in all (putative) α-glucan synthases (Table 3). Other aspergillus genomes harbour two or three Agt homologues (Fig. S1A; Table S2), all sharing the aberrant conserved regions and predicted GPI-anchoring. In the A. niger genome, both agtB and agtC are located next to genes encoding putative α-glucan synthases, and this arrangement of genes is conserved in other aspergilli. The agtA gene is constitutively expressed in both the wild type strain N402 and the ΔamyR strain under all growth conditions examined (Fig. 2). Expression of agtB was only detected 8 h after transfer, regardless of the carbon source and independent of AmyR, while expression of agtC was not detected.
Group III consists of two putative, intracellular α-glucosidases, named AgdC (An02g13240) and AgdD (An13g03710). The protein sequences contained all residues commonly conserved in the α-amylase superfamily (Table 3). The predicted intracellular proteins lack clear similarity to any previously characterized fungal protein, although similar enzymes are predicted in A. oryzae, A. nidulans and A. fumigatus (Fig. S1A). Their most related functionally characterized homologue is an α-glucosidase from Bacillus sp. SAM1606 (Nakao et al. 1994). Expression of agdC was low and not induced on maltose, and expression of agdD was not observed (Fig. 2).
Group IV contains two putative intracellular α-amylases, named AmyD (An01g13610) and AmyE (An09g03110), which share 55% identity. AmyD was recently characterized as an α-amylase with low-hydrolyzing activity on starch and related substrates (Van der Kaaij et al. 2007b). AmyD and AmyE are similar to a recently identified protein Amy1p from H. capsulatum, which was shown to be involved in cell wall α-glucan synthesis. The latter protein has not been characterized biochemically. Functionally characterized enzymes with similarity to this cluster therefore included only bacterial enzymes, of which maltohexaose-forming α-amylase of alkalophilic Bacillus sp. #707 (Tsukamoto et al. 1988) had the highest similarity. No significant expression of amyD or amyE was detected in our experiments (Fig. 2). Predicted enzymes highly similar to A. niger AmyD and AmyE were also present in other Aspergillus species (Fig. S1A; Table S2). Like the A. niger amyE gene, the orthologues in A. nidulans (AN3309.3) and in A. oryzae (AO003001497) are clustered in the genome with genes encoding α-glucanotransferases and α-glucan synthases (Table S2).
The two A. niger proteins in Group V could be reliably annotated as enzymes involved in glycogen metabolism: glycogen branching enzyme (GbeA) and glycogen debranching enzyme (GdbA). A homologue for each of these enzymes is present in the other Aspergillus genome sequences. Transcriptional analysis in A. niger showed that both genes were expressed both on xylose and maltose, and that their expression was unaffected in the ΔamyR strain (Fig. 2).
Group VI contains the five predicted α-glucan synthase genes (Damveld et al. 2005). The derived proteins are highly similar (66–83%) to each other, and all contain the two catalytic domains (GT5 and GH13) characteristic for these proteins. Both agsB and agsC are clustered in the genome with genes encoding the aforementioned α-glucanotransferases agtC and agtB, respectively. In both cases, the direction of transcription of the pair of genes is such that they can be transcribed from their intergenic region. Expression analysis in our microarray data collection, which includes data from various time points on several carbon sources (glucose, maltose, xylose, inulin, sucrose) indicate that agsC and agtB are co-expressed at a later growth stage, independent of the carbon source (data not shown). Expression of both agsB and its neighbouring gene agtC was not detected in any of the growth conditions. The expression levels of agsA and agsD were very low, or not detectable. AgsE was highly expressed in all experiments independent of AmyR (Fig. 2). As expected, several proteins with high similarity to the A. niger α-glucan synthases are predicted from the other Aspergillus genomes, although the number of homologues present is highest in A. niger (Fig. S1A; Table S2).
Identification and transcriptional regulation of GH15 family members
An HMMer search for GH15 family members in the A. niger genome returned two predicted proteins: the previously described glucoamylase GlaA (Boel et al. 1984) and an unknown predicted protein named GlaB (An12g03070). The GlaB protein lacks both an N-terminal signal sequence and an SBD, and displays a low similarity of 26% with GlaA. These two types of family GH15 enzymes are also recognized in other aspergilli studied (Fig. S1B). All predicted proteins similar to GlaB lack both a signal for secretion and an SBD, features typically present in the fungal GH15 enzymes described to date. A. niger glaA was expressed on xylose and strongly induced on maltose, as described previously (Fowler et al. 1990). The induction of glaA was AmyR dependent. Expression of glaB was not detected in any of the conditions tested (Fig. 2).
Identification and transcriptional regulation of GH31 family members
HMMer searches in the genome of A. niger revealed the presence of seven GH31 family members, of which only one was previously identified as an α-glucosidase (aglA) (Nakamura et al. 1997). We propose to name all (putative) α-glucosidases in A. niger Agd enzymes, similar to the nomenclature in A. nidulans, and to rename AglA as AgdA, to prevent confusion with α-galactosidases (den Herder et al. 1992). The phylogenetic tree (Fig. 1b) shows the presence of (at least) four subgroups within family GH31. In the group of the α-glucosidases, AgdA clusters with AgdB, a predicted extracellular protein. AgdB has some similarity to two A. nidulans enzymes: AN7345.2 (62% identity) with both α- and β-glucosidase activity (Bauer et al. 2006) and α-glucosidase B (AndAgdB/ AN8953.3, 53% identity), with strong transglycosylation activity (Kato et al. 2002b). However, none of the true orthologues of AgdB has been characterized (Fig. S1C). Transcript analysis revealed that agdB was regulated similar to agdA, as the strong induction of both genes in the presence of maltose was dependent on the presence of amyR. However, where most AmyR-regulated genes (aamA, glaA and agdA) reached their highest level of induction after 8 h growth on maltose, the expression level of agdB was decreased after 8 h compared to 2 h (Fig. 2).
