Abstract
The protein encoded by SNM1 in Saccharomyces cerevisiae has been shown to act specifically in DNA interstrand crosslinks (ICL) repair. There are five mammalian homologs of SNM1, including Artemis, which is involved in V(D)J recombination. Cells from mice constructed with a disruption in the Snm1 gene are sensitive to the DNA interstrand crosslinker, mitomycin (MMC), as indicated by increased radial formation following exposure. The mice reproduce normally and have normal life spans. However, a partial perinatal lethality, not seen in either homozygous mutant alone, can be noted when the Snm1 disruption is combined with a Fancd2 disruption. To explore the role of hSNM1 and its homologs in ICL repair in human cells, we used siRNA depletion in human fibroblasts, with cell survival and chromosome radials as the end points for sensitivity following treatment with MMC. Depletion of hSNM1 increases sensitivity to ICLs as detected by both end points, while depletion of Artemis does not. Thus hSNM1 is active in maintenance of genome stability following ICL formation. To evaluate the epistatic relationship between hSNM1 and other ICL repair pathways, we depleted hSNM1 in Fanconi anemia (FA) cells, which are inherently sensitive to ICLs. Depletion of hSNM1 in an FA cell line produces additive sensitivity for MMC. Further, monoubiquitination of FANCD2, an endpoint of the FA pathway, is not disturbed by depletion of hSNM1 in normal cells. Thus, hSNM1 appears to represent a second pathway for genome stability, distinct from the FA pathway.
Keywords: SNM1, genome stability, radials, interstrand crosslink repair, Fanconi anemia
Introduction
Mammalian SNM1 protein is a member of a family of proteins that contain a conserved β—lactamase domain [1–3]. The group includes at least four other homologs: Artemis, SNM1B, CPSF73, and ELAC2 [2,4–7]. The original identification of the SNM1 (PSO2) gene was in Saccharomyces cerevisiae by selection of mutants showing increased sensitivity to DNA crosslinking agents [8,9]. In yeast the gene is non-essential and the repair function is strikingly specific for interstrand crosslinks (ICLs), with mutants showing normal UV and methylmethane sulfonate (MMS) resistance. The yeast enzyme, which contains the conserved β-lactamase motif, is known to have a 5’-exonuclease activity which is required for its function in ICL repair [1,10]. The hSNM1 protein is also a 5’-exonuclease [11] and the SNM1 protein belongs to the β-CASP motif-containing family, which includes proteins that function in DNA repair, V(D)J recombination and RNA processing [12,13]. Hydrophobic cluster analysis of amino acid sequences reveals additional members of this family [12]. Overproduction of SNM1 protein appears harmful to cells since over-expression is not stable [11,14], suggesting the exonuclease activity may need to be tightly regulated in its DNA processing activity.
ICLs in DNA result from the action of bifunctional crosslinking agents such as mitomycin C (MMC), psoralen with long-wave length UV, or cisplatin (CDDP). The repair of ICLs is complex, and in yeast is known to depend on three distinct pathways [15], with SNM1 being the herald gene for the ‘excision repair’ pathway. The function of the SNM1 protein is distinct from, and non-epistatic to, the post-replication repair (PRR) or homologous recombination (HR) pathways [15]. The repair of ICLs may be cell cycle dependent [16], however, snm1 mutants of S. cerevisiae have a normal S-phase after MMC treatment, with a prolonged G2 [17]. Interestingly, yeast snm1 mutants repair some forms of double strand breaks (DSB) normally [1], indicating that the breaks arising as intermediates during ICL repair must differ from those caused by other means, such as the mating type switch.
A homolog of SNM1 in Drosophila has been identified, and mutants (mus322) are hypersensitive to nitrogen mustard [18]. Thus in the fly, the SNM1 protein does serve a role in genome maintenance. The role of the mammalian SNM1 protein is less clear. The homologous protein Artemis acts in V(D)J recombination; its deficiency results in a severe combined immunodeficiency (SCID) phenotype [19–21]. A mouse model has been constructed, also manifesting SCID [22]. Artemis is a 5’-exonuclease with a cryptic endonuclease activity that is activated by phosphorylation, cleaving putative recombination intermediates [23–25]. Thus, a function in DNA recombination and immunity is clear for one member of the mammalian SNM1 family. However, patients with a defect in SNM1 and associated clinical indications of genome instability have not been identified. Disruption of the murine Snm1 gene has been reported to cause only a modest increase in sensitivity to MMC [4] in ES cells, with an apparent D37 ratio of about 2 for cell survival after MMC treatment. SNM1-deficient mice also have been reported to have decreased survival and increased tumorigenicity as well as increased susceptibility to infection in the C57BL6 background [26]. It also appears that SNM1 is a component of a mitotic stress checkpoint [27]. Thus the role, if any, of SNM1 in genome stability is not clear.
Fanconi anemia (FA) is a genome instability syndrome which results from a defect in any of at least thirteen genes [28–33]. The disease manifests pancytopenia of marrow cells, developmental abnormalities, altered pigmentation and increased incidence of leukemia and squamous cell cancers [34]. At the cellular level, FA shows increased chromosomal breakage and radial formation as well as decreased cell survival following exposure to DNA crosslinkers such as MMC. The FA pathway relies on BRCA1 for normal function [35], with evidence of direct interaction between FANC proteins and the Brca1/2 proteins (reviewed in [36–39]). Indeed, the FANCD1 gene is BRCA2 [31]. Thus there is a FA/BRCA pathway for genome stability in response to ICL formation.
