Abstract
Recurrent submicroscopic genomic copy number changes are the result of nonallelic homologous recombination (NAHR). Nonrecurrent aberrations, however, can result from different nonexclusive recombination-repair mechanisms. We previously described small microduplications at Xq28 containing MECP2 in four male patients with a severe neurological phenotype. Here, we report on the fine-mapping and breakpoint analysis of 16 unique microduplications. The size of the overlapping copy number changes varies between 0.3 and 2.3 Mb, and FISH analysis on three patients demonstrated a tandem orientation. Although eight of the 32 breakpoint regions coincide with low-copy repeats, none of the duplications are the result of NAHR. Bioinformatics analysis of the breakpoint regions demonstrated a 2.5-fold higher frequency of Alu interspersed repeats as compared with control regions, as well as a very high GC content (53%). Unexpectedly, we obtained the junction in only one patient by long-range PCR, which revealed nonhomologous end joining as the mechanism. Breakpoint analysis in two other patients by inverse PCR and subsequent array comparative genomic hybridization analysis demonstrated the presence of a second duplicated region more telomeric at Xq28, of which one copy was inserted in between the duplicated MECP2 regions. These data suggest a two-step mechanism in which part of Xq28 is first inserted near the MECP2 locus, followed by breakage-induced replication with strand invasion of the normal sister chromatid. Our results indicate that the mechanism by which copy number changes occur in regions with a complex genomic architecture can yield complex rearrangements.
With the introduction of array comparative genomic hybridization (array-CGH), high-resolution detection of microdeletions and microduplications became possible. This resulted in the identification of many disease-associated genomic submicroscopic aberrations (Pinkel and Albertson 2005; Vissers et al. 2005; Lockwood et al. 2006). In our screen of a large cohort of patients with X-linked mental retardation (XLMR) by full coverage X-chromosome-specific array-CGH (Froyen et al. 2007) and real-time quantitative PCR (qPCR), we identified small duplications at Xq28 in four unrelated male patients with severe to profound mental retardation and additional clinical features (Van Esch et al. 2005), referred to as the Lubs X-linked mental retardation syndrome (XLMRL; OMIM 300260) (http://www.ncbi.nlm.nih.gov/omim/) (Lubs et al. 1999). Delineation of the minimal critical region and detection of a twofold increased expression of MECP2 mRNA in the patient-derived cell lines compared with controls pointed to an increased dosage of MECP2 as the cause of the MR phenotype, thereby demonstrating a new disease mechanism in mental retardation (Van Esch et al. 2005). Subsequently, other groups reported (del Gaudio et al. 2006; Friez et al. 2006; Lu et al. 2007; Madrigal et al. 2007) or communicated on additional patients with a gain of the MECP2 locus. Since all reported duplications seem to be different in size and location, this duplication entity is defined as a nonrecurrent event. However, the mechanism by which this apparent frequent rearrangement occurs has not been resolved so far, and potential mechanisms deduced from breakpoint studies of other nonrecurrent rearrangements are still speculative.
Recurrent rearrangements are mediated by nonallelic homologous recombination (NAHR) between low-copy repeats (LCRs), also referred to as segmental duplications, or between highly similar Alu repeats. Such an event can result in deletions, duplications, or inversions of the intermediate genomic segments, generating aberrations of equal size and location (Shaw and Lupski 2004; Lupski 2006). In nonrecurrent rearrangements on the other hand, the breakpoints are scattered throughout a genomic region and thus the aberrations are variable in size. Although the precise underlying mechanism(s) remain(s) elusive, genomic architectural features have been associated with the generation of these copy number differences (Shaw and Lupski 2004; Lupski 2006). Studies of nonrecurrent PLP1 duplications and deletions at Xq22 in patients with Pelizaeus-Merzbacher disease (PMD) implied that the presence of several LCRs and other smaller repeats seems to render the region unstable and, thus, more susceptible to rearrangements (Woodward et al. 2005; Lee et al. 2006). In such cases, the DNA repair mechanism is not always a simple event. Several groups recently reported complex rearrangements that at first glance seem to be generated in a mechanistically simple way but after detailed molecular analysis revealed more complex rearrangements potentially due to alternative DNA repair mechanisms (Balciuniene et al. 2007; Gotter et al. 2007; Potocki et al. 2007; Sheen et al. 2007) or replication errors (Lee et al. 2007).
We present a comprehensive analysis of 16 unique duplications at Xq28. We found the region to be highly repetitive, which likely contributed to chromosomal breakage at one or more locations and subsequent DNA misrepair. Analysis of the junctions demonstrated that the recombination in two patients resulted from an insertion of a noncontiguous neighboring region preceding the MECP2 duplication event.
Results
Identification of male patients with MECP2 duplications
In collaboration with several international groups, we screened for duplications of the MECP2 gene by qPCR in patients selected based on the clinical features of our initially reported patients with MECP2 duplications. In addition to the four male patients reported earlier (Van Esch et al. 2005), we identified four new positive patients, two from France (E316, X04), one from Germany (326037), and one sporadic patient from Belgium (HT). Additionally, we utilized DNA from previously published male patients: seven from Greenwood, SC (K8210, K8300, K8315, K9227, K9228, K9244, and K9423) (Friez et al. 2006) and one from Brazil (15982) (Rosenberg et al. 2006). Together with the four initially reported by our group (Van Esch et al. 2005), we analyzed the breakpoints of MECP2 microduplications in a total of 16 patients. Clinical details of 12 of these are described elsewhere (Van Esch et al. 2005; Friez et al. 2006; Rosenberg et al. 2006).