From the remaining five GH31 family members, AgdE (An09g05880) shows similarity to Trichoderma reesei glucosidase II (TrAguII) (Geysens et al. 2005). This type of α-glucosidases is located in the ER where it is involved in the trimming of α-(1,3)-linked glucose residues from N-glycan core structure Glc3Man9GlcNAc2, which may be attached to proteins designated to be secreted (Geysens et al. 2005). AgdE contains a predicted signal sequence which might serve to direct the protein to the ER. The agdE gene was constitutively expressed in all conditions tested (Fig. 2), consistent with its predicted function as an α-glucosidase II. Two additional GH31 family members, AgdF (An18g05620) and AgdG (An07g0350), lack similarity to any functionally described proteins. Expression of both genes was not detected in any of the tested growth conditions.
The two final family GH31 members, named AxlA and AxlB, are highly similar to AN7505.2 from A. nidulans, which was recently characterized as an α-xylosidase (Bauer et al. 2006). Gene axlA was highly expressed in the presence of xylose, whereas no expression was detected in maltose grown cultures (Fig. 2). The high expression of axlA on xylose further supports its putative function as a xylanolytic enzyme. The gene encoding the putatively intracellular AxlB was expressed at a very low level independent of the carbon source (Fig. 2).
A BLASTP search in the genomes of A. fumigatus, A. nidulans and A. oryzae for family GH31 enzymes resulted in a similar collection of enzymes as identified in A. niger (Fig. S1C). Several clusters of orthologous proteins are distinguishable, but the assignment of an enzymatic activity to these clusters is not yet possible due to a lack of well studied homologues for these enzymes, thus requiring biochemical studies in our future work.
Genome-wide analysis of AmyR dependent maltose induced genes using microarrays
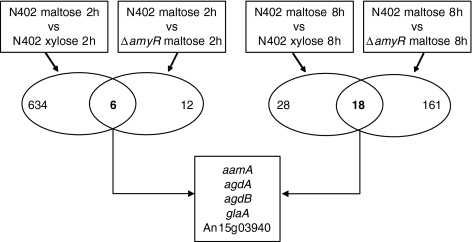
The expression analysis of genes encoding family GH13, GH15 and GH31 enzymes in the A. niger genome revealed that the expression of only a limited number of genes was induced by maltose in an AmyR dependent way. In fact, only four genes (aamA, glaA, agdA, and agdB) showed the predicted expression pattern for genes encoding enzymes involved in the breakdown of starch (Fig. 2). To identify additional genes with a possible role in starch metabolism, the expression of all 14,509 predicted open reading frames on the Affymetrix microarray chips was analysed. Comparison of the transcriptome of the wild type strain (N402), grown for 2 h after transfer on either xylose or maltose, identified 634 genes that were significantly induced by maltose (One-way ANOVA analysis P < 0.08 and >2-fold change in expression level). A set of 12 genes was expressed >2-fold higher in N402 compared to the ΔamyR strain (One-way ANOVA analysis P < 0.08) after the transfer of these strains from xylose to maltose. Combining the two gene sets resulted in a collection of six genes that were >2-fold induced on maltose in an AmyR-dependent manner (Fig. 3, Tables 4, 5). Five of these were assigned to the category of carbohydrate transport and metabolism according to FunCat (Ruepp et al. 2004), including the three genes encoding known extracellular starch degrading enzymes (aamA, glaA and agdA) and a putative α-glucosidase (agdB) (Table 4). Gene An15g03940, encoding a protein with high similarity (68%) to a Candida intermedia glucose/xylose symporter (Leandro et al. 2006), was 2.3-fold induced by maltose; this induction was not observed in the ΔamyR strain. The sixth induced gene (An11g02550) encodes a protein highly similar (72%) to Kluyveromyces lactis phosphoenolpyruvate carboxykinase (Kitamoto et al. 1998) which functions in gluconeogenesis by catalyzing the conversion of oxaloacetate into phosphoenolpyruvate. It should be noted that the pH of the growth medium remained 6.2 during the 2 h growth period after transfer.
Fig. 3.
Results of microarray analysis for maltose induced and AmyR dependent genes. Venn-diagram showing the number of genes induced on maltose compared to xylose in A. niger N402, and the number of genes induced in N402 compared to the ΔamyR deletion strain. The number of genes both induced by maltose and dependent on AmyR is indicated in bold. The maltose induced and AmyR dependent genes which are present in both 2 and 8 h after transfer from a preculture are shown in the box
Table 4.
