Abstract
Diversification of mammalian species began more than 160 million years ago when the egg-laying monotremes diverged from live bearing mammals. The duck-billed platypus (Ornithorhynchus anatinus) and echidnas are the only potential contemporary witnesses of this period and, thereby, provide a unique insight into mammalian genome evolution. It has become clear that small RNAs are major regulatory agents in eukaryotic cells, and the significant role of non-protein-coding (npc) RNAs in transcription, processing, and translation is now well accepted. Here we show that the platypus genome contains more than 200 small nucleolar (sno) RNAs among hundreds of other diverse npcRNAs. Their comparison among key mammalian groups and other vertebrates enabled us to reconstruct a complete temporal pathway of acquisition and loss of these snoRNAs. In platypus we found cis- and trans-duplication distribution patterns for snoRNAs, which have not been described in any other vertebrates but are known to occur in nematodes. An exciting novelty in platypus is a snoRNA-derived retroposon (termed snoRTE) that facilitates a very effective dispersal of an H/ACA snoRNA via RTE-mediated retroposition. From more than 40,000 detected full-length and truncated genomic copies of this snoRTE, at least 21 are processed into mature snoRNAs. High-copy retroposition via multiple host gene-promoted transcription units is a novel pathway for combining housekeeping function and SINE-like dispersal and reveals a new dimension in the evolution of novel snoRNA function.
Small nucleolar RNAs (snoRNAs) are essential for RNA maturation, guiding 2′-O-ribose methylation (C/D-box snoRNAs) and pseudouridylation (H/ACA-box snoRNAs) of chiefly ribosomal RNAs, the central components of the protein synthesis machinery. Such an important housekeeping function implies high genomic conservation. However, in nematodes, we recently found an extraordinary degree of plasticity in the generation of snoRNA paralogs as well as in the selection of their modified targets (Zemann et al. 2006). In a process termed cis-duplication, we found that some snoRNAs duplicated from one intronic location to a neighboring intron of the same gene. Others distributed via trans-duplication to distant locations in other genes or chromosomes. One mechanism of trans-duplication is retrotransposition (Vitali et al. 2003). In mammals, nonautonomous retroposons such as short interspersed elements (SINEs) are predominantly dispersed by an autonomous long interspersed element 1 (LINE1; L1)-mediated mechanism. The autonomous, non-LTR (long terminal repeat), L1 retroposons provide the machinery needed for the reverse transcription and integration of the associated nonautonomous retroposons. In mammals the activity of L1 elements is restricted to therians (placentals and marsupials). To date, no L1 activity has been demonstrated for monotremes (Kordis et al. 2006); thus, L1-mediated co-retroposition of RNAs is also expected to be restricted to therian mammals. In monotremes, a different retropositional landscape, mediated by the L2 LINE machinery, dominates. Representative mobile elements retroposed in this way include the L2-mobilized mammalian-wide interspersed elements (MIRs) and the monotreme-specific (Mon1) SINEs. Another class of retroposon-like non-LTR transposable elements (RTEs) facilitating retroposition, originally discovered in Caenorhabditis elegans (Youngman et al. 1996), is occasionally represented in mammals (Malik and Eickbush 1998), comprising, for example, ∼2.3% of the Monodelphis domestica (a small South American opossum) genome (Gentles et al. 2007). snoRNA distribution associated with nonautonomous retroposition usually occurs only in few copies (Weber 2006; Luo and Li 2007). However, this is remarkably different in platypus, where we have identified about 40,000 genomic copies of a platypus-specific snoRNA retroposon we have called a snoRTE, reflecting its hybrid nature consisting of a snoRNA and an RTE-like sequence. Here we provide evidence for a platypus-specific processing pathway that differentially leads to H/ACA snoRNAs or full-length snoRTE retroposons.