Position and orientation of the duplications
We investigated the position and orientation of the duplication by fluorescent in situ hybridization (FISH) in four patients from whom we had an Epstein Barr virus-transformed peripheral blood lymphocyte-derived (EBV-PBL) cell line (L36, T33, T88, and K9228), using the differentially labeled adjacent BAC clones RP11-314B3 (152.63 Mb) and RP11-119A22 (152.90 Mb). Positions on the X chromosome are based on the UCSC genome browser build hg18 (http://genome.ucsc.edu/). For L36, T33, and T88, both BAC clones are present within the duplication as indicated in Figure 1A. With all samples tested the green and red probe signals were detected on metaphase spreads at the very end of the Xq arm and not at other positions in the genome (Supplemental Fig. S1A–C), demonstrating that the duplication occurred at Xq28. On interphase nuclei of these patients, alternating green-red-green-red signals were detected (Supplemental Fig. S1A–C), a pattern strongly suggestive for a tandem duplication event. For patient K9228, a green-red-red order was obtained because RP11-314B3 is not within the duplication (Fig. 1B). However, the signals on metaphase spreads showed that the duplication also occurred at Xq28 (Supplemental Fig. S1D).
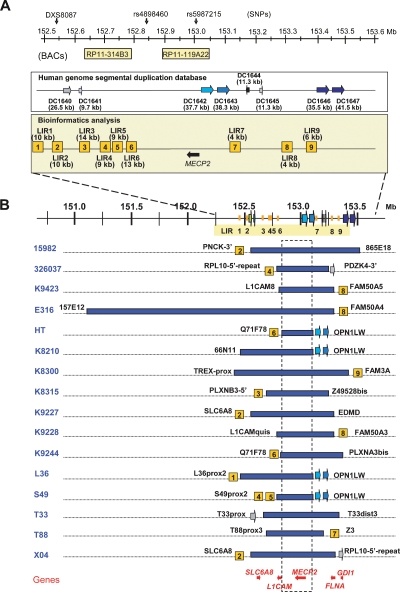
Figure 1.
Bioinformatics analysis of the breakpoint regions and mapping of the duplications in the 16 patients. (A) Schematic overview of the repeat structures that coincide with duplication breakpoints in a 1-Mb region flanking MECP2 (152.55–153.55 Mb). Position and size of interchromosomal (DC1640 and DC1641) and intrachromosomal (DC1642, DC1643, DC1644, DC1645, DC1646, and DC1647) segmental duplications are depicted as reported in the Human genome segmental duplication database. In the Bioinformatics analysis box, LIR1 to LIR9 represent the breakpoint regions enriched for interspersed repeats with their lengths in brackets. The position of the BAC clones RP11-314B3 (152.63 Mb) and RP11-119A22 (152.90 Mb) used for FISH analysis is shown, as well as the position of DXS8087 (152.54 Mb) and the SNPs rs4898460 (152.83 Mb) and rs5987215 (153.05 Mb), used to investigate the origin of the duplications. (B) Duplication breakpoint mapping in the 16 patients (indicated on the left). Horizontal blue bars represent duplicated regions at Xq28. Size and location of each duplication were determined by iterative rounds of qPCR. Positions and sizes of all duplications were different. The colocalization of the breakpoint regions with the repeat structures, if present, is indicated with the name of the last duplicated qPCR primer set mentioned next to it (primer sequences can be found in Supplemental Table S1). The genes for which mutations are known to result in XLMR that also map in this region are shown at the bottom. The commonly duplicated region is boxed and includes only one XLMR gene, MECP2. Except for E316, all duplications lie within a 1.2-Mb region.
Mapping of the duplications at Xq28
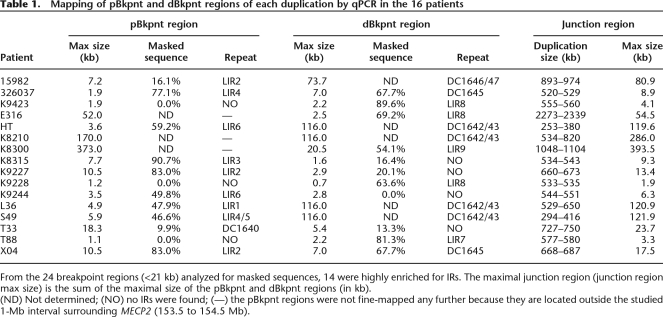
We mapped the location and extent of the 16 duplications as narrowly as possible by iterative rounds of qPCR using a total of 103 primer pairs (Supplemental Table S1). The common overlap of 0.3 Mb coincides with the smallest duplication (152.73–153.02 Mb) found in patient HT, and except for E316 with a duplication encompassing 2.3 Mb, all other aberrations are smaller than 1.2 Mb (Fig. 1B). The common interval contains eight genes (AVPR2, ARHGAP4, ARD1A, RENBP, HCFC1, TMEM187, IRAK1, and MECP2) and possibly one or more genes of the opsin gene cluster. Interestingly, besides MECP2, other genes for which mutations are known to result in XLMR are also duplicated in several of our patients. Seven patients have a duplication of the SLC6A8 gene (OMIM 300036), 14 had a duplication of L1CAM (OMIM 308840), 10 had a duplication of FLNA (OMIM 300017), and seven had a duplication of GDI1 (OMIM 300104) (Fig. 1B). Although the highly similar phenotype of all patients is mainly due to overexpression of MECP2, we cannot exclude dosage effects from any of the other genes within the duplication. Except for the largest duplication in E316, the proximal breakpoints (pBkpnt) are scattered throughout a 0.45-Mb region (152.25 to 152.73 Mb) while the distal breakpoints (dBkpnt) lie in a 0.44-Mb interval (153.02 to 153.45 Mb). In Figure 2, we schematically show a genomic microduplication at Xq28 with the terminology that is used to map and identify the junctions presented here. Fine-mapping by qPCR revealed that the pBkpnt regions of the 16 patients could be narrowed to less than 8 kb in 10 cases and between 8 and 20 kb in three patients, while in the remaining three the delimited breakpoint regions were still significantly larger than 20 kb (Table 1). On the distal side, 10 breakpoint regions were smaller than 8 kb, one was between 8 and 20 kb, and five were larger than 20 kb. When combined, the theoretical junction region was smaller than 8 kb in four patients (T88, K9228, K9244, and K9423). Inability to further narrow down the regions that were larger than 10 kb was due to the presence of repetitive sequences of LCRs and the interspersed repeats (IRs) also known as transposable elements (long interspersed elements [LINEs], short interspersed elements [SINEs], long terminal repeats [LTRs], and DNA elements) (Smit 1999), which precluded the design of unique primer sets.