Overview of six maltose induced and AmyR dependent genes in A. niger at 2 h (pH 6.2) after transfer from preculture
| ORF no.a | N402 maltoseb | Callc | N402 xylose | Call | ΔamyR maltose | Call | Fold induction | P-value | Putative function | |
|---|---|---|---|---|---|---|---|---|---|---|
| Maltose vs xylose | N402 vs ΔamyR | |||||||||
| Carbohydrate transport and metabolism | ||||||||||
| An11g03340 | 3.15 ± 0.12 | P | 0.08 ± 0.04 | A | 0.13 ± 0.09 | A | 38.6 | 24.2 | 0.080 | Acid alpha-amylase AamA |
| An04g06920 | 85.96 ± 22.54 | P | 6.24 ± 0.98 | P | 5.03 ± 0.03 | P | 13.8 | 17.1 | 0.064 | Extracellular alpha-glucosidase AgdA |
| An01g10930 | 96.50 ± 31.50 | P | 10.47 ± 2.59 | P | 8.35 ± 2.13 | P | 9.2 | 11.5 | 0.062 | Extracellular alpha-glucosidase AgdB |
| An03g06550 | 117.80 ± 43.16 | P | 13.78 ± 4.14 | P | 5.85 ± 0.47 | P | 8.5 | 20.1 | 0.072 | Glucoamylase GlaA |
| An15g03940 | 152.30 ± 19.00 | P | 65.74 ± 3.04 | P | 70.59 ± 2.88 | P | 2.3 | 2.2 | 0.074 | Putative monosaccharide transporter |
| Energy | ||||||||||
| An11g02550 | 58.87 ± 3.86 | P | 10.13 ± 1.41 | P | 25.27 ± 5.35 | P | 5.8 | 2.3 | 0.033 | Putative phosphoenolpyruvate carboxykinase |
aGene name in bold indicates that the gene is >2-fold induced on maltose in an AmyR dependant way after both 2 and 8 h
bThe expression level was based on the geometric mean value of the duplicate samples, and the deviation values between the duplicate samples are indicated. The P-value was based on 1-way ANOVA analysis
cP, M or A represent detection calls for present, marginal or absent, respectively. Genes were divided over different functional groups according to FunCat (Ruepp et al. 2004)
Table 5.
Overview of the 18 maltose induced and AmyR dependent genes in A. niger at 8 h (pH4.0) after transfer from preculture
| ORF no.a | N402 maltoseb | Callc | N402 xylose | Call | ΔamyR maltose | Call | Fold induction | P-value | Putative function | |
|---|---|---|---|---|---|---|---|---|---|---|
| Maltose vs xylose | N402 vs ΔamyR | |||||||||
| Carbohydrate transport and metabolism | ||||||||||
| An11g03340 | 65.26 ± 7.48 | P | 0.58 ± 0.57 | A | 0.08 ± 0.01 | A | 113.0 | 808.4 | 0.004 | Acid alpha-amylase AamA |
| An01g10930 | 49.46 ± 3.42 | P | 2.37 ± 0.08 | P | 6.84 ± 1.99 | P | 20.9 | 7.2 | 0.006 | Extracellular alpha-glucosidase AgdB |
| An04g06920 | 179.10 ± 13.90 | P | 9.89 ± 3.63 | P | 3.36 ± 0.46 | P | 17.6 | 51.7 | 0.012 | Extracellular alpha-glucosidase AgdA |
| An02g03540 | 62.79 ± 8.36 | P | 8.81 ± 0.61 | P | 0.96 ± 0.82 | P, A | 6.7 | 61.9 | 0.012 | Putative hexose transport protein MstC |
| An15g03940 | 144.40 ± 6.80 | P | 35.93 ± 2.72 | P | 3.04 ± 0.03 | P, M | 4.0 | 47.6 | 0.003 | Putative monosaccharide transporter |
| An03g06550 | 239.30 ± 2.05 | P | 83.03 ± 4.35 | P | 19.03 ± 1.75 | P | 2.9 | 12.6 | 0.012 | Glucoamylase GlaA |
| An04g06930 | 4.36 ± 0.50 | P | 1.50 ± 0.16 | P | 1.40 ± 0.03 | P | 2.9 | 3.1 | 0.062 | Extracellular alpha-amylase AmyC |
| An09g04810 | 18.47 ± 0.44 | P | 6.46 ± 0.35 | P | 2.52 ± 0.02 | P | 2.9 | 7.4 | 0.003 | Putative hexose transporter |
| An12g07450 | 137.60 ± 13.00 | P | 51.57 ± 11.82 | P | 52.50 ± 6.31 | P | 2.7 | 2.6 | 0.064 | Sugar/H+ symporter MstA |
| Energy | ||||||||||
| An03g06270 | 4.72 ± 0.29 | P | 1.59 ± 0.14 | A | 0.20 ± 0.25 | A | 3.3 | 25.6 | 0.035 | Putative isoamyl alcohol oxidase |
| An16g06010 | 5.18 ± 1.55 | P | 1.80 ± 0.39 | P | 0.22 ± 0.02 | A | 2.9 | 23.8 | 0.032 | Putative phosphoglycerate mutase |
| Amino acid transport and metabolism | ||||||||||
| An03g00280 | 8.35 ± 0.90 | P | 3.57 ± 0.74 | P, A | 0.12 ± 0.06 | A | 2.3 | 68.0 | 0.050 | Similarity to tyrosinase protein |
| Cell rescue, defence and virulence | ||||||||||
| An14g05730 | 3.99 ± 0.59 | P | 0.23 ± 0.03 | A | 1.37 ± 0.73 | P, A | 18.8 | 3.2 | 0.037 | Similarity to integral membrane protein |
| An03g00290 | 5.96 ± 0.89 | P | 1.68 ± 0.47 | P | 0.64 ± 0.21 | P | 3.5 | 9.3 | 0.068 | Similarity to integral membrane protein |
| An06g00490 | 8.14 ± 0.89 | P | 3.81 ± 0.30 | P, A | 2.23 ± 0.44 | A | 2.1 | 3.7 | 0.066 | Similarity to integral membrane protein |
| Protein fate | ||||||||||
| An18g04260 | 5.44 ± 0.40 | P | 2.57 ± 0.07 | P | 1.77 ± 0.28 | M, A | 2.1 | 3.1 | 0.056 | Similarity to UDP-galactose transporter |
| Unclassified | ||||||||||
| An16g01290 | 15.04 ± 0.28 | P | 5.22 ± 1.69 | P, A | 1.97 ± 0.26 | A | 2.6 | 6.9 | 0.027 | Unknown |
| An09g06130 | 12.62 ± 2.51 | P | 5.41 ± 0.93 | P | 2.69 ± 0.30 | P, A | 2.3 | 4.7 | 0.059 | Unknown |
a,b,cSee footnotes of Table 4
A similar comparative analysis was performed for samples taken for cultures that had grown for 8 h after the transfer. To prevent differences in pH between the N402 and the ΔamyR strain after 8 h of growth, the pH of the medium was buffered to 4.0 using 100 mM of citric acid/sodium citrate to allow comparison between the N402 and the ΔamyR strain at this time point, as the medium of the ΔamyR strain did not acidify as quickly as the medium of the N402 strain. A total of 28 genes were significantly induced by maltose in comparison with xylose, and 161 genes were 2-fold higher expressed in N402 compared with the ΔamyR strain (one-way ANOVA analysis P < 0.08). By combining the two sets, we identified 18 genes which were induced by maltose and whose induction was AmyR dependent (Fig. 3, Table 5). Nine of these genes encode proteins involved in carbohydrate transport and metabolism, from which five were also identified as differentially expressed after 2 h (aamA, glaA, agdA, agdB, and An15g03940). Additionally identified genes included amyC, encoding