Results and Discussion
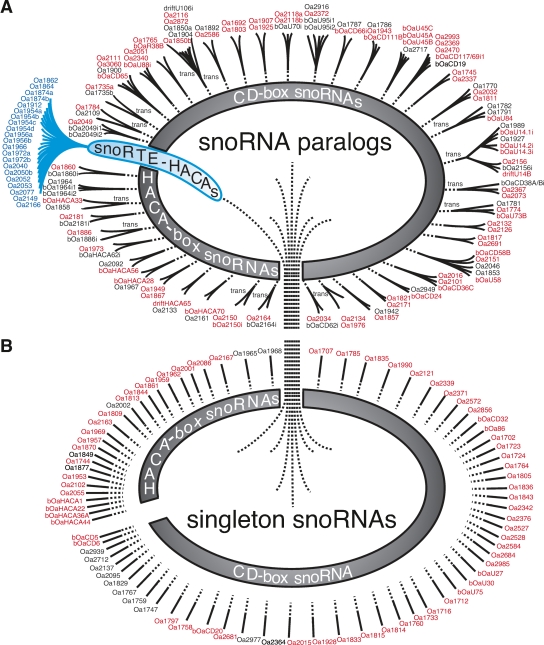
In a platypus brain cDNA library generated from small non-protein-coding RNAs (npcRNAs), we identified 166 individual snoRNAs among more than 20,000 cloned sequences. Furthermore, BLAST searches of platypus genomic sequences revealed an additional 51 such snoRNAs (for a complete description, see Fig. 1; Supplemental Fig. S1; Supplemental material). Seventy-three of the snoRNAs are H/ACA snoRNAs, and 144 are C/D snoRNAs. A comparative analysis of these snoRNAs with the genomic information from other key vertebrate species revealed that 49 are platypus/monotreme-specific (Supplemental Material 2). Of the remaining 168, we detected 146 orthologs in non-mammalian vertebrates, and six were present in all mammals but were clearly absent in birds and other non-mammalian species (Fig. 2). The remaining 16 snoRNAs may belong to either of the last two groups but could not be clearly assigned because of a lack of corresponding sequence information in non-mammalian species. The high degree of conservation among vertebrate snoRNAs is indicative of the purifying selection on their function in modifying housekeeping RNAs. At the same time, snoRNAs also show a high degree of plasticity among their paralogs (Fig. 2; Supplemental material). This dualism of conservation levels is similar to that of certain vertebrate populations of miRNAs (Tanzer and Stadler 2006).
Figure 1.
snoRNA representation in the genome of platypus. (A) snoRNAs with corresponding paralogs distributed by cis- or trans-duplication (trans). (Red lettering) snoRNAs conserved in human. snoRNAs are grouped as CD-box snoRNAs, H/ACA-box snoRNAs, and snoRTE-H/ACAs, the last group was generated by retroposition. (B) Nonduplicated snoRNAs. (Oa) Experimentally identified snoRNAs; (bOa) snoRNAs identified computationally. Diverged snoRNAs (drift) show no canonical structural requirements necessary for function.
Figure 2.
Conservation and plasticity of platypus snoRNAs. Phylogenetic tree of mammals modified after Kriegs et al. (2006). (Black vertical bars) The conservation (platypus-specific, mammalian-specific, and presence in at least mammals plus birds) of the platypus snoRNAs. The numbers above the black bars represent the number of experimentally identified snoRNA orthologs that show clear presence or absence in the corresponding groups. The numbers in parentheses denote mammalian conserved loci for which no non-mammalian sequence information was accessible. (Gray bars) Examples of shifts from one potentially active paralog (a) to another potentially active paralog (b) at different genomic locations (the details are presented as Supplemental material). The snoRNA designations in the cDNA library are Oa1973, Oa2916, and Oa2364.