To evaluate the function of the SNM1 protein in genome stability we constructed a mouse model with disruption of the Snm1 gene in the second exon. Using cells from the mouse model, as well as siRNA depletion of hSNM1 in human fibroblasts, we found that absence of SNM1 protein causes genome instability as manifest by aberrant chromosome forms after ICL formation. Survival of human cells lacking hSNM1 also is decreased following MMC treatment, reflecting a lack of normal processing of ICLs. Snm1 −/− mice are viable and have normal growth parameters, but a perinatal semi-lethality was observed in the mixed 129/SvJ/C57BL/6J background when the Snm1 gene disruption was crossed with a Fancd2 disruption model mouse. By using depletion in FA cells, the SNM1 pathway was found to be non-epistatic with Fanconi anemia for genome stability following ICL formation. The results of these studies establish at least two separate pathways responding to ICL damage in mammalian cells and show that the SNM1 function in maintaining genome stability is distinct from the FA pathway for ICL response.
Materials and Methods
Generation of Snm1 Mutant Mice
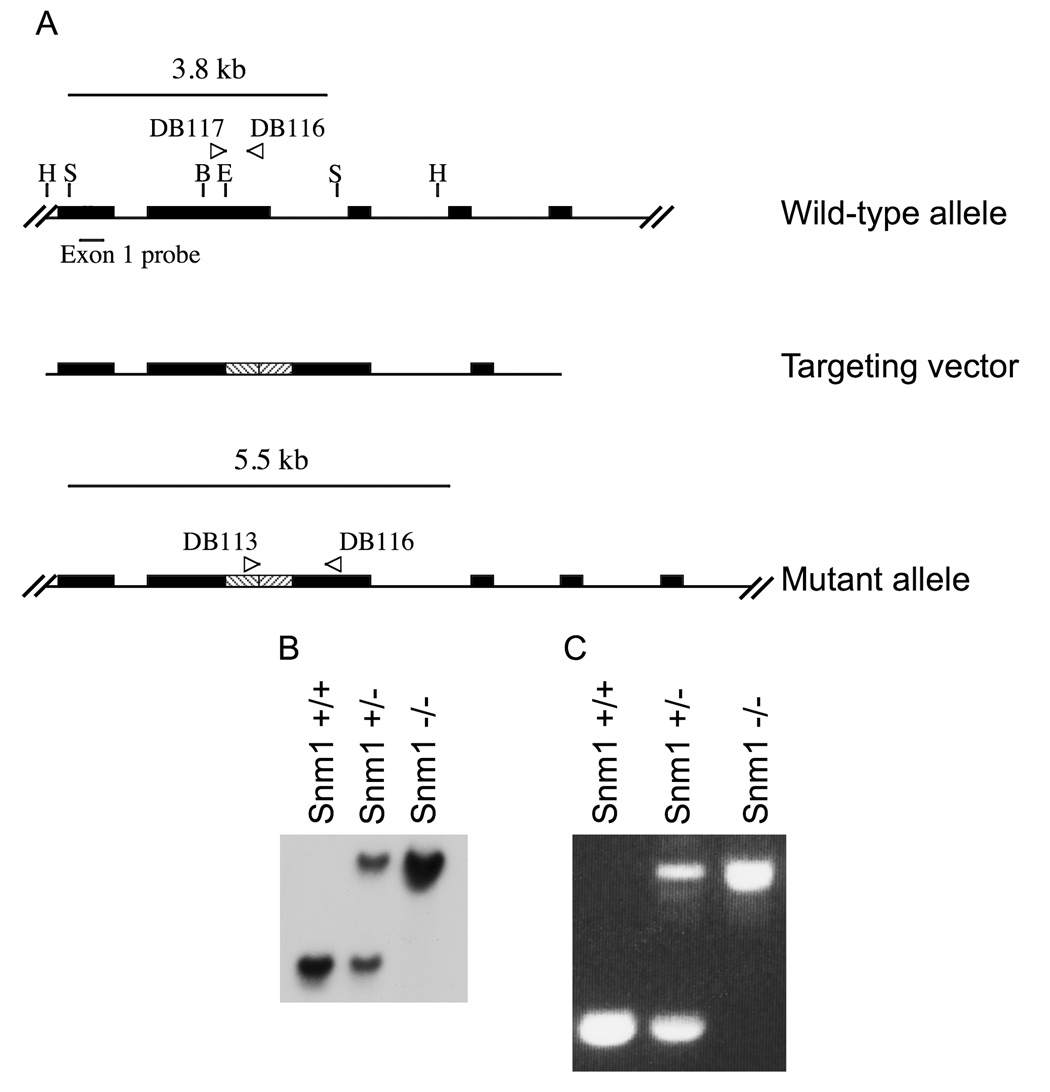
A genomic mouse 129/SvJBAC library (Invitrogen) was screened with human SNM1 specific primers. A positive clone was identified and isolated. A 5kb HindIII fragment containing exons 1–4 of mSnm1 was subcloned into pBluescript SK (Stratagene). The construct was then cut with BglII and Eco47III, removing 623 bases of exon 2. A 2.5kb fragment containing a neo-ura cassette was excised from pRAY-1 by digestion with HindIII, which was filled in with Klenow (Boehringer Mannhein), then with BglII and was ligated into the pBluescript-mSnm1, creating an out of frame insertion in exon 2. The disrupted mSnm1 construct was linearized with HindIII and transfected into embryonic stem (ES) cells from a 129/SvJ background, which were then inserted into blastocysts from the C57BL/6J background. Germ line insertions were screened by Southern blot and PCR (Fig. 1).
Figure 1.
A) Generating a disrupted allele of Snm1. A 623 bp cassette (stripes) was inserted into Exon 2 of Snm1, creating an out of frame disruption of the gene. The figure does not show all nine exons. H = HindIII, B = BGLII, E = Eco47III. B) Southern blot of Snm1, displaying a single insertion of the targeting vector into cells. The wild-type allele produces a band of 3.8 kb, while the mutant allele creates a band of 5.5 kb. C) PCR genotyping of wild-type and mutant mice.