an extracellular α-amylase, three genes encoding putative sugar transporters, and several other genes belonging to various functional categories (Tables 4, 5). At this stage we cannot exclude that additional overlapping genes have escaped our attention, because of the different pH values of the medium after transfer, which also could affect gene expression. The results from the microarray experiments were validated using Northern blot analysis for a selected number of amylolytic genes. These genes include glaA, aamA, agdB and amyC. As shown in supplementary Fig. 3, the results of the Northern hybridizations are in good agreement with the microarray data (Supplementary Table 1). The Northern analysis also revealed an additional mRNA of a larger size for the amyC gene, 8 h after transfer. The detection of two different-sized mRNAs suggests two different mRNA start sites or different polyadenylation sites, but the exact sequence of the different mRNAs and possible consequences for the gene model have not been addressed so far.
Interestingly, the transcription factor gene amyR was also induced 2 and 8 h after transfer to maltose compared to the transfer to xylose (2.6-fold, P-value 0.014; 2.8-fold, P-value 0.027, respectively), indicating transcriptional regulation of the amyR gene itself. As the AmyR transcription factor is per se missing in the deletion strain, it is not appropriate to include the amyR gene in the group of maltose induced, AmyR dependent genes. However, the 1-kb promoter region of the amyR gene contains two AmyR binding motifs, allowing for the possibility that AmyR can induce its own expression.
Discussion
The full inventory of glycoside hydrolases belonging to families GH13, GH15 and GH31 in A. niger strain CBS 513.88 has recently been described (Pel et al. 2007). Members of these protein families in aspergilli, including A. niger, have been studied extensively mainly because of their industrial relevance. Nevertheless, several groups of new alpha-glucan acting enzymes that had not been identified previously were identified. These novel enzymes are conserved among several Aspergillus species as well as other Ascomycetes, indicating that they may play an important role in fungal metabolism. In the present study, we have combined transcriptional analysis and more detailed phylogenetic analysis to further understand their function.
The GH13 family in A. niger contains six separate groups of amylase-type enzymes (Fig. 1), of which three groups had not been described thus far. The best-described group comprises the extracellular α-amylases, and is now extended with AmyC in addition to the three known α-amylases. Overexpression of the amyC gene in A. niger resulted in increased levels of endo-α-amylase activity in the medium, indicating that this gene indeed encodes an extracellular α-amylase (R. M. Van der Kaaij and X. L. Yuan, unpublished results). The relatively low expression of amyC compared to other starch degrading enzymes may explain why this protein has not been identified previously. The localization of the amyC gene in the genome is noteworthy, as it is part of a small cluster of amylolytic genes (with agdA) and their transcriptional regulator gene amyR. A similar organization is observed in the genomes of other aspergilli such as A. nidulans and A. oryzae RIB40 (Gomi et al. 2000). In A. fumigatus the same cluster is extended with a glucoamylase (similar to GlaA). Gene clusters of transcriptionally co-regulated genes in filamentous fungi are often involved in the same process, e.g. secondary metabolite production (Woloshuk et al. 1995) or catabolism of amino acids (Hull et al. 1989). A possible function of AmyC might be to act as a scouting enzyme for the presence of starch, resulting in the subsequent activation of AmyR by starch-derived molecules (maltose or isomaltose). Alternatively, AmyC might be regulated by another system additional to AmyR, e.g. pH-regulated expression, or its expression might be upregulated locally, e.g. in hyphal tips.
The lack of detectable expression of amyA and amyB in A. niger N402 is puzzling. Both genes were expressed in A. niger strain CBS 513.88 in a fed-batch fermentation using glucose as a carbon source (Pel et al. 2007), but batch cultivation of A. niger N402 on glucose did not result in detectable expression in microarray studies (E. Martens, H. Kools, P. Schaap, personal communication). These results indicate a difference between both strains with regard to either the presence of amyA/B in A. niger strain N402 or in the transcriptional regulation of amyA/B. In the recently released genome sequence of A. niger strain ATCC 1015 from the Joint Genome Institute (Baker, 2006) we could not identify orthologues of the AmyA/AmyB proteins suggesting that these genes might be actually absent from the N402 strain. The absence, or lack of expression of amyA/amyB, is consistent with the observation that the A. niger MGG029ΔaamA strain (N402 background) with gene disruptions for both aamA and glaA grows very poorly on starch (Weenink et al. 2006), confirming that AmyA and AmyB do not contribute significantly to the amylolytic potential of N402. The microarray analysis revealed that in addition to the amyA/B genes, five more amylolytic genes (agtC, amyD, agdD, agdG and glaB) were not detected (Absent calls) in any of the conditions tested. However, orthologous genes were identified in the genome of A. niger strain ATCC1015, suggesting that these genes are not absent from the N402 genome, but rather not expressed under the tested conditions.