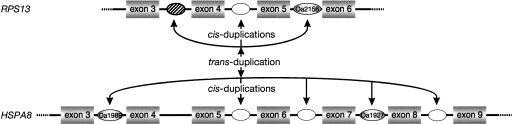
Forty-five of the snoRNAs had undergone one or more duplications in platypus, yielding a total of 138 paralogs (the number of paralogs refers to the total number of related forms with a common origin), 91 of which were C/D-box and 47 were H/ACA-box snoRNAs (Fig. 1). The majority of C/D and H/ACA snoRNAs in our study were generated via cis-duplication (119 paralogs). This is somewhat different in nematodes—the only other species for which such analysis is available—in which only H/ACA snoRNAs were dispersed in this way (Zemann et al. 2006). If the immediate neighboring introns were already occupied by other snoRNAs, the next free intron was often targeted. In nine of 138 cis-duplications, we observed the birth of novel platypus-specific expressed C/D snoRNA paralogs into neighboring introns, evidenced by the fact that the orthologous loci in other mammals clearly lacked the snoRNA paralogs. Among the cDNA sequences, we found only a few cases of paralogs generated via trans-duplication (nine C/D and four H/ACA snoRNAs). A combination of cis- and trans-duplication was observed for the eight C/D snoRNA paralogs hosted by the ribosomal protein S13 (RPS13) and the heat-shock protein 8 (HSPA8) genes in platypus (Fig. 3). In this case, three paralogs (including one highly diverged form) reside in neighboring introns in the RPS13 gene, and five closely related paralogs are located in adjacent introns of the HSPA8 gene. Based on sequence similarities, we propose that a single trans-duplication event initiated further cis-dispersal within one of the two genes. In nematodes, we found only single duplication events (Zemann et al. 2006), while in one particular H/ACA platypus snoRNA, at least 20 duplications occurred leading to 21 paralogs in our cDNA library (Supplemental material). Compared to known duplication patterns, this extraordinarily high number is rather unusual. Alignment of these sequences to platypus genomic data revealed a highly conserved 229-nt 3′-flank (Supplemental material). Parts of the conserved flank exhibit ∼70% sequence similarity to the 3′-region of the RTE-mobilized marsupial MAR1b-SINEs and ∼90% sequence similarity to BovB_Plat, a recently characterized LINE-related RTE retroposon (Arian Smit, unpubl.; Fig. 4). RTE retroposons described for ruminants (Okada and Hamada 1997), afrotherians (Gogolevsky et al. 2007), and marsupials (Gentles et al. 2007) show a mosaic distribution in mammals and are proposed to be dispersed by horizontal transfer (Zupunski et al. 2001; Piskurek and Okada 2007). A genomic BLAST of the 372-nt consensus sequence in platypus, including the snoRNA plus the conserved flanking region, revealed 14,844 full-length (∼27% with perfect direct repeats ≥10 nt) and 27,332 5′- and/or 3′-truncated copies. One thousand nine hundred and five of the full-length forms and 346 of the truncated forms (but including the entire snoRNA) were located in currently annotated introns (out of a total of about 142,000 introns), with a significant preference for the antisense orientation relative to the transcribed direction of the host gene (1286 of 2251; χ2-test, P < 0.0001).
Figure 3.
Cis- and trans-duplication of C/D snoRNA paralogs in platypus. Cis-duplications of three paralogs of a C/D snoRNA were detected in the gene for ribosomal protein S13 (RPS13). One of these was found in the cDNA library (snoRNA Oa2156); the second was detected by BLAST in the neighboring intron and is probably functional (oval between exons 4 and 5; intact box motifs, conserved between platypus and humans), and the third, also detected by BLAST, is diverged and most likely nonfunctional (based on structural requirements) in platypus but functional in human, mouse, and cow (oval between exons 3 and 4). Trans-duplication of one of these paralogs occurred to the 70-kDa heat-shock protein 8 gene (HSPA8) or vice versa. Within the HSPA8 gene, we also found two paralogs in our cDNA library that evolved via cis-duplications (snoRNAs Oa1989 and Oa1927). Three additional, potentially functional paralogs were found by BLAST search and are conserved between platypus and humans. (Filled ovals) snoRNAs in our platypus cDNA library; (open ovals) snoRNAs located by BLAST search with the library sequences; (hatched oval) a nonfunctional snoRNA in platypus.
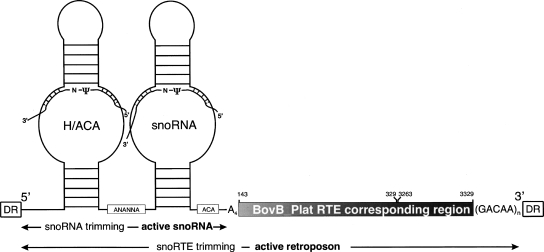
Figure 4.