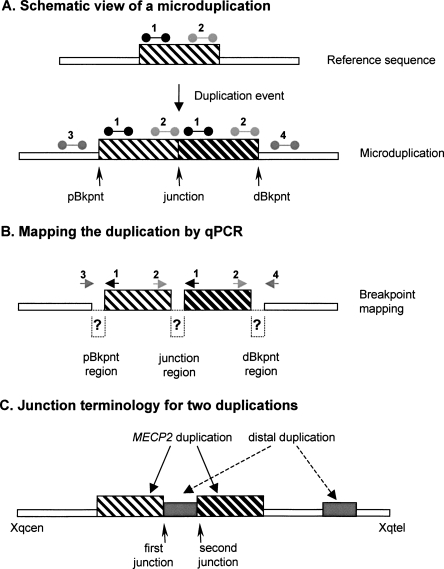
Figure 2.
Terminology used for Xq28 duplication breakpoint mapping and cloning. (A) Schematic of a duplicated segment in the genome. The reference genomic sequence (top), with the region that is duplicated in the lower scheme, is indicated as a thin-striped box. qPCR primer pairs used for duplication size mapping are shown above this box at the proximal (1) and distal (2) side of this region. When duplicated in tandem (bottom), the proximal position where the duplication starts is called the proximal breakpoint (pBkpnt). Similarly, the most distal position where the duplication stops is called the distal breakpoint (dBkpnt). The position where the first copy is followed by the second one is the junction. In qPCR analysis, primer pairs 1 as well as 2 will yield a double dose while the primer pairs for 3 and 4 will give a single copy (relative to the reference genome). (B) For mapping purposes the pBkpnt and dBkpnt regions lie in between the last “normal” forward (3) and the first “duplicated” reverse primer (1), and the last “duplicated” forward (2) and the first “normal” reverse primer (4), respectively. To amplify over the junction, the first “duplicated” forward primer of the proximal breakpoint (1) was combined with the last “duplicated” reverse primer of the distal breakpoint (2). The region in between both primer pairs is called the junction region. Normal sequences are represented by thin open boxes; duplicated sequence by thin-striped and thick-striped boxes, which represent the first and second copy, respectively. (C) Junction terminology for the two duplications.
Table 1.
Mapping of pBkpnt and dBkpnt regions of each duplication by qPCR in the 16 patients
From the 24 breakpoint regions (<21 kb) analyzed for masked sequences, 14 were highly enriched for IRs. The maximal junction region (junction region max size) is the sum of the maximal size of the pBkpnt and dBkpnt regions (in kb).
(ND) Not determined; (NO) no IRs were found; (—) the pBkpnt regions were not fine-mapped any further because they are located outside the studied 1-Mb interval surrounding MECP2 (153.5 to 154.5 Mb).
Bioinformatics analysis of recombination breakpoint regions
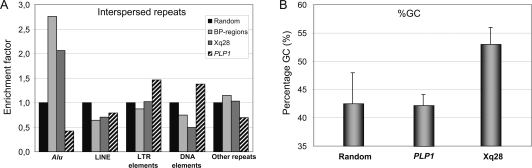
Our mapping data obtained by qPCR revealed at least 14 different pBkpnt and 10 different dBkpnt regions in the 16 patients (Fig. 1B), and each duplication appeared to be unique. Analysis (http://projects.tcag.ca/humandup/) of the 32 mapped breakpoint regions revealed that eight of them coincide with segmental duplications (Fig. 1B). These include the interchromosomal LCR DC1640, which is also found twice on chromosome 16 (DC3591 and DC3600), at the pBkpnt of the duplication in T33; the tandem LCRs DC1642/1643 at the opsin gene locus, which are present distally of the duplications in L36, S49, K8210, and HT; the intrachromosomal LCR DC1645 located at the distal side of the duplications of X04 and 326037; and the intrachromosomal LCRs DC1646/1647 at the distal end of 15982. Irrespective of their location within LCRs, 24 breakpoint regions with a size smaller than 21 kb (Table 1) were checked for enrichment in IRs with RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) in comparison with randomly selected sequences (total of 1.1 Mb) spread over the entire X chromosome (available upon request). Strikingly, the Alu repeat content of the breakpoint regions was at least 2.5-fold enriched compared with the random sequences (Fig. 3A). When compared with the gene-dense and highly recombinogenic region around PLP1 at Xq22.2, in which many rearrangements have been described (OMIM 300401), the Alu enrichment was even fivefold. All other classes of IRs did not show significant differences. Interestingly, sequence analysis of a 1-Mb region surrounding MECP2 revealed a mean GC content of 53% (±3) (Fig. 3B), which is substantially higher than that of the whole genome (41.0%), randomly selected loci on the X chromosome (42.4% ± 5.5), and the gene-rich and recombinogenic PLP1 region (42% ± 1.9).
Figure 3.
Interspersed repeat analysis of the MECP2 region. (A) Comparison of several regions on the X chromosome for potential differential enrichment in an interspersed repeat class (Alu, LINE, LTR elements, DNA elements, other repeats). (Random) Random DNA sequence on chromosome X (total length: 1140 kb); (BP-regions) sum of all breakpoint regions (total length of sequence is 113 kb); (Xq28) Xq28 region ranging from 152.5 to 153.6 Mb chopped into 100-kb fragments (total length, 1133 kb); (PLP1) BAC/PAC clones covering 231 kb of the PLP1 region. The Alu repeat content of all breakpoint regions (BP-regions) was significantly enriched compared with other analyzed sequences on the X chromosome. The other classes of interspersed repeats did not show significant differences. (B) GC content of the different genomic regions.