In this study we have identified three new groups within the GH13 family that had not been described before in aspergilli: α-amylase-like proteins predicted to be GPI-anchored enzymes (group II) (subfamily GH13_1, as defined by Stam et al. 2006), intracellular α-glucosidases (group III) (unknown subfamily) and intracellular α-amylases (group IV) (subfamily GH13_5). Detailed biochemical studies of two GPI-anchored proteins revealed that they are α-glucanotransferases, acting mainly on maltooligosaccharides (van der Kaaij et al. 2007a). The expression of these genes was not regulated in response to maltose metabolism or AmyR. The Agt enzymes are homologues of a group of four GPI-anchored amylase-like proteins recently identified in S. pombe (Aah1-4p), which have similarly aberrant conserved regions (Table 3). A deletion of one of these homologues resulted in aberrant morphology of the cell and an increased sensitivity towards cell wall degrading enzymes (Morita et al. 2006). A similar phenotype was observed for an A. niger knockout of agtA (van der Kaaij et al. 2007a). Such a phenotype can be explained if α-glucan synthesis is affected. The clustering of agt genes with genes encoding α-glucan synthases in all four Aspergillus species studied, and the expression pattern of the A. niger agt genes both support the hypothesis that Agt enzymes play a role in α-glucan synthesis.
Aspergillus niger contains two putative intracellular α-amylases, AmyD and AmyE, that belong to the recently identified subfamily GH13_5 (Stam et al. 2006). A homologous gene in H. capsulatum, AMY1, was shown to be essential for the formation of α-(1,3)-glucan in the cell wall (Marion et al. 2006). The possibility of a role for subfamily GH13_5 enzymes in the cell wall of Aspergilli is strengthened by their location in the genome: A. niger, A. nidulans and A. oryzae, all contain a cluster of genes, containing a homologue of AMY1 as well as ags and agt genes (Table S2). Both the GPI-anchored α-glucanotransferases and the family GH13_5 α-amylase-like proteins are only present in fungi with cell wall α-glucan and not in the true yeasts, which lack this type of cell wall glucans. However, a study with heterologously expressed AmyD did not confirm a role for this enzyme in α-(1,3)-glucan formation, as the enzyme only showed a (low) hydrolyzing activity on substrates with α-(1,4) glycosidic bonds in vitro (van der Kaaij et al. 2007b). The expression levels of amyD and amyE were below detection level, and thus could not confirm nor refute the possible involvement of these proteins in cell wall α-glucan synthesis in A. niger.
At this moment, no function can be assigned to the predicted intracellular α-glucosidases AgdC and AgdD, and their homologues identified in other fungi. The enzymes show similarity to a Bacillus α-glucosidase as well as to a maltase from C. albicans, and might therefore be involved in the intracellular hydrolysis of maltose. Alternatively, these enzymes could play a role in trehalose metabolism, as they show some similarity to bacterial trehalose-6-phosphate hydrolases. Trehalose is known to be a common reserve carbohydrate in fungi including A. niger (Wolschek and Kubicek 1997; Arguelles 2000).
A surprising finding from the genome mining of A. niger was the high number of GH enzymes with a predicted intracellular location. This group included not only the starch-acting enzymes described here, but also other predicted intracellular glycoside hydrolases such as β-glucosidase, chitinase, β-mannosidase, β-xylosidase, rhamnosidase and invertase, present in various Aspergillus genomes (Pel et al. 2007; Goosen et al. 2007). Most of these proteins were predicted to be exo-acting, i.e. releasing terminal mono- or disaccharide residues from the non-reducing end of the substrate. In A. niger, the genes encoding these proteins are expressed at a relatively low level compared to the extracellular starch degrading enzymes, sometimes even below the detection limit. In a recent publication, expression of intracellular chitinases in A. nidulans was found to be induced during the autolysis phase of the culture (Yamazaki et al. 2007). A role for these and other predicted intracellular proteins during the autolysis phase would explain their low expression levels observed in A. niger during vegetative growth, as well as their lack of secretion signals.
The genome-wide expression analysis identified only five genes that were >2-fold induced by maltose in an AmyR dependent way, 2 and 8 h after the transfer to maltose. Among them are genes encoding known starch degrading enzymes (aamA, glaA), two (putative) α-glucosidases (agdA, agdB), and a putative sugar transporter protein (An15g03940). As described in the Results section, the samples taken after 2 and 8 h after transfer were grown at different pH values (pH 6.2 and 4.0, respectively), which might influence our results and lead to an underestimation of the number of genes that are induced by maltose in an AmyR dependant way at both 2 and 8 h as additional pH-controlled gene expression might also occur. To fully explore this, both strains should be cultivated at controlled conditions at identical pH.
The specificity of the putative sugar transporter encoded by the An15g03940 gene has not been examined, but its expression profile suggests that it might function as a glucose or a maltose transporter. Three additional sugar transporters were expressed higher on maltose than on xylose after 8 h (Table 5). One of them, An12g07450/MstA has been functionally characterized as a high-affinity glucose transporter (Vankuyk et al. 2004) which is in-line with its expression profile. The two additional sugar transporters encoding genes (An02g03540/mstC and An09g04810) showed an interesting expression pattern, with induced expression on maltose but also relatively high expression levels on xylose (Table 5), suggesting that these transporters might have a broad sugar transporting activity. In the ΔamyR strain both their expression levels were strongly reduced, also relative to the xylose conditions, indicating that the deletion of AmyR might have had an indirect effect. A possible explanation is that disruption of the AmyR transcription factor resulted in low levels of extracellular enzymes converting maltose to glucose, and consequently stress due to low glucose levels. The low availability of glucose might be a signal to downregulate mstC and An09g04810 if these genes encode low-affinity sugar transporters. To further address these observations additional array analysis should be performed using the ΔamyR strain grown on xylose. Interestingly, the amyR gene itself was also induced by the presence of maltose suggesting that its regulation takes place, at least partly, at the transcriptional level. Furthermore, conserved AmyR binding motifs are present in the promoters of all co-regulated genes, including the 1-kb amyR promoter region, with the exception of aamA (Table 1). Possibly the aamA promoter contains a currently unknown AmyR binding motif. Alternatively, an AmyR binding site might be present in the promoter region of aamA in the N402 background.