Generating Snm1−/−, Fancd2−/− mice
The construction of the Fancd2−/+ mouse in C57BL/6J background has been described [40]. Mice heterozygous for the Snm1 allele in a 129/SvJ background were crossed with the Fancd2−/+ mice. The Fancd2 and Snm1 mice were bred to produce mice heterozygous for both mutations. Double heterozygous mice were then bred to produce mice homozygous for both mutations in a mixed C57BL/6J×129/SvJ background.
Mouse genotyping
The Snm1 allele was genotyped by PCR using the following primers: DB115, 5’-TCGCCAATGACAAGACGCTG; DB116, 5’-GTGGTAGTCCAAAAAACACGCC; DB117, 5’-CTGCCGTTACTTACAAGAGCCAG. The Fancd2 allele was genotyped as previously described [40].
Cell lines and culture
Transformed fibroblasts were cultured in α-MEM medium (Mediatech) supplemented with 5% fetal bovine serum (Hyclone), 5% calf serum (Hyclone) and 0.1% gentamicin (Gibco) in a humidified incubator with 5% CO2 at 37°C. GM639 cells were obtained from the NIGMS Human Genetic Cell Repository. Immortalized Fanconi anemia cell lines GM6914 (FANCA) were from NIGMS and PD331 (FANCC) were provided by the OHSU Fanconi Anemia Cell Repository. Mouse primary fibroblasts were derived from the adult ears for each of the genotypes of interest. The ears were soaked in 70% ethanol, rinsed several times with PBS, minced in RPMI medium (Gibco) containing collagenase (Gibco) and incubated at 37°C for three hours. The collagenase-containing medium was then replaced with DMEM medium containing 20% fetal calf serum (Hyclone) and the cells were grown at 37°C in a humidified incubator with 5% CO2.
Transfection
Transfections were performed as previously described [35]. Final volumes of 1 ml for a T25 flask and 3.2 ml for a 100 mm dish were used. Controls were transfected with either a non-functional siRNA duplex or an siRNA specific to lamin. The SNM1, ARTEMIS, and ERCC1 siRNA were SMARTpools (Dharmacon). A search of the NCBI expressed sequence database sowed that for the four siRNAs comprising the SNM1 SMARTpool, all had two or more mismatches with any known sequence.
Cell survival assay
Twenty-four hours following transfection, cells were plated on 100 mm dishes at 300 cells per dish and treated with MMC. Each data point was plated in duplicate. Cells were allowed to grow for 10 days in MMC, then fixed in a solution of 50% MeOH, 1% new methylene blue (Sigma). Colonies were counted, and standard errors of the means, denoted by bars, were derived from the duplicate data sets.
Immunoblotting
Cells were transfected on 100 mm dishes as described above, using a 3.2 ml final solution volume, then washed with PBS, trypsinized, pelleted, and frozen at −80°C at the indicated time points (Fig. 1). Cell lysates were prepared as previously described [35]. 50 µg whole protein extract from each lysate was run on a 7.5% acrylamide gel and then transferred to Immobilon-P PVDF membranes (Millipore). Membranes were blocked overnight in TBST (TBS plus 0.1% Tween-20) and 5% non-fat dry milk. The β-tubulin blots were probed with a rabbit polyclonal antibody (Santa Cruz) at a 1:3000 dilution in TBST. β-tubulin blots were then incubated with goat anti-rabbit IgG HRP antibody (Bio-Rad) at a 1:10,000 dilution in TBST with milk. FANCD2 blots were probed with a monoclonal antibody (Santa Cruz) at a 1:200 dilution in TBST with 5% non-fat milk. FANCD2 blots were incubated with goat anti-mouse (Bio-Rad) in the same conditions. Anti-ERCC1 (Santa Cruz) was diluted 1:400 in PBST (0.1% Tween-20) with 5% non-fat dry milk, and goat anti-mouse secondary antibody was diluted 1:5000 in PBST (0.1% Tween-20) with 5% non-fat dry milk. FANCD2-L was quantitated using Photoshop. Anti-Artemis (Novus Biologicals) was diluted 1:1000 in TBST (0.1% Tween-20) with 5% non-fat dry milk. The membrane was washed with TBST (0.15% Tween-20), followed by goat anti-rabbit secondary antibody diluted 1:10,000 in TBST (0.1% Tween-20) with 5% non-fat dry milk. Detection was performed using a chemiluminescence kit (Perkin Elmer).
Quantification of SNM1 mRNA by qRT-PCR
Cells were harvested twenty-four hours after transfection and RNA was stabilized using RNAlater (Ambion). Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and was quantified. Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) using 1 µg of starting RNA material. Real Time quantitative PCR was performed using the iCycler iQ Detection System (BioRad) with 10 ng of starting cDNA material using the TaqMan Gene Expression Assay specific to SNM1 (Applied Biosystems). Endogenous Control primers specific to β-Actin were used for the housekeeping gene (Applied Biosystems). Each sample was tested in triplicate. An amplification plot was created for each sample. Threshold values were calculated from the amplification plots correlating to the cycle number where fluorescence was detected above a calculated threshold. mRNA concentrations for each sample were calculated with the Ct value and were normalized versus. β-Actin expression. Negative controls for each primer were included in each experiment.
Chromosome stability
For chromosome breakage studies, cells were treated with MMC (5–40 ng/ml) or DEB (50–150 ng/ml) 24 hours after transfection or mock transfection. Following 48 hours incubation with the clastogens, cells were harvested as previously described [35]. Slides were stained with Wright’s stain, and 50 metaphases from each culture were scored for breaks and radial formation.