To define the repetitive nature of the sequences directly flanking the breakpoint regions, we arbitrarily extended the theoretical breakpoint regions by 10 kb at either side and again analyzed those by RepeatMasker. A 4- to 12-kb region with an IR content higher than 75% was present entirely or at least partially within the breakpoint region in 14 out of 24 breakpoint regions investigated. We have marked those structures as local IRs (LIR1 to LIR9 in Fig. 1 and Table 1). Position, length, and percentage of masking of each enriched LIR can be found in Supplemental Table S2.
In a search to look for hairpin secondary structures that might predispose to recombination, we performed M-fold analysis (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html) on the pBkpnt and dBkpnt regions of the duplications in the four patients (T88, K9423, K9244, and K9228) with the smallest maximal sizes of junction regions <7 kb (Table 1). We detected hairpin structures at both breakpoints in three of them (K9228, K9244, and T88) but similar structures were observed at about the same frequency in randomly selected sequences (data not shown). Similarly, the search for matrix association regions (MARs) (http://www.futuresoft.org/MAR-Wiz/), indicative of a highly destabilized DNA duplex, did not detect differences between breakpoint and control regions (data not shown).
Mapping and sequence analysis of junctions
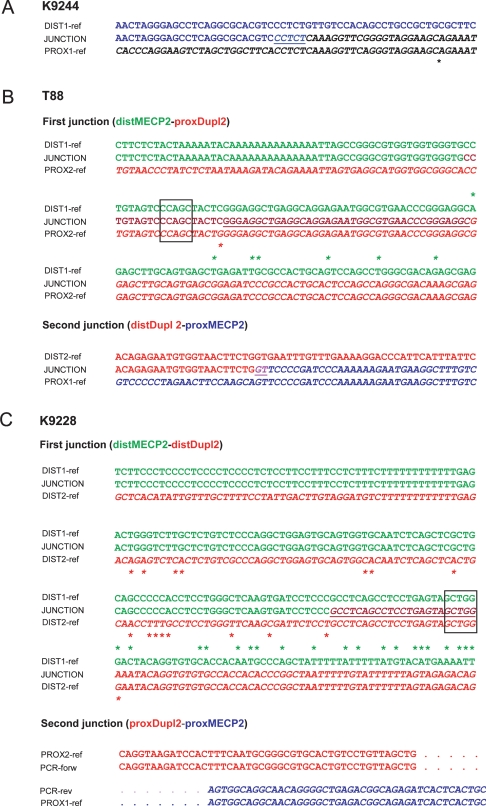
Next, we attempted to identify by expand long template PCR (ELT-PCR) the junctions of the four patients (K9423, K9228, K9244, and T88) in whom the maximal size of the junction region (column “junction region max size” in Table 1) was <7 kb. Although these junction regions theoretically were 4.1, 1.9, 6.3, and 3.3 kb, respectively, we were only able to amplify the junction in K9244. We did not obtain any ELT-PCR products for the remaining three cases. Potential inversion duplication in these three patients was excluded since the sequences at both the pBkpnt and the dBkpnt regions were equal to that of the reference genome (Supplemental Fig. S2). Sequence analysis of the junction in K9244 revealed a tandem duplication with its proximal and distal flanking segments sharing 5-bp microhomology (Fig. 4A), thus suggesting a repair mechanism of nonhomologous end joining (NHEJ). Bioinformatics analysis detected an ERV9-LTR (LTR12B) (located in LIR5) at the pBkpnt and a Chi sequence (GCTGGTGG) at the dBkpnt located 202 bp from the junction. Recombination motifs were searched for via the Genamics expression software (http://genamics.com/expression/index.htm), but no significant enrichment was found compared with random sequences on the X chromosome (data not shown).
Figure 4.
Alignment of the sequenced junctions in patients K9244, T88, and K9228 with the reference genome sequence. Proximal and distal reference sequences are printed in normal font and italics, respectively, and colored differently. All junctions are underlined and in the merge color of proximal and distal reference sequence. Asterisks represent sequence mismatches. (A) K9244: Proximal (top) and distal (bottom) sequence were aligned against the junction sequence (middle), demonstrating the tandem orientation of the duplication. A microhomology of 5 bp (CCTCT) is found at the junction between the distal and proximal sequences, characteristic for NHEJ. (B) T88: Proximal (top) and distal (bottom) sequence of the first and second junction were aligned against the respective junction sequences (middle). The first junction occurs within an AluY present at both dBkpnt and pBkpnt of the MECP2 and the distal duplication, respectively. There is a perfect match of 39 bp. The Alu pentanucleotide motif (CCAGC) is boxed. These data are consistent with homology-assisted NHEJ. A microhomology of 2 bp (GT) is found at the second junction, in agreement with a collapse of the BIR-fork at this locus and search for microhomology on the sister chromatid. (C) K9228: Alignment of the reference sequence against the respective junction sequences (middle). The first junction occurs within an AluJo at the dBkpnt of the MECP2 duplication and an AluSg at the dBkpnt of the distal duplication. The homology-assisted NHEJ at this junction results in the insertion of the distal duplication in between the MECP2 duplications in an inverted orientation. The 23-bp junction terminates in the Alu pentanucleotide motif (boxed). Boundaries of the PCR-fragment containing the second junction were sequenced and revealed sequences of the pBkpnt of the MECP2 duplication at one side (PCR-forw) while at the other end (PCR-rev) sequences of LCR DC1649 were obtained, coinciding with the pBkpnt of the distal duplication. Dots represent predicted reference sequence.