The results presented in this paper suggest that A. niger can metabolize maltose by inducing the expression of a limited amount of enzymes and sugar transporters. Other members of the GH13, GH15 and GH31 protein families might function in starch metabolism in different conditions (solid state growth, or influenced by pH, nitrogen metabolism, etc.), or they may be involved in other processes such as the synthesis of cell wall α-glucan, or the glycosylation of proteins. To determine the exact function of these enzymes will require further biochemical characterization in combination with the detailed analysis of gene deletion mutants.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Harrie Kools (Wageningen University, The Netherlands) for performing the HMM searches, Patricia vanKuyk (Wageningen University, The Netherlands) for the amyR plasmid, Marie-Francoise Coeffet-Le Gal and David Archer for sharing unpublished results. This work was financially supported by SenterNovem in the framework of the Dutch IOP-Genomics program (project IGE1021).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ER
Endoplasmic reticulum
- GH
Glycoside hydrolase
- GPI
Glycosylphosphatidylinositol
- HMM
Hidden Markov model
- SBD
Starch-binding domain
Footnotes
Xiao-Lian Yuan and Rachel M. van der Kaaij are the authors who contributed equally to this study.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-008-0332-7) contains supplementary material, which is available to authorized users.
Contributor Information
Xiao-Lian Yuan, Email: xyuan@science.uva.nl.
Arthur F. J. Ram, Phone: +31-71-5274914, FAX: +31-71-5274999, Email: A.F.J.Ram@biology.leidenuniv.nl
References
- Arguelles JC (2000) Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol 174:217–224 [DOI] [PubMed]
- Baker SE (2006) Aspergillus niger genomics: past, present and into the future. Med Mycol 44:17–21 [DOI] [PubMed]
- Barker SA, Carrington TR (1953) Studies of Aspergillus niger. Part II. Transglycosylation by Aspergillus niger. J Chem Soc 1953:3588–3593 [DOI]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR (2006) Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci USA 103:11417–11422 [DOI] [PMC free article] [PubMed]
- Beauvais A, Latgé JP (2001) Membrane and cell wall targets in Aspergillus fumigatus. Drug Resist Updat 4:38–49 [DOI] [PubMed]
- Bendtsen JD, Nielsen H, Von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795 [DOI] [PubMed]
- Bennett JW, Lasure LL (1991) More gene manipulations in fungi. Academic Press, San Diego
- Birney E, Clamp M, Durbin R (2004) GeneWise and Genomewise. Genome Res 14:988–995 [DOI] [PMC free article] [PubMed]
- Boel E, Brady L, Brzozowski AM, Derewenda Z, Dodson GG, Jensen VJ, Petersen SB, Swift H, Thim L, Woldike HF (1990) Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus. Biochemistry 29:6244–6249 [DOI] [PubMed]
- Boel E, Hjort I, Svensson B, Norris F, Norris KE, Fiil NP (1984) Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNAs. EMBO J 3:1097–1102 [DOI] [PMC free article] [PubMed]
- Bos CJ, Debets AJ, Swart K, Huybers A, Kobus G, Slakhorst SM (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet 14:437–443 [DOI] [PubMed]
- Chang CT, Lo HF, Chi MC, Yao CY, Hsu WH, Lin LL (2003) Identification of essential histidine residues in a recombinant alpha-amylase of thermophilic and alkaliphilic Bacillus sp. strain TS-23. Extremophiles 7:505–509 [DOI] [PubMed]
- Coutinho PM, Henrissat B (1999) Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies GJ, Henrissat B, Svensson B (eds) Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, pp 3–12
- Damveld RA, Vankuyk PA, Arentshorst M, Klis FM, van den Hondel CA, Ram AF (2005) Expression of agsA, one of five 1,3-alpha-d-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet Biol 42:165–177 [DOI] [PubMed]
- de Ruiter-Jacobs YM, Broekhuijsen M, Unkles SE, Campbell EI, Kinghorn JR, Contreras R, Pouwels PH, van den Hondel CA (1989) A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr Genet 16:159–163 [DOI] [PubMed]
- De Vries RP, Visser J (2001) Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev 65:497–522 [DOI] [PMC free article] [PubMed]
- den Herder IF, Rosell AM, van Zuilen CM, Punt PJ, van den Hondel CA (1992) Cloning and expression of a member of the Aspergillus niger gene family encoding alpha-galactosidase. Mol Gen Genet 233:404–410 [DOI] [PubMed]
- Durbin R, Eddy S (1998) Biological sequence analysis: probabilistic models of proteins and nucleic acids. A tutorial introduction to hidden Markov models and other probabilistic modelling approaches in computational sequence analysis. Cambridge University Press, Cambridge
- Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F (2004) A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Mol Biol 337:243–253 [DOI] [PubMed]
- Fontaine T, Simenel C, Dubreucq G, Adam O, Delepierre M, Lemoine J, Vorgias CE, Diaquin M, Latgé JP (2000) Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J Biol Chem 275:41528 [PubMed]
- Fowler T, Berka RM, Ward M (1990) Regulation of the glaA gene of Aspergillus niger. Curr Genet 18:537–545 [DOI] [PubMed]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, d’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed]