Cell cycle analysis
Cells were transfected in 100 mm dishes as described above. Duplicate plates were transfected to assure appropriate cell numbers. Twenty-four hours post transfection, cells were treated with the indicated amount of MMC and grown an additional 48 hours in the dark. Cells were harvested and fixed in ice cold 70% EtOH. Before analysis, cells were spun at 200 × g for 5 minutes, and the ethanol removed. They were then incubated for 2 hours at 37°C in 200 µl DNA extraction buffer (0.2 M phosphate citrate buffer, pH 7.8). The cells were then spun for 10 minutes at 1500 × g, and the supernatant was removed. Cells were stained in a 20 µg/ml solution of propidium iodide in PBS with 0.1% Triton X-100 and 200 µg/ml DNase-free RNase, and then incubated for 30 minutes at room temperature in the dark. The stained cells were sorted on a Becton Dickinson FACSCalibur machine, using CellQuest software for analysis. Approximately 20,000 cells per sample were analyzed.
Results
The Snm1−/− Mouse is viable and has normal life expectancy
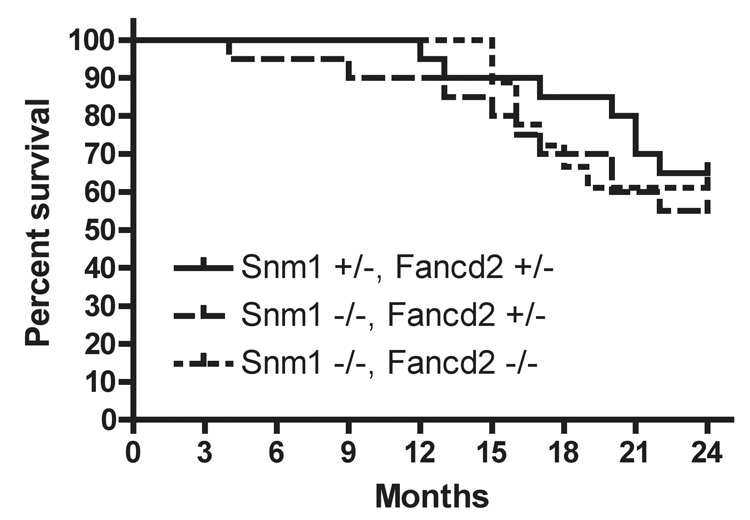
Snm1 gene disruption in mice leads to no obvious abnormalities, but the mice have been reported to die sooner than heterozygous mice when injected with large doses of MMC (50% mortality in eight days at 10mg/kg of MMC) [4]. We constructed mice (Fig. 1) with a frame-shifting disruption of most of exon 2. Homozygous Snm1−/− mice were born at the expected frequency and there was no imbalance regarding sex of offspring. Long-term survival of the mice in the 129Sv background also was not changed by the homozygous Snm1 mutation (Fig 2), with a p-value of 0.59 at twenty four months compared to heterozygous mice. Thus, in contrast to reports of a shortened life span in C57BL/6J background [26], we do not observe increased spontaneous mortality in the SNM1 knockout construct mice, using the 129SV background.
Figure 2.
Kaplan-Meier survival curve of Snm1−/− and Snm1/Fancd2−/− mice. The doubly heterozygous cohort was 20 animals, the Snm1−/−,Fand2+/− cohort was 20 animals and the doubly homozygously defective cohort was 18 animals.
Fibroblasts from the Snm1−/− mice show genome instability after MMC treatment
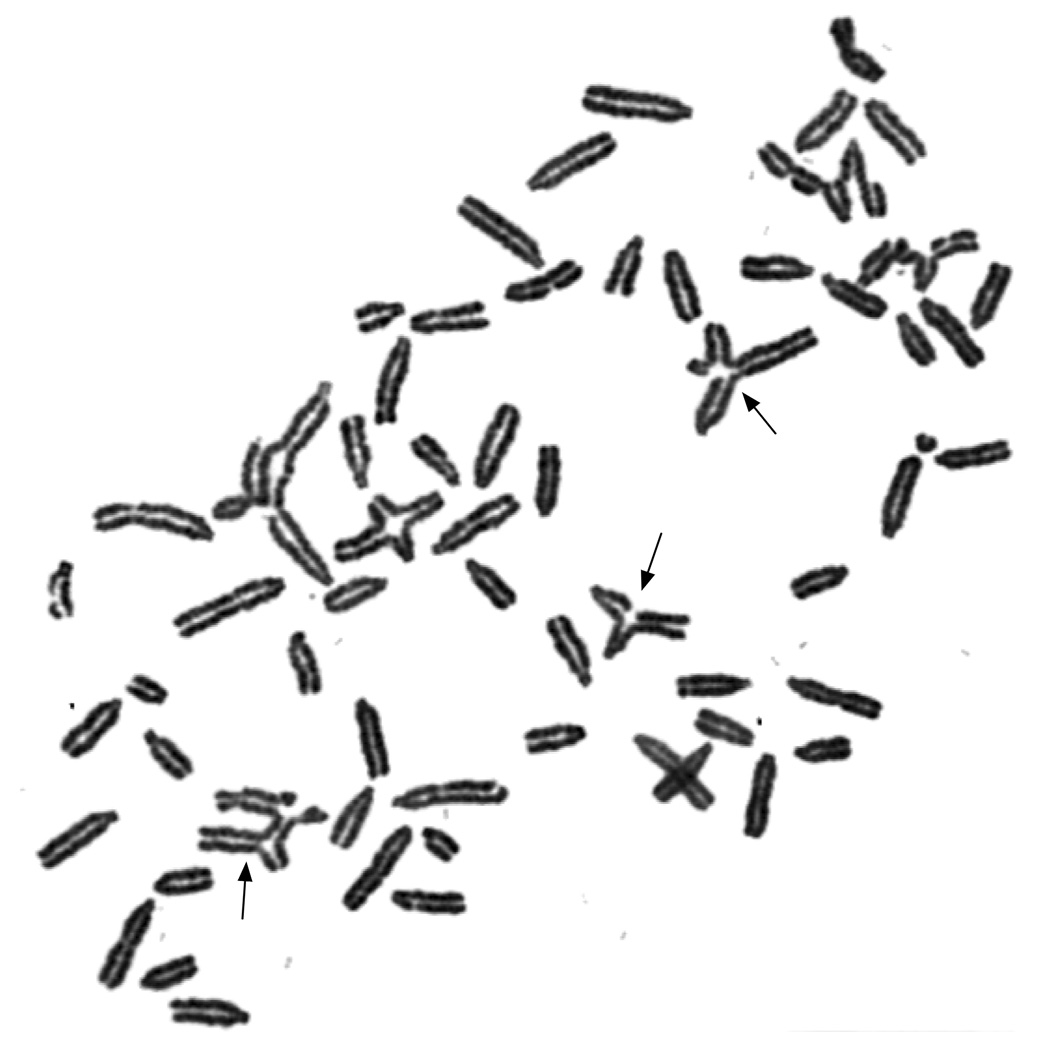
To evaluate the effect of the loss of Snm1 protein on genomic stability, murine fibroblast cell lines were established from ear flaps and tested for sensitivity to MMC, using chromosome stability as the end point by scoring breaks and radials in the telocentric chromosomes (Fig 3). Fibroblasts from Snm1−/− mice were significantly more sensitive to MMC than fibroblasts from their normal littermates (Table 1). The increase in the number of radials per cell was approximately 2-fold in normal cells after MMC, but 7-fold in Snm1−/− cells. These observations establish a role for Snm1 protein in maintenance of genome stability.