Because we were unable to identify the junctions in the three remaining patients (T88, K9228, and K9423) by ELT-PCR, we hypothesized that the rearrangement would be more complex. Therefore, we performed inverse PCR (iPCR) on at least one breakpoint region, for which we found appropriate restriction sites, for all three patients. In this way, we obtained patient-specific PCR products for the pBkpnt region in T88 and the dBkpnt region in K9228. Surprisingly, sequence analysis demonstrated that both breakpoints were associated with DNA sequences from more telomeric regions at Xq28. The exact pBkpnt in T88 was located at nucleotide position 152,637,734 on chromosome X and was preceded by sequence of RP11-95M2 (junction at 154,371,862). For sake of clarity, throughout the paper, we defined the more centromeric located junction as the first junction and the more telomeric junction as the second junction (Fig. 2C). Two base pairs of microhomology were identified at the second junction in T88. The dBkpnt in K9228 was located at nucleotide 153,333,324 bp, which was then followed by an inverted sequence (compared with the reference genome) starting at nucleotide 154,431,416 within the TMLHE gene. The dBkpnt of the MECP2 duplication was located in an AluJo repeat, which is 83% identical to the AluSg repeat present at the dBkpnt of the distal duplication located in TMLHE. A perfect match of 23 base pairs within these highly similar Alu repeats defines this first junction in K9228. The 23-bp junction itself terminates in the Alu pentanucleotide motif (5′-CCAGC), which is part of the consensus Alu core sequence (5′-CCTGTAATCCCAGCACTTTGGG AGGC). Both junctions were confirmed by regular PCR, which did not yield products from control DNA samples (data not shown). The iPCR strategy did not allow us to identify a patient-specific PCR product for K9423, at least not with the restriction enzymes tested.
Next, we performed X-chromosome-specific array-CGH on DNA samples from probands of T88, K9228, and K9423. In T88, we detected in addition to the MECP2 duplication, a second microduplication of a segment 800 kb telomeric (153.9–154.3 Mb) of the MECP2 duplication (Supplemental Fig. S3A). This copy number change was confirmed by qPCR with primers located in RP11-143H17 (154.02 Mb) and CLIC2 (154.21 Mb), while normal copy number values were obtained for primer pairs in F8 (153.86 Mb) and TMLHE (154.39 Mb) (data not shown; primer sequences in Supplemental Table S1). This second duplication comprises VBP1, RAB39B, CLIC2, H2AFB1-3, and two F8A genes and harbors the intrachromosomal LCRs DC1649 (154.21 Mb) and (partially) DC1650 (154.33 Mb). Fine-mapping of the duplication size by qPCR revealed that the dBkpnt region of this second duplication was in the direct neighborhood of the distal sequence obtained at the pBkpnt of the MECP2 duplication (second junction). From these data we assumed that the second duplicated segment was inserted in between both MECP2 duplicons. Fine-mapping the other side of this second duplication by qPCR, allowed amplifying the junction between the dBkpnt of the MECP2 duplication and the pBkpnt of the second duplication by ELT-PCR (first junction). For this PCR, the last forward “duplicated” primer at the dBkpnt of the MECP2 duplication was combined with the first “duplicated” reverse primer of the second duplication. A 1.5-kb fragment was obtained for the patient but not for controls. Sequence analysis revealed the first junction at the nucleotide level (Fig. 4B). The dBkpnt of the MECP2 duplication and the pBkpnt of the distal duplication coincide with an AluY repeat, with a microhomology of 39 bp, including part of the 26-bp Alu core sequence. Taken together, our data on T88 demonstrated that the distal duplicated fragment (355 kb) is located in a direct orientation in between both MECP2 duplicons. Similarly, array-CGH on DNA from K9228 revealed an additional microduplication of a more distal region at Xq28 encompassing the clones RP11-405N23 (154.24–154.31 Mb) and RP11-218L14 (154.33–154.48 Mb) (Supplemental Fig. S3B). Using a similar mapping approach as for T88, we amplified the second junction region in K9228 (Fig. 4C). Sequence analysis of both ends of this PCR fragment, which was absent in controls, revealed sequences at the pBkpnt of the MECP2 region at one side, while at the other end sequences of LCR DC1649 were obtained. Although we could as yet not obtain the junction sequence, our data demonstrated that the second duplicated fragment (206 kb) was located in an inverted orientation in between both MECP2 duplicons.
By qPCR, we also showed that the mothers of both patients carried the distal Xq28 duplication, too (data not shown). X-array-CGH on DNA from the fourth patient with a small theoretical junction region, K9423, did not reveal an additional duplicated region on the X chromosome (data not shown). Finally, analysis of all 14 other MECP2 duplication patients (all except T88 and K9228) for an additional duplication at distal Xq28 with qPCR primers sets in F8, RP11-143H17, CLIC2, and TMLH did not identify additional copy number changes for these loci (data not shown).
Based on the sequenced junctions in K9244 (one junction), T88 (two junctions), and K9228 (two junctions), we also sequenced the pBkpnt and dBkpnt regions that determine the boundaries between single copy and duplicated DNA in these patients. All 10 sequences were identical to the reference genome sequence.
Chromosomal origin of the duplications
We analyzed the carrier status of all mothers and three grandmothers for whom DNA was available by qPCR and found three copies in all of them demonstrating that all duplications were inherited (data not shown). We evaluated the X-inactivation pattern based on the methylation status of the AR locus and demonstrated that a skewed pattern (>85%) was obtained in all carriers (data not shown) who were all diagnosed as asymptomatic. To assess the type of chromosomal recombination, we first analyzed the repeat length of the one polymorphic marker (http://genome.ucsc.edu/cgi-bin/hgGateway) present in the duplicated region (DXS8087; 152.54 Mb) (indicated on Fig. 1A). The four patients in whom DXS8087 was located within the duplicated region (E316, K8210, K8300, and L36) were homozygous for repeat lengths of 380 bp, 386 bp, or 388 bp (Supplemental Table S3). Furthermore, all 16 probands were homozygous for both tested SNPs (indicated on Fig. 1A) in the common duplicated region (rs4898460 and rs5987215) (Supplemental Table S3). From the carrier mothers three out of four tested are heterozygous for DXS8087 (K8210, K8300, E316) and three out of 16 for both SNPs (K8210, K8300, X04). Although these data are not informative, they are not in favor of an interchromosomal duplication event.