- Geber A, Williamson PR, Rex JH, Sweeney EC, Bennett JE (1992) Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol 174:6992–6996 [DOI] [PMC free article] [PubMed]
- Geysens S, Pakula T, Uusitalo J, Dewerte I, Penttila M, Contreras R (2005) Cloning and characterization of the glucosidase II alpha subunit gene of Trichoderma reesei: a frameshift mutation results in the aberrant glycosylation profile of the hypercellulolytic strain Rut-C30. Appl Environ Microbiol 71:2910–2924 [DOI] [PMC free article] [PubMed]
- Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y (2000) Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci Biotechnol Biochem 64:816–827 [DOI] [PubMed]
- Goosen C, Yuan XL, van Munster JM, Ram AF, van der Maarel MJ, Dijkhuizen L (2007) Molecular and biochemical characterization of a novel intracellular invertase from Aspergillus niger with transfructosylating activity. Eukaryot Cell 6:674–681 [DOI] [PMC free article] [PubMed]
- Grün CH, Hochstenbach F, Humbel BM, Verkleij AJ, Sietsma JH, Klis FM, Kamerling JP, Vliegenthart JF (2005) The structure of cell wall alpha-glucan from fission yeast. Glycobiology 15:245–257 [DOI] [PubMed]
- Hochstenbach F, Klis FM, Van Den EH, van DE, Peters PJ, Klausner RD (1998) Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc Natl Acad Sci USA 95:9161–9166 [DOI] [PMC free article] [PubMed]
- Horisberger M, Lewis BA, Smith F (1972) Structure of a (1, 3)-α-d-glucan (pseudonigeran) of Aspergillus niger NNRL 326 cell wall. Carbohydr Res 23:183–188 [DOI] [PubMed]
- Hull EP, Green PM, Arst HN Jr, Scazzocchio C (1989) Cloning and physical characterization of the l-proline catabolism gene cluster of Aspergillus nidulans. Mol Microbiol 3:553–559 [DOI] [PubMed]
- Inoue H, Nojima H, Okayama H (1990) High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28 [DOI] [PubMed]
- Ito T, Tani S, Itoh T, Tsukagoshi N, Kato M, Kobayashi T (2004) Mode of AmyR binding to the CGGN8AGG sequence in the Aspergillus oryzaetaaG2 promoter. Biosci Biotechnol Biochem 68:1906–1911 [DOI] [PubMed]
- Johnston IR (1965) The composition of the cell wall of Aspergillus niger. Biochem J 96:651–658 [DOI] [PMC free article] [PubMed]
- Kato N, Murakoshi Y, Kato M, Kobayashi T, Tsukagoshi N (2002a) Isomaltose formed by alpha-glucosidases triggers amylase induction in Aspergillus nidulans. Curr Genet 42:43–50 [DOI] [PubMed]
- Kato N, Suyama S, Shirokane M, Kato M, Kobayashi T, Tsukagoshi N (2002b) Novel alpha-glucosidase from Aspergillus nidulans with strong transglycosylation activity. Appl Environ Microbiol 68:1250–1256 [DOI] [PMC free article] [PubMed]
- Kitamoto HK, Ohmomo S, Iimura Y (1998) Isolation and nucleotide sequence of the gene encoding phosphoenolpyruvate carboxykinase from Kluyveromyces lactis. Yeast 14:963–967 [DOI] [PubMed]
- Korman DR, Bayliss FT, Barnett CC, Carmona CL, Kodama KH, Royer TJ, Thompson SA, Ward M, Wilson LJ, Berka RM (1990) Cloning, characterization, and expression of two alpha-amylase genes from Aspergillus niger var. awamori. Curr Genet 17:203–212 [DOI] [PubMed]
- Leandro MJ, Goncalves P, Spencer-Martins I (2006) Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochem J 395:543–549 [DOI] [PMC free article] [PubMed]
- Leemhuis H, Wehmeier UF, Dijkhuizen L (2004) Single amino acid mutations interchange the reaction specificities of Cyclodextrin Glycosyltransferase and the acarbose-modifying enzyme Acarviosyl Transferase. Biochemistry 43:13204–13213 [DOI] [PubMed]
- MacGregor EA, Janecek S, Svensson B (2001) Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochim Biophys Acta 1546:1–20 [DOI] [PubMed]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:W327–W331 [DOI] [PMC free article] [PubMed]
- Marion CL, Rappleye CA, Engle JT, Goldman WE (2006) An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol Microbiol 62:970–983 [DOI] [PubMed]
- Morita T, Tanaka N, Hosomi A, Giga-Hama Y, Takegawa K (2006) An alpha-amylase homologue, aah3, encodes a GPI-anchored membrane protein required for cell wall integrity and morphogenesis in Schizosaccharomyces pombe. Biosci Biotechnol Biochem 70:1454–1463 [DOI] [PubMed]
- Nakajima T, Imanaka T, Aiba S (1986) Comparison of amino acid sequences of eleven different alpha-amylases. Appl Microbiol Biotechnol 23:355–360 [DOI]
- Nakamura A, Nishimura I, Yokoyama A, Lee DG, Hidaka M, Masaki H, Kimura A, Chiba S, Uozumi T (1997) Cloning and sequencing of an alpha-glucosidase gene from Aspergillus niger and its expression in A. nidulans. J Biotechnol 53:75–84 [DOI] [PubMed]
- Nakamura T, Maeda Y, Tanoue N, Makita T, Kato M, Kobayashi T (2006) Expression profile of amylolytic genes in Aspergillus nidulans. Biosci Biotechnol Biochem 70:2363–2370 [DOI] [PubMed]
- Nakao M, Nakayama T, Kakudo A, Inohara M, Harada M, Omura F, Shibano Y (1994) Structure and expression of a gene coding for thermostable alpha-glucosidase with a broad substrate specificity from Bacillus sp. SAM1606. Eur J Biochem 220:293–300 [DOI] [PubMed]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latgé JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O’Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de CS, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed]