Figure 3.
Cytogenetic analysis of Snm1 mutant mouse fibroblasts reveals an increase in chromosomal breaks and radials (indicated by arrows) following MMC damage.
Table 1.
Radial formation in Snm1 mouse fibroblasts
| Cell Line | % Radials | Std. Error | p-value |
|---|---|---|---|
| Snm1 +/+ | 4.0 | 4.0 | |
| Snm1 −/− | 4.0 | 7.2 | 1.0 |
| Snm1 +/+ MMC | 9.0 | 1.4 | |
| Snm1 −/− MMC | 28.5 | 8.2 | 0.03 |
p-values were determined using a two-tailed paired t-test. MMC was 20 ng/ml.
Mice with a compound defect in Snm1 and Fancd2 show perinatal lethality
Since a defect in Snm1 introduces chromosomal instability similar to that seen in FA, mice with a combined defect in the pivotal Fancd2 protein and Snm1 were constructed to see if the defects were additive. The breeding program was to cross mice heterozygously defective for either one of the proteins. While there was no indication of lack of reproductive fitness in either of the constructs alone, in the backgrounds used, there was a significant shortfall in the number of expected compound homozygously defective mice born (Table 2). Since partial perinatal lethality has been reported for Fanc−/− in C57BL mice [40,41], this observation is not surprising. Beginning with a different breeding stock, with a higher percentage of 129/SvJ background, we observed expected ratios of offspring. Thus the partial perinatal lethality seems to be an epigenetic effect of the C57BL background. The semi-lethality in the combination mutants is compatible with Snm1 and Fancd2 acting in different pathways for genome stability. This suggests non-epistasis, since partial perinatal lethality was not observed for Fancd2 homozygous mutants in our breeding stocks, in contrast to others [40,41].
Table 2.
Results of breeding Snm1 +/−, Fancd2 +/− mice.
| Snm1 −/− | ||||
|---|---|---|---|---|
| Genotype | ||||
| Fancd2 +/+ | Fancd2 +/− | Fancd2 −/− | ||
| First 190a | 21 (12) | 22 (24) | 8 (12) | |
| Observed (Expected) | ||||
| Next 220 | 15 (14) | 26 (27) | 14 (14) | |
The p-value for the variance from predicted values for the first 190 is less than 0.02, calculated by the chi-squared test. Expected numbers are in parentheses.
Depletion of hSNM1 produces sensitivity to ICLs in human fibroblasts
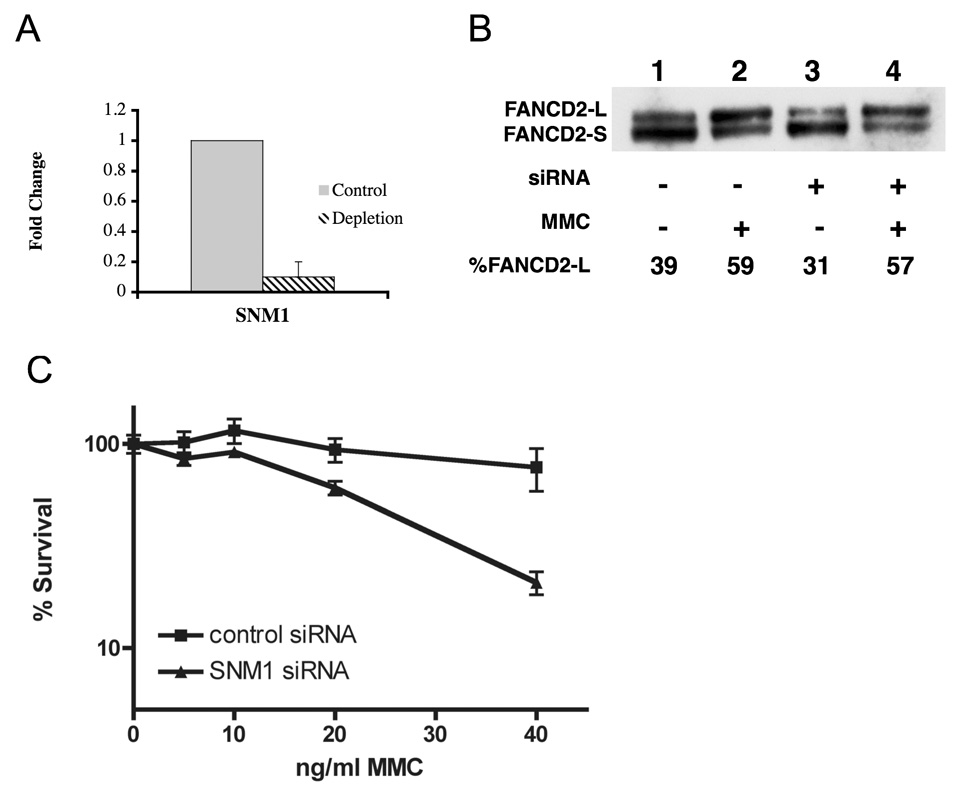
In order to obtain human cells deficient in SNM1, siRNA depletion was used. siRNA-mediated depletion of hSNM1 protein leads to an increase in radial formation after exposure to MMC or diepoxybutane (DEB) (data not shown) (Table 3). This result agrees with the finding of decreased survival of human fibroblasts depleted for hSNM1 after exposure to MMC (Fig. 4). Thus by two standards, radial formation and cell survival, depletion of hSNM1 increases sensitivity to ICLs. Depletion of hERCC1, another NER pathway protein known to act in ICL repair [42] also increases sensitivity to ICLs ( Figure 5B and Table 3). However, depletion of XPC, a NER enzyme known not to act in the repair of ICLs, did not cause an increase in radial formation following ICL formation (data not shown).