Structure of the novel class of platypus snoRTEs. snoRTEs are composed of a 5′-H/ACA-snoRNA with the H-box consensus sequence ANANNA and ACA-box sequence (structured part). The sequences that potentially interact with ribosomal or spliceosomal RNA by base complementarities guiding pseudouridylation are indicated (N-ψ). The (gray bar) 3′ part is similar to RTE-elements. Coordinates are given for the BovB_Plat RTE LINE corresponding 145-nt 5′ and 67-nt 3′ sequence regions. [(GACAA)n] Terminal simple repeats; (A4) a spacer sequence between the H/ACA snoRNA and BovB_Plat region. The flanks of the entire snoRTE exhibit (DR) direct repeats. The snoRTE is distributed via retroposition (retro-duplication), is dependent on the host gene RNA polymerase II promoter, and is probably subsequently trimmed in the presence of RTE proteins to a retropositionally active RNA, absence to an H/ACA-box snoRNA.
The high copy number, flanking direct repeats, and the high similarity and temporal overlap in activity to BovB_Plat RTE-elements (Supplemental Fig. S2) are indications of a novel retroposon class (termed snoRTE) associated with RTE-autonomous retroposition. snoRTEs are platypus/monotreme-specific and could not be detected in other vertebrates (Supplemental Material 2). A typical vertebrate snoRNA cotranscription embedded in an RNA polymerase II-transcribed host gene RNA with subsequent trimming of the flanking regions (Cavaillé and Bachellerie 1996) and an associated SINE-like retroposition via an RTE-mechanism represents a novel dual “symbiotic” evolutionary pathway of snoRNA and retroposon propagation manifested as a snoRTE. Splicing and differential trimming might lead to either snoRNAs or snoRTEs (Fig. 4). RNAs of both forms were identified by Northern blot analysis (Supplemental Fig. S3). We propose that in the absence of active snoRTE retroposition, the classical snoRNA trimming process (Cavaillé and Bachellerie 1996) removes the 3′-RTE part. The remaining processed sequence constitutes the mature H/ACA-snoRNA. Alternatively, in the presence of an active RTE retroposition machinery, the RTE part of the snoRTE is associated with proteins forming a ribonucleoprotein complex. This association might protect the 3′-snoRNA part against further trimming. Consequently, the partially processed RNA constitutes the retropositionally active snoRTE. These elements are a source for new snoRNA isoforms and potentially new functions apart from originally modifying a specific 28S rRNA target sequence (Supplemental Fig. S4). Interestingly, the significantly lower number of sense-oriented snoRTEs (potential retropositionally active; 965 copies) compared with the antisense-oriented snoRTEs (potential retropositionally inactive; 1286 copies) indicates selection that might prevent excessive intronic persistence of these particular snoRTEs. Recently Weber described a small number of mammalian-specific, L1 reverse-transcribed snoRNAs with random locus-specific 3′-regions termed snoRTs (Weber 2006). It was proposed that the inefficient retropseudogene-like distribution of such elements is due to lack of the cytoplasmic phase of nucleolar RNAs in their retroposition cycle. However, we previously demonstrated that, in the case of mammalian-specific tailless retropseudogenes, a cytoplasmatic phase is not necessary for efficient retroposition (Schmitz et al. 2004). Hence, we rather propose that the retropositionally inefficient 3′-tail of snoRTs is responsible for the low copy number in the L1-mediated retroposition. On the other hand, the highly efficient RTE-mediated platypus snoRTE retroposition is favored by a BovB RTE LINE-like 3′-end, fortuitously acquired after trans-duplication of the snoRNA.
A BLAST search for this new snoRTE in all major vertebrate clades revealed the snoRNA’s original intronic locus in the RPL32 gene, which is conserved from human to lizard. At that locus in platypus, however, a diverged, nonfunctional copy (lacking the ACA-box) was identified. In contrast, for most of all 21 retroposed paralogs identified in the cDNA library, the predicted secondary structures characteristic for snoRNAs are highly conserved, further indicating their possible functionality.