- Orlean P (1997) Biogenesis of yeast wall and surface components. In: Pringle J, Broach JR, Jones EW (eds) Cell cycle and cell biology. Cold Spring Harbor Laboratory Press, New York, pp 229–362
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, De Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, De Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231 [DOI] [PubMed]
- Peters D (2006) Carbohydrates for fermentation. Biotechnol J 1:806–814 [DOI] [PubMed]
- Petersen KL, Lehmbeck J, Christensen T (1999) A new transcriptional activator for amylase genes in Aspergillus. Mol Gen Genet 262:668–676 [DOI] [PubMed]
- Punt PJ, van den Hondel CA (1992) Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol 216:447–457 [DOI] [PubMed]
- Rappleye CA, Engle JT, Goldman WE (2004) RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol Microbiol 53:153–165 [DOI] [PubMed]
- Rappleye CA, Goldman WE (2006) Defining virulence genes in the dimorphic fungi. Annu Rev Microbiol 60:281–303 [DOI] [PubMed]
- Robyt JF (1998) Essentials of carbohydrate chemistry. Springer, New York
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes HW (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32:5539–5545 [DOI] [PMC free article] [PubMed]
- Sasangka P, Matsuno A, Tanaka A, Akasaka Y, Suyama S, Kano S, Miyazaki M, Akao T, Kato M, Kobayashi T, Tsukagoshi N (2002) Structural features of the glycogen branching enzyme encoding genes from Aspergilli. Microbiol Res 157:337–344 [DOI] [PubMed]
- Sauer J, Sigurskjold BW, Christensen U, Frandsen TP, Mirgorodskaya E, Harrison M, Roepstorff P, Svensson B (2000) Glucoamylase: structure/function relationships, and protein engineering. Biochim Biophys Acta 1543:275–293 [DOI] [PubMed]
- Southall SM, Simpson PJ, Gilbert HJ, Williamson G, Williamson MP (1999) The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett 447:58–60 [DOI] [PubMed]
- Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Des Sel 19:555–562 [DOI] [PubMed]
- Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, Gomi K, Kobayashi T, Tsukagoshi N (2001) Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr Genet 39:10–15 [DOI] [PubMed]
- Teste MA, Enjalbert B, Parrou JL, Francois JM (2000) The Saccharomyces cerevisiae YPR184w gene encodes the glycogen debranching enzyme. FEMS Microbiol Lett 193:105–110 [DOI] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed]
- Tsukagoshi N, Kobayashi T, Kato M (2001) Regulation of the amylolytic and (hemi-)cellulolytic genes in Aspergilli. J Gen Appl Microbiol 47:1–19 [DOI] [PubMed]
- Tsukamoto A, Kimura K, Ishii Y, Takano T, Yamane K (1988) Nucleotide sequence of the maltohexaose-producing amylase gene from an alkalophilic Bacillus sp. #707 and structural similarity to liquefying type alpha-amylases. Biochem Biophys Res Commun 151:25–31 [DOI] [PubMed]
- Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW (1999) X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. Nat Struct Biol 6:432–436 [DOI] [PubMed]
- van der Kaaij RM, Yuan XL, Franken A, Ram AFJ, Punt PJ, Maarel MJEC, Dijkhuizen L (2007a) Characterization of two novel, putatively cell wall associated and GPI-anchored, alpha-glucanotransferase enzymes of Aspergillus niger. Eukaryot Cell 6:1178–1188 [DOI] [PMC free article] [PubMed]
- Van der Kaaij RM, Janecek S, van der Maarel MJEC, Dijkhuizen L (2007b) Phylogenetic and biochemical characterization of a novel cluster of intracellular fungal α-amylase enzymes. Microbiology 153:4003–4015 [DOI] [PubMed]
- van Dijck PW, Selten GC, Hempenius RA (2003) On the safety of a new generation of DSM Aspergillus niger enzyme production strains. Regul Toxicol Pharmacol 38:27–35 [DOI] [PubMed]
- van Hartingsveldt W, Mattern IE, van Zeijl CM, Pouwels PH, van den Hondel CA (1987) Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet 206:71–75 [DOI] [PubMed]
- Vankuyk PA, Diderich JA, MacCabe AP, Hererro O, Ruijter GJ, Visser J (2004) Aspergillus niger mstA encodes a high-affinity sugar/H+ symporter which is regulated in response to extracellular pH. Biochem J 379:375–383 [DOI] [PMC free article] [PubMed]
- Weenink XO, Punt PJ, van den Hondel CA, Ram AF (2006) A new method for screening and isolation of hypersecretion mutants in Aspergillus niger. Appl Microbiol Biotechnol 69:711–717 [DOI] [PubMed]
- Woloshuk CP, Yousibova GL, Rollins JA, Bhatnagar D, Payne GA (1995) Molecular characterization of the afl-1 locus in Aspergillus flavus. Appl Environ Microbiol 61:3019–3023 [DOI] [PMC free article] [PubMed]
- Wolschek MF, Kubicek CP (1997) The filamentous fungus Aspergillus niger contains two “differentially regulated” trehalose-6-phosphate synthase-encoding genes, tpsA and tpsB. J Biol Chem 272:2729–2735 [DOI] [PubMed]
- Yamazaki H, Yamazaki D, Takaya N, Takagi M, Ohta A, Horiuchi H (2007) A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr Genet 51:89–98 [DOI] [PubMed]
- Yuan XL, Goosen C, Kools H, van der Maarel MJ, van den Hondel CA, Dijkhuizen L, Ram AF (2006) Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology 152:3061–3073 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.