Discussion
Identification of MECP2 duplication patients
We recently identified small duplications at Xq28 containing MECP2 in male patients with a distinct syndromic form of X-linked mental retardation (Van Esch et al. 2005). In this study, we analyzed the duplications in these four reported and 12 additional male patients to reveal the mechanism that mediates the formation of these duplications. All aberrations were maternally inherited, and the three investigated grandmothers were also carriers. Therefore, the parental origin of the duplications could not be determined. All 16 mothers showed a skewed pattern of X-inactivation (>85%), as frequently found in carriers of mutations on the X chromosome associated with mental retardation (Plenge et al. 2002). Inactivation of the X chromosome with the MECP2 duplication, therefore, will lead to a normal MECP2 dosage, without clinical implications.
Overall genomic instability at Xq28
Several groups already identified Xq28 as a region at high risk for genomic instability (Aradhya et al. 2002; Sanlaville et al. 2005). Next to large aberrations in Xq28, a multitude of microrearrangements have been reported in a relatively small region at Xq28 (152.5–154.0 Mb). Microrearrangements that lead to disease are reported as the result of NAHR or NHEJ in adrenoleukodystrophy (OMIM 300100) (Kutsche et al. 2002), X-linked hydrocephalus (OMIM 307000) (Kutsche et al. 2002), red–green color-blindness (OMIM 303800) (Deeb and Kohl 2003), Emery-Dreifuss Muscular Dystrophy (OMIM 310300) (Small and Warren 1998), incontinentia pigmenti (OMIM 308300) (Aradhya et al. 2002), and hemophilia A (OMIM 306700) (Bagnall et al. 2005). The presence of multiple LCRs (LC1640 to LC1650) in this interval (Lee and Lupski 2006) could render this region prone to these subtle rearrangements. Moreover, the region between ABCD1 (152.66 Mb) and IKBKG (153.44 Mb) shows linkage disequilibrium (Taillon-Miller et al. 2000), a mark that has been related to unstable regions in the genome (Locke et al. 2006). Our data provide additional proof for the genomic instability at Xq28.
Breakpoint analysis revealed high Alu repeat and GC content
Mapping of 32 breakpoints of 16 unique MECP2 duplications by iterative rounds of qPCR allowed us to locate 20 breakpoints in a region smaller than 10 kb (24 were smaller than 21 kb) (Table 1). In our search for colocalization of these breakpoints with segmental duplications, we found that the pBkpnt region of one patient and the distal regions of seven patients coincide with an LCR, but not a single patient had LCRs at both sides. We also looked for IRs, short homologous DNA stretches (100–3000 bp), and specific recombination motifs using online bioinformatics tools. When compared with random sequences on the X chromosome, a 2.5-fold enrichment of Alu sequences was detected in 14 out of 24 breakpoint regions. Alu repeats are frequently involved in recombination because they may serve as binding sites for proteins operative in homologous recombination (HR) (Kolomietz et al. 2002). An evolutionary search for Alu recombination-mediated deletions (ARMDs) demonstrated that ARMDs tend to occur in regions with high GC content (>45%) (Sen et al. 2006). The high mean GC content (53%) in the region around MECP2 therefore could contribute to the genomic instability. In neuroblastoma, the translocation breakpoints were often found in regions with high GC content (Stallings et al. 2007). Instead, the Alu repeat and GC content of the highly recombinogenic PLP1 region at Xq22 was not above average. Analysis of microduplications in the PLP1 region revealed the presence of Alu segments at seven junctions, although only one junction had Alu repeats at both sides (Woodward et al. 2005). For the PLP1 duplications, the authors suggested a homology-assisted NHEJ, based on the model of coupled homologous and nonhomologous recombination that implies homologous single-strand invasion and completion of the event by NHEJ (Richardson and Jasin 2000). In our study, we identified the bilateral presence of an Alu repeat at the first junction in T88 as well as K9228.
We did not find a higher frequency of short homologous DNA stretches or specific recombination motifs at the breakpoints compared with control regions.
Model for a complex rearrangement based on breakpoint analyses
Although we tried to identify the junction in the four cases (K9228, K9244, T88, and K9423) for which we narrowed down the theoretical size of the junction region to smaller than 7 kb, we succeeded in amplifying the junction by ELT-PCR in just patient K9244. Microhomology was present at the pBkpnt and dBkpnt regions resulting in a 5-bp deletion at the junction, thus revealing a NHEJ mechanism. Similarly, efforts to obtain the junctions in PMD patients with nonrecurrent duplications at Xq22 were unsuccessful in 17 out of 30 cases (Woodward et al. 2005). At least nine did not fit a simple tandem duplication event. In two patients, the authors demonstrated that the duplication was associated with the inversion of a region near the PLP1 locus. In their model of coupled homologous and nonhomologous recombination, deletions and duplications would have arisen from LCR-driven double-strand breaks (DSBs) followed by one-sided single-strand invasion at the homologous position, and repair by NHEJ (Woodward et al. 2005; Lee et al. 2006). Multiple DSBs in close proximity prior to, or in trans after strand invasion, may thus result in inversion, deletion, or insertion events. LCRs thus can also stimulate genomic rearrangements without being physically involved. Recent data on 13 patients with nonrecurrent duplications in the Smith-Magenis Syndrome (OMIM 182290) region at 17p11 (Potocki et al. 2007) corroborate on this model. It is of interest to note that in three patients with PLP1 duplications, and in one of the complex Smith-Magenis rearrangements, a second duplication of a neighboring region was present (Woodward et al. 2005; Potocki et al. 2007). We also found an additional duplication in patients T88 and K9228.