Table 3.
Radial formation in human fibroblasts with siRNA depletion.
| Cell Line | % Radials | Std. Error | p-value |
|---|---|---|---|
| GM639 + control siRNA | 0.7 | 1.2 | |
| GM639 + SNM1 siRNA | 0.0 | 0.0 | 0.4 |
| GM639 + ARTEMIS siRNA | 0.0 | 0.0 | |
| GM639 + ERCC1 siRNA | 2.0 | 1.0 | 0.4 |
| GM639 + control siRNA + MMC | 20.0 | 15.6 | |
| GM639 + SNM1 siRNA + MMC | 41.3 | 11.6 | 0.01 |
| GM639 + ARTEMIS siRNA + MMC | 18.0 | 11.3 | |
| GM639 + ERCC1 siRNA + MMC | 28.2 | 4.5 | 0.003 |
| GM6914 + control siRNA | 6.4 | 2.2 | |
| GM6914 + SNM1 siRNA | 3.2 | 1.8 | 0.1 |
| GM6914 + control siRNA + MMC | 48.0 | 4.0 | |
| GM6914 + SNM1 siRNA + MMC | 65.5 | 6.4 | 0.002 |
p-values were determined using a two-tailed paired t-test. MMC was 5 ng/ml for GM6914 and 40 ng/ml for GM639.
Figure 4.
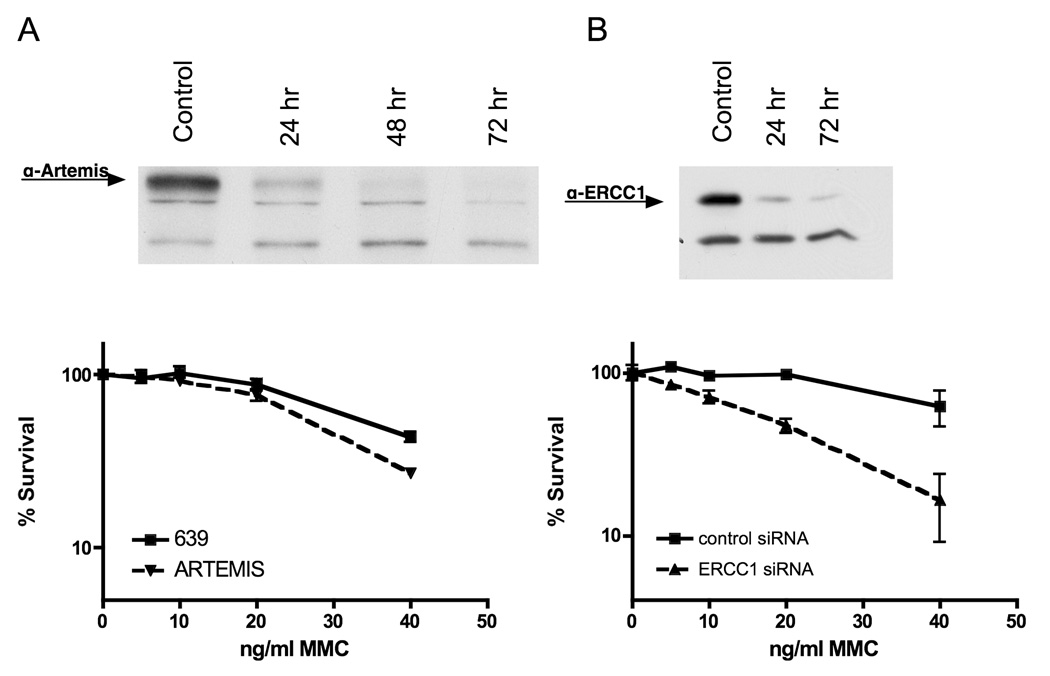
Depletion of SNM1 in human fibroblasts by siRNA results in increased sensitivity to MMC and increased radial formation. A) An siRNA specific to SNM1 results in over 90% reduction of SNM1 mRNA compared to a control siRNA in wildtype human fibroblasts (GM639). B) Depletion of SNM1 by siRNA in GM639 fibroblasts. Following depletion, and extraction the immunoblot was probed with anit-FANCD2 antibody. The percent of FAND2-L form is indicated for each blot. C) Depletion of SNM1 by siRNA results in decreased survival of fibroblasts after MMC compared to a control siRNA.
Figure 5.
Survival of human fibroblasts after depletion of ARTEMIS (A) or ERCC1 (B) with MMC treatment. Immunoblots demonstrate depletion.