Conservation and diversity are two major opposing forces shaping the evolution of genomes. As an example, the interplay of these forces is clearly observed in the presence of both constitutive and alternative splicing (Xing and Lee 2006; Krull et al. 2007) or of both genes and pseudogenes (Long et al. 2003). The duplication processes of highly conserved npcRNAs, like snoRNAs, are important mechanisms for maintaining advantageous functions, and at the same time for evolving potentially novel ones. This fundamental mechanism of duplication accompanied by the functional change of one of the paralogs has been described in relatively simple multicellular nematodes (Zemann et al. 2006), and the analysis of platypus snoRNAs clearly demonstrates that a similar diversification occurs in vertebrates. It is interesting that, although jumping snoRNAs represent an unusual pathway to generate genomic and functional diversity, the high degree of amplification inherent in the “strategy” followed by sense-oriented intronic insertion appears to be limited by unknown regulatory mechanisms. This is clearly noticeable in the restricted amount of such snoRTEs compared to the number of antisense-oriented, retropositionally inactive elements. In general, the birth of new retropositionally active master genes is quite rare. New master genes require that the internal RNA polymerase III promoter elements in the corresponding SINE be fortuitously juxtaposed at the right distance to sequences at the locus of integration that act as external RNA polymerase III promoter elements (Ludwig et al. 2005; Khanam et al. 2007). Hence, most SINE copies are transcriptionally silent. The existence of a snoRTE that can hitch a ride on virtually any intron-containing RNA polymerase II transcript might pose a great challenge to the organism, as, unchecked, and owing to the extremely high number of potential active snoRTE “mastergenes” compared with the temporary activity of one or a few mastergenes (e.g., for SINE retroposition), it might develop into a dangerous runaway situation of retroposon amplification. It was shown in Drosophila that an increased genomic copy number of transposable elements had deleterious effects on organismal fitness (Pasyukova et al. 2004). In the case of the significant dominance of antisense- over sense-oriented intronic snoRTEs, which are cotranscribed but not capable of being retroposed, the platypus genome might already have developed countermeasures, whose mechanisms will be interesting to reveal.
Methods
Brain tissue was obtained from an adult male duck-billed platypus (Ornithorhynchus anatinus) collected at the upper Barnard river, New South Wales (Animal ethics permit no. R.CG.07.03 to F.G.). Total RNA was prepared using the TRIzol method as previously described (Zemann et al. 2006) and fractionated on denaturing PAGE gels. The 10–60-nt (small) and 60–500-nt (large) RNA fractions were excised, passively eluted, ethanol-precipitated, and then C-tailed (DeChiara and Brosius 1987). Full-length cDNAs were generated using the adapter ligation protocol (Chen et al. 2007) with a SalI restriction site and T4 RNA ligase. First-strand cDNA was synthesized with a Thermoscript cDNA synthesis kit (Invitrogen) using a NotI primer. The RNA–cDNA hybrid strand was amplified using a SalI primer. After SalI and NotI double digestion, the cDNAs were cloned into the pSPORT1 vector (Invitrogen) and transformed. cDNAs were sequenced using M13 standard primers (detailed methodologies are included in the Supplemental Methods).
A total of 11,521 and 10,369 cDNA clones were sequenced from the small and large RNA fractions, respectively. Vectors were trimmed and assembled with a modified version of the DNASTAR Lasergene 7.1 package. For mammalian comparative analyses, we used NCBI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi), the Ensembl Platypus-BLAST (http://www.ensembl.org/Multi/blastview), and the UCSC BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat); for RNA structures, the RNAfold program, mfold (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi); for scanning additional snoRNAs, the Ribosomal Protein Genes Database (http://ribosome.med.miyazaki-u.ac.jp/); and for analysis of transposed elements, RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker).