Very recently, a novel DNA repair mechanism called replication Fork Stalling and Template Switching (FoSTeS) was proposed to explain nonrecurrent rearrangements based on junction analysis studies of PLP1 duplications (Lee et al. 2007). In this model, the LCR-associated genomic instability at the PLP1 locus negatively affects the smooth progression of the replication fork and, upon stalling, introduces switching from one active replication fork to another for which only microhomology is required. This FoSTeS model was hypothesized upon oligo-array analysis and junction sequencing, which revealed complex rearrangements of highly variable-sized fragments of noncontiguous duplicated, normal copy-number, or deleted sequences. Indeed, such a replication-based recombination mechanism can readily explain many complex rearrangements (Lee et al. 2007). As such, FoSTeS could explain the nonrecurrent MECP2 duplications too, but in contrast to Lee et al. (2007), we did not find similar highly complex junctions thus far. Therefore, our findings allow alternative (simpler) recombination events too.
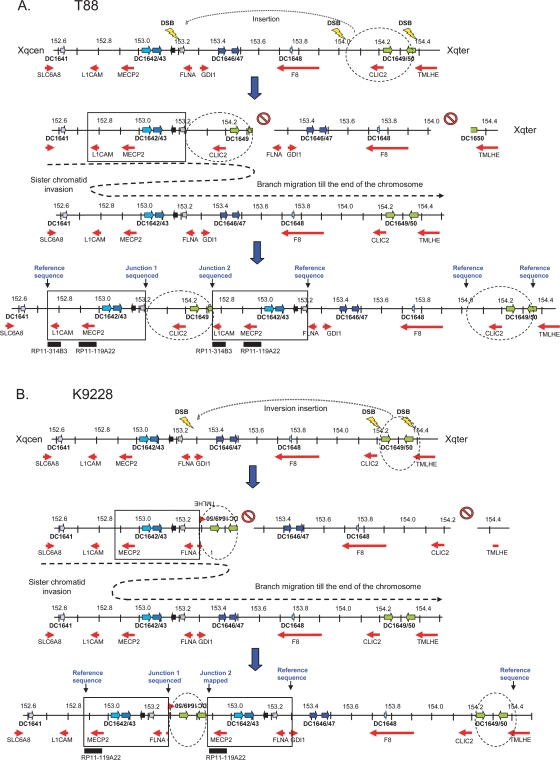
The mechanism of break-induced-replication (BIR) is an alternative repair model that results in recombination (Kraus et al. 2001). It is a DSB repair process in which the broken chromosome end invades another chromosome or chromatid based on only a few nucleotides of homology and replicates the sequence from there on. BIR was initially reported in yeast to repair unprotected or elongate shortened telomeres (McEachern and Haber 2006). Microhomology-dependent BIR events with strand invasion on the sister chromatid will result in genomic deletions or duplications (Ricchetti et al. 2003; Koszul et al. 2004). Recently, Sheen et al. (2007) reinvested this model based on a complex rearrangement involving F8 and FUNDC2 at Xq28 in a patient with hemophilia A. In another recent study, BIR was also suggested for repair of breaks in subtelomeric rearrangements in which unusual DNA structures were found (Rooms et al. 2007). The rearrangements in patients T88 and K9228 can be readily explained by the BIR model (schematically represented in Fig. 5). The DSBs could have been stimulated by the high repeat content of the MECP2 region via the complex set of LCRs and the significant enrichment in Alu repeats. Also, the high average GC-content could be an additional critical factor. We propose that at least three DSBs occurred simultaneously in patients K9228 and T88. Subsequently, the region corresponding to the second duplicated fragment in these patients would have been excised and attached in a direct (T88) or inverted (K9228) orientation to a DSB site distal to MECP2 most likely by Alu-driven homology-assisted NHEJ. Alu sequences were present at either breakpoint side of the first junction in both patients. Repair of the remaining DSB would then initiate BIR, in which the BIR fork, because of sterical constraints, would locally invade the sister chromatid using minor or microhomology. The continuation of BIR following strand invasion remains to be elucidated but both models, replication until the end of the chromosome (McEachern and Haber 2006) and size-limited replication with subsequent reannealing to the original strand, have been proposed (Sheen et al. 2007). The absence of microaberrations at the boundaries between single-copy and duplicated sequences in T88 and K9228 argue against the latter. Moreover, elongation until the end of the chromosome is more likely at the MECP2 locus, which is near the telomere, than, for example, at the PLP1 locus that is located far more centromeric. The BIR model is in agreement with our PCR, FISH, and sequencing data. Assuming this model, the template for replication is likely provided by the intact sister chromatid on which DNA synthesis continued to the end of the X chromosome (Fig. 5). This conservative DNA synthesis process will result in homozygosity for the duplicated regions. Indeed, marker (DXS8087) and SNP analysis revealed homozygous calls in all 16 patients. Our data thus are not suggestive for an interchromosomal recombination event. Intrachromosomal recombination in meiosis has been proposed as the mechanism giving rise to deletions and duplications of the PLP1 locus at Xq22.2, and duplications in the dystrophin gene (Hu et al. 1991; Mimault et al. 1999; Inoue et al. 2002). We hypothesize that both duplications occur as a single event in the germ line lineage. The carrier status of the mothers of T88 and K9228 for both duplications is in line with this assumption, but since we did not have access to samples of any maternal ancestor, we were unable to study this any further.
Figure 5.