In contrast to hSNM1, depletion of Artemis does not lead to increased radials or decreased cell survival after exposure to MMC (Figure 5A and Table 2). Rather, Artemis deficiency has been shown to lead to sensitivity to ionizing radiation (IR) damage [21]. Thus homologs in the β-CASP family have strikingly different functions in response to, and repair specificity for, DNA damage.
SNM1 is non-epistatic with the FA pathway for genome stability
To test whether or not SNM1 acts in the FA pathway, we depleted SNM1 in GM6914 cells, a cell line deficient for FANCA. If SNM1 functions in the FA pathway, it would be expected that FA cells depleted of SNM1 would not show an increase in radial formation following treatment with DNA interstrand crosslinker. However, as in the normal cell line, there was an increase in radial formation in FANCA cells following treatment with ICL agents in cells depleted for SNM1 (Table 3). Activation of the FA pathway following DNA damage is reflected in mono-ubiquitination of FANCD2 (FANCD2-Ub), resulting in a FANCD2 of greater molecular weight (FANCD2-L). Thus, if SNM1 functioned in the FA pathway in a step required for this activation, FANCD2-L should not be formed in cells depleted for SNM1. Depletion of hSNM1 did not reduce FANCD2-L formation after MMC treatment (Figure 4 B). This is compatible with SNM1 acting in an entirely separate pathway from FA. Taken together the results support SNM1 being non-epistatic with the FA pathway.
Discussion
SNM1, a member of the β-CASP family, is specific for repair of ICLs in yeast. In order to study the role of SNM1 in mammalian DNA repair, we created a mouse model knockout of the gene. Mice with a deletion in Snm1 are born at expected frequencies, and show no gross anatomical abnormalities. They also have a normal life expectancy. However, fibroblast cell lines derived from Snm1 mutant mice show sensitivity to ICLs, as shown by increased chromosomal breaks and radial formation as well as decreased cell survival after MMC treatment. Also, Snm1 mice crossed with a Fancd2 deficient mouse show a strain-specific decrease in doubly homozygous mutant mice. Thus it appears that there is a defect in genome stability in Snm1 mutants that is not sufficient to cause decreased survival unless combined with another defect. This observation implies a defect in two separate pathways. Because of the homology with Artemis protein and its role in the immune response, with defects being associated with a SCID phenotype, mice defective in Snm1 were inspected for immune system function. T-cell numbers were identical between Snm1−/− mice and wild-type littermates as determined by CD4 FITC-labeled antibody and FACS analysis.
Like the mouse cells deficient in Snm1, normal human fibroblasts depleted for SNM1 show sensitivity to ICLs, as measured by decreased survival and increased breaks and radials in the presence of mitomycin C. Depleting SNM1 in a cell line with a mutation in the Fanconi anemia protein FANCA increased the amount of radial formation. This, along with the decrease in perinatal survival of Snm1/Fancd2 double mutant mice, indicates that SNM1 is non-epistatic to the FA pathway of ICL response.
The results indicate that there are at least two pathways for maintenance of genome stability following ICL formation in mammalian cells, and that components of the NER pathway function in one such response. This is not surprising. There are three known pathways for the repair of ICLs in yeast [15], which do not have any of the known FA homologs. The pathways define three epistasis groups for ICL repair. Thus, the HR pathway does not function in the PRR pathway or the NER pathway. The same picture is emerging for mammalian cells.
Recently a patient has been identified with a severe XP-F defect [43]. The patient manifests a progeroid syndrome and cellular sensitivity to ICL, implicating the ERCC1/XP-F dyad in ICL repair in humans. As noted above, depletion of ERCC1 in human fibroblasts results in increased radial formation after MMC treatment. Since ERCC1, XP-F and SNM1 are all in the NER pathway, it might be expected that hSNM1 defects would result in a similar phenotype, if compatible with life. This remains to be shown, since there are no patients identified lacking hSNM1 activity.
A striking finding is that the closely related members of the β-CASP family appear to serve different roles in response to DNA damage. Since the active site for the exonuclease is conserved and several members are demonstrated to be 5’-exonucleases, it might have been expected that the function would be the same. However, hSNM1, a 5’-exonuclease, does not show detectable endonuclease activity, whereas Artemis does. As reported here, depletion of hSNM1, but not Artemis, leads to cellular sensitivity to MMC. Thus the presence of a 5’-exonuclease does not reflect the subtle variation in DNA processing the proteins demonstrate. The activity of Artemis in V(D)J rejoining appears to be via the NHEJ mechanism. The lack of increased sensitivity to ICLs we observe suggests that the Artemis activity does not contribute ICL repair, implying that NHEJ function of the Artemis type does not play a role in acting on intermediates of ICL repair.