The 21 paralogs of the snoRTE were BLASTed against platypus genomic sequences and the 3′-conserved RTE-like parts extended. Full-length snoRTEs were aligned and preliminary consensus snoRTE sequences derived and used to create a RepeatMasker library. The Ornithorhynchus_anatinus-5.0.1 chromosomes were then locally screened with this library. A novel C-script was used to extract about 40,000 sequences that were aligned by MAFFT (Katoh et al. 2002) and used to define snoRTE consensus sequences. Intronic snoRTEs were identified from http://www.ensembl.org/Ornithorhynchus_anatinus/index.html. All experimentally obtained snoRNA sequences were submitted to the NCBI database.
Acknowledgments
We thank Russell Jones (Newcastle University) for help with the tissue collection. Facilities were provided by Macquarie Generation and Glenrock Station, New South Wales. Approval to collect animals was granted by the New South Wales National Parks and Wildlife Services, New South Wales Fisheries and the Animal Experimentation and Ethics Committee, Australian National University. We thank Jean Marie Smith for optimizing the DNASTAR package Version 7.1 to perform high-throughput data analysis of our cDNA library sequences, Marsha Bundman for editorial assistance, and Janina Thiel and Sven Klages for excellent technical assistance. This work was supported by the Nationales Genomforschungsnetz (NGFN, 0313358A) to J.B. and J.S., the Deutsche Forschungsgemeinschaft (DFG, SCHM1469) to J.S. and J.B., the Max-Planck Society to R.R., and the Australian Research Council to F.G.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to NCBI GenBank under accession nos. EU093990–EU094198.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7177908.
References
- Cavaillé J., Bachellerie J.-P., Bachellerie J.-P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: An exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- Chen X.S., Rozhdestvensky T.S., Collins L.J., Schmitz J., Penny D., Rozhdestvensky T.S., Collins L.J., Schmitz J., Penny D., Collins L.J., Schmitz J., Penny D., Schmitz J., Penny D., Penny D. Combined experimental and computational approach to identify non-protein-coding RNAs in the deep-branching eukaryote Giardia intestinalis. Nucleic Acids Res. 2007;35:4619–4628. doi: 10.1093/nar/gkm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T.M., Brosius J., Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc. Natl. Acad. Sci. 1987;84:2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Wakefield M.J., Kohany O., Gu W., Batzer M.A., Pollock D.D., Jurka J., Wakefield M.J., Kohany O., Gu W., Batzer M.A., Pollock D.D., Jurka J., Kohany O., Gu W., Batzer M.A., Pollock D.D., Jurka J., Gu W., Batzer M.A., Pollock D.D., Jurka J., Batzer M.A., Pollock D.D., Jurka J., Pollock D.D., Jurka J., Jurka J. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolevsky K.P., Vassetzky N.S., Kramerov D.A., Vassetzky N.S., Kramerov D.A., Kramerov D.A. Bov-B-mobilized SINEs in vertebrate genomes. Gene. 2007;407:75–85. doi: 10.1016/j.gene.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K.-I., Miyata T., Misawa K., Kuma K.-I., Miyata T., Kuma K.-I., Miyata T., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanam T., Rozhdestvensky T.S., Bundman M., Galiveti C.R., Handel S., Sukonina V., Jordan U., Brosius J., Skryabin B.V., Rozhdestvensky T.S., Bundman M., Galiveti C.R., Handel S., Sukonina V., Jordan U., Brosius J., Skryabin B.V., Bundman M., Galiveti C.R., Handel S., Sukonina V., Jordan U., Brosius J., Skryabin B.V., Galiveti C.R., Handel S., Sukonina V., Jordan U., Brosius J., Skryabin B.V., Handel S., Sukonina V., Jordan U., Brosius J., Skryabin B.V., Sukonina V., Jordan U., Brosius J., Skryabin B.V., Jordan U., Brosius J., Skryabin B.V., Brosius J., Skryabin B.V., Skryabin B.V. Two primate-specific small non-protein-coding RNAs in transgenic mice: Neuronal expression, subcellular localization and binding partners. Nucleic Acids Res. 2007;35:529–539. doi: 10.1093/nar/gkl1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordis D., Lovsin N., Gubensek F., Lovsin N., Gubensek F., Gubensek F. Phylogenomic analysis of the L1 retroposons in Deuterostomia. Syst. Biol. 2006;55:886–901. doi: 10.1080/10635150601052637. [DOI] [PubMed] [Google Scholar]