Acknowledgment
This work was supported by NHLBI Program Project Grant 1PO1HL48546.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li X, Moses RE. The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair (Amst) 2003;2:121–129. doi: 10.1016/s1568-7864(02)00192-1. [DOI] [PubMed] [Google Scholar]
- 2.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 3.Poinsignon C, Moshous D, Callebaut I, de Chasseval R, Villey I, de Villartay JP. The metallo-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J Exp Med. 2004;199:315–321. doi: 10.1084/jem.20031142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dronkert ML, de Wit J, Boeve M, Vasconcelos ML, van Steeg H, Tan TL, Hoeijmakers JH, Kanaar R. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol Cell Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science. 1996;274:1514–1517. doi: 10.1126/science.274.5292.1514. [DOI] [PubMed] [Google Scholar]
- 6.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, Desrochers M, Dumont M, Farnham JM, Frank D, Frye C, Ghaffari S, Gupte JS, Hu R, Iliev D, Janecki T, Kort EN, Laity KE, Leavitt A, Leblanc G, McArthur-Morrison J, Pederson A, Penn B, Peterson KT, Reid JE, Richards S, Schroeder M, Smith R, Snyder SC, Swedlund B, Swensen J, Thomas A, Tranchant M, Woodland AM, Labrie F, Skolnick MH, Neuhausen S, Rommens J, Cannon-Albright LA. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 7.Bonatto D, Brendel M, Henriques JA. The eukaryotic Pso2p/Snm1p amily revisited: in silico analyses of Pso2p A, B and Plasmodium groups. Comput Biol Chem. 2005;29:420–433. doi: 10.1016/j.compbiolchem.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Brendel M, Bonatto D, Strauss M, Revers LF, Pungartnik C, Saffi J, Henriques JA. Role of PSO genes in repair of DNA damage of Saccharomyces cerevisiae. Mutat Res. 2003;544:179–193. doi: 10.1016/j.mrrev.2003.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Henriques JA, Moustacchi E. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics. 1980;95:273–288. doi: 10.1093/genetics/95.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Hejna J, Moses RE. The yeast Snm1 protein is a DNA 5′-exonuclease. DNA Repair (Amst) 2005;4:163–170. doi: 10.1016/j.dnarep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Hejna J, Philip S, Ott J, Faulkner C, Moses R. The hSNM1 protein is a DNA 5′-exonuclease. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonatto D, Revers LF, Brendel M, Henriques JA. The eukaryotic Pso2/Snm1/Artemis proteins and their function as genomic and cellular caretakers. Braz J Med Biol Res. 2005;38:321–334. doi: 10.1590/s0100-879x2005000300002. [DOI] [PubMed] [Google Scholar]
- 13.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase old within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Richie C, Legerski RJ. Translation of hSNM1 is mediated by an internal ribosome entry site that upregulates expression during mitosis. DNA Repair (Amst) 2002;1:379–390. [PubMed] [Google Scholar]
- 15.Grossmann KF, Ward AM, Matkovic ME, Folias AE, Moses RE. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat Res. 2001;487:73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- 16.Barber LJ, Ward TA, Hartley JA, McHugh PJ. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol. 2005;25:2297–2309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossmann KF, Ward AM, Moses RE. Saccharomyces cerevisiae lacking Snm1, Rev3 or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment. Mutat Res. 2000;461:1–13. doi: 10.1016/s0921-8777(00)00035-5. [DOI] [PubMed] [Google Scholar]
- 18.Laurencon A, Orme CM, Peters HK, Boulton CL, Vladar EK, Langley SA, Bakis EP, Harris DT, Harris NJ, Wayson SM, Hawley RS, Burtis KC. A large-scale screen for mutagen-sensitive loci in Drosophila. Genetics. 2004;167:217–231. doi: 10.1534/genetics.167.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, Hu D, de Villartay JP, Cowan MJ. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J Immunol. 2002;168:6323–6329. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 20.Moshous D, Callebaut I, de Chasseval R, Poinsignon C, Villey I, Fischer A, de Villartay JP. The V(D)J recombination/DNA repair factor artemis belongs to the metallo-beta-lactamase family and constitutes a critical developmental checkpoint of the lymphoid system. Ann N Y Acad Sci. 2003;987:150–157. doi: 10.1111/j.1749-6632.2003.tb06043.x. [DOI] [PubMed] [Google Scholar]
- 21.Noordzij JG, Verkaik NS, van der Burg M, van Veelen LR, de Bruin-Versteeg S, Wiegant W, Vossen JM, Weemaes CM, de Groot R, Zdzienicka MZ, van Gent DC, van Dongen JJ. Radiosensitive SCID patients with Artemis gene mutations show a complete B-cell differentiation arrest at the pre-B-cell receptor checkpoint in bone marrow. Blood. 2003;101:1446–1452. doi: 10.1182/blood-2002-01-0187. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Salido E, Zhou Y, Bhattacharyya S, Yannone SM, Dunn E, Meneses J, Feeney AJ, Cowan MJ. Targeted disruption of the Artemis murine counterpart results in SCID and defective V(D)J recombination that is partially corrected with bone marrow transplantation. J Immunol. 2005;174:2420–2428. doi: 10.4049/jimmunol.174.4.2420. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 24.Poinsignon C, de Chasseval R, Soubeyrand S, Moshous D, Fischer A, Hache RJ, de Villartay JP. Phosphorylation of Artemis following irradiation-induced DNA damage. Eur J Immunol. 2004;34:3146–3155. doi: 10.1002/eji.200425455. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahkter S, Richie CT, Zhang N, Behringer RR, Zhu C, Legerski RJ. Snm1-deficient mice exhibit accelerated tumorigenesis and susceptibility to infection. Mol Cell Biol. 2005;25:10071–10078. doi: 10.1128/MCB.25.22.10071-10078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akhter S, Richie CT, Deng JM, Brey E, Zhang X, Patrick C, Jr, Behringer RR, Legerski RJ. Deficiency in SNM1 abolishes an early mitotic checkpoint induced by spindle stress. Mol Cell Biol. 2004;24:10448–10455. doi: 10.1128/MCB.24.23.10448-10455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Andrea AD. The Fanconi road to cancer. Genes Dev. 2003;17:1933–1936. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 29.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell. 2007 doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 31.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 32.Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G, de Winter JP, Wang W, Joenje H. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 33.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101:2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- 35.Bruun D, Folias A, Akkari Y, Cox Y, Olson S, Moses R. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst) 2003;2:1007–1013. doi: 10.1016/s1568-7864(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 36.Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, de Winter JP, Ashworth A, Jones NJ, Mathew CG. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 37.Folias A, Matkovic M, Bruun D, Reid S, Hejna J, Grompe M, D'Andrea A, Moses R. BRCA1 interacts directly with the Fanconi anemia protein FANCA. Hum Mol Genet. 2002;11:2591–2597. doi: 10.1093/hmg/11.21.2591. [DOI] [PubMed] [Google Scholar]
- 38.Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12:2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 42.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]