- Kriegs J.O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Kiefmann M., Jordan U., Brosius J., Schmitz J., Jordan U., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull M., Petrusma M., Makalowski W., Brosius J., Schmitz J., Petrusma M., Makalowski W., Brosius J., Schmitz J., Makalowski W., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–1145. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Betran E., Thornton K., Wang W., Betran E., Thornton K., Wang W., Thornton K., Wang W., Wang W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Ludwig A., Rozhdestvensky T.S., Kuryshev V.Y., Schmitz J., Brosius J., Rozhdestvensky T.S., Kuryshev V.Y., Schmitz J., Brosius J., Kuryshev V.Y., Schmitz J., Brosius J., Schmitz J., Brosius J., Brosius J. An unusual primate locus that attracted two independent Alu insertions and facilitates their transcription. J. Mol. Biol. 2005;350:200–214. doi: 10.1016/j.jmb.2005.03.058. [DOI] [PubMed] [Google Scholar]
- Luo Y., Li S., Li S. Genome-wide analyses of retrogenes derived from the human box H/ACA snoRNAs. Nucleic Acids Res. 2007;35:559–571. doi: 10.1093/nar/gkl1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H., Eickbush T.H. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 1998;15:1123–1134. doi: 10.1093/oxfordjournals.molbev.a026020. [DOI] [PubMed] [Google Scholar]
- Okada N., Hamada M., Hamada M. The 3′ ends of tRNA-derived SINEs originated from the 3′ ends of LINEs: A new example from the bovine genome. J. Mol. Evol. 1997;44:S052–S056. doi: 10.1007/pl00000058. [DOI] [PubMed] [Google Scholar]
- Pasyukova E.G., Nuzhdin S.V., Morozova T.V., Mackay T.F., Nuzhdin S.V., Morozova T.V., Mackay T.F., Morozova T.V., Mackay T.F., Mackay T.F. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J. Hered. 2004;95:284–290. doi: 10.1093/jhered/esh050. [DOI] [PubMed] [Google Scholar]
- Piskurek O., Okada N., Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Churakov G., Zischler H., Brosius J., Churakov G., Zischler H., Brosius J., Zischler H., Brosius J., Brosius J. A novel class of mammalian-specific tailless retropseudogenes. Genome Res. 2004;14:1911–1915. doi: 10.1101/gr.2720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer A., Stadler P.F., Stadler P.F. Evolution of microRNAs. Methods Mol. Biol. 2006;342:335–350. doi: 10.1385/1-59745-123-1:335. [DOI] [PubMed] [Google Scholar]
- Vitali P., Royo H., Seitz H., Bachellerie J.P., Hüttenhofer A., Cavaille J., Royo H., Seitz H., Bachellerie J.P., Hüttenhofer A., Cavaille J., Seitz H., Bachellerie J.P., Hüttenhofer A., Cavaille J., Bachellerie J.P., Hüttenhofer A., Cavaille J., Hüttenhofer A., Cavaille J., Cavaille J. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M.J. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;3:e36. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C., Lee C. Alternative splicing and RNA selection pressure evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Youngman S., van Luenen H.G.A.M., Plasterk R.H.A., van Luenen H.G.A.M., Plasterk R.H.A., Plasterk R.H.A. Rte-1, a retrotransposon-like element in Caenorhabditis elegans. FEBS Lett. 1996;380:1–7. doi: 10.1016/0014-5793(95)01525-6. [DOI] [PubMed] [Google Scholar]
- Zemann A., de op Bekke A., Kiefmann M., Brosius J., Schmitz J., de op Bekke A., Kiefmann M., Brosius J., Schmitz J., Kiefmann M., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupunski V., Gubensek F., Kordis D., Gubensek F., Kordis D., Kordis D. Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 2001;18:1849–1863. doi: 10.1093/oxfordjournals.molbev.a003727. [DOI] [PubMed] [Google Scholar]