Proposed model for the recombination/repair mechanism originating both duplications in patients T88 and K9228. Illustration of the Xq28 region from 152.45 Mb to 154.65 Mb. The LCRs are represented by differently colored block arrows and genes by red block arrows. The MECP2 duplication is boxed while the additional distal duplication is circled (broken line). (A) Patient T88. Several DSBs occur simultaneously resulting in an excised fragment that is attached through Alu homology, in a direct orientation to the DSB generated proximally from FLNA. Repair of the remaining DSB is performed by BIR in which the BIR fork invaded the DNA duplex of the sister chromatid at a proximal location with microhomology of a few bp, and DNA synthesis proceeded to the telomere. (B) Patient K9228. Similar recombination/repair event in K9228 with minor differences. Several DSBs occur simultaneously resulting in an excised fragment at LCRs DC1649 and DC1650. This fragment is then rejoined, via Alu homology, in an inverted way to the DSB generated at GDI1. Then, the BIR fork invaded the sister chromatid at a more proximal locus, and DNA synthesis continued to the end of the X chromosome.
In conclusion, we report the breakpoint mapping of 16 unique MECP2 duplications and strongly suggest that the high content of segmental duplications around the MECP2 locus renders the region prone to DSBs or replication fork stalling, and thus rearrangements. We also showed that this region has a much higher Alu repeat and GC content compared with random sequences in the genome. In patient K9244, the DSB seemed to be repaired by NHEJ resulting in a simple tandem duplication. In T88 and K9228, the repair mechanism seemed to be a complex two-step process in which the broken strand was first linked to a free noncontiguous fragment with subsequent strand invasion from the other end onto the sister chromatid, followed by DNA replication to the end of the chromosome (BIR model). As a result, the second duplicated fragment was inserted in between the MECP2 duplicated sequences generating a complex rearrangement. Alternatively, FoSTeS can explain our findings, too, although absence of microaberrations at the switches from normal to duplicated sequence could argue against this model. Analysis of additional breakpoints in patients with MECP2 duplications is required to support one or the other model. Nevertheless, our data suggest that distinct complex genomic architectures can trigger the common NAHR and NHEJ mechanisms but can also activate a combination of common and alternative repair processes to restore genomic integrity, yielding either a simple or complex rearrangement.
Methods
Patient samples
The protocol was approved by the appropriate Institutional Review Boards of the different hospitals, and informed consent was obtained from the parents of the affected patients and their healthy family members. Genomic DNA from patients as well as from healthy controls was isolated from peripheral blood according to standard procedures.
Based on the clinical characteristics present in the four initially reported patients with severe MR, hypotonia, limited speech, and possibly recurrent infections, most other patients were selected for screening of the MECP2 duplication and collected from Genetic Institutes in Belgium, France, Germany, Greenwood (SC), and Brazil. The screening method used was either multiplex ligation-dependent probe amplification (MLPA) (Greenwood) or qPCR real-time quantitation (all others). DNA from the mothers and three grandmothers was equally available. In all except two cases (S49 and HT), the probands investigated in this report belong to families in whom one or more affected males are present. All duplications segregate with the disease in the family and carrier females are asymptomatic (Van Esch et al. 2005; del Gaudio et al. 2006; Friez et al. 2006).
Array-CGH, qPCR, and MLPA
We have developed an X chromosome-specific BAC array (X-array) with a theoretical resolution of 80 kb (Bauters et al. 2005). X-array-CGH was performed essentially as described elsewhere (Froyen et al. 2007). For qPCR, we used the comparative ddCt method (SDS User bulletin #2; Applied Biosystems) with the qPCR Mastermix Plus for SYBR Green I (Eurogentec) as described previously (Van Esch et al. 2005). Screening for Xq28 duplications was performed with the MECP2 primer set (Supplemental Table S1). Mapping the duplications was done with primer pairs in this region for which the sequences and locations are given in Supplemental Table S1. All samples were run in duplicate on an ABI7000 instrument (Applied Biosystems), and two independent runs were performed for all samples. MLPA of the MECP2 gene was performed as described elsewhere (Friez et al. 2006).
FISH analysis
FISH was performed as described elsewhere (Menten et al. 2006). Briefly, degenerate oligonucleotide-primed (DOP) amplified BAC fragments were labeled in a direct PCR reaction with SpectrumOrange or SpectrumGreen dNTPs (Vysis) and purified with Qiaquick PCR Purification kit (QIAGEN). Metaphase chromosomes and interphase nuclei from patient lymphocyte cell lines were obtained according to standard protocols. After hybridization for 18 h at 37°C and several washing steps, chromosomes were counterstained with DAPI and visualized by digital imaging microscopy with Cytovision capturing software (Applied Imaging).
Breakpoint mapping
For mapping the exact position and extent of the duplications in the 16 MECP2 duplication patients, we designed, in an iterative way, qPCR primers, using the PrimerExpress software (Applied Biosystems), at the proximal as well as distal side of the MECP2 gene. The copy number was then determined for each tested primer set in the patient relative to control samples by qPCR. In case of a double dosage for one primer set and a normal dosage for the next one, additional primer sets were designed in between these two loci. DNA of a patient with a known duplication at the tested locus was used as a positive control for each primer set. In total, 103 primer sets (Supplemental Table S1) were designed to fine-map the duplications. Efforts to amplify over the junctions were started whenever the combined physical distance between the pBkpnt and dBkpnt regions was <10 kb. Both breakpoint regions were defined as the sequence between two consecutive primer pairs for which copy number values of (theoretically) 1 and 2 were found. We performed ELT-PCR (Roche) with the forward primer of the most distally duplicated locus combined with the reverse primer of the most proximally duplicated locus (see Fig. 2). Reaction buffer and PCR conditions were first tested on control amplifications using the same primers but in combinations that allow amplification of control DNA. PCR conditions were: 95°C for 2 min, followed by 10 cycles of 95°C for 10 s, 65°C for 30 s, 68°C for 1 to 5 min (depending on the maximum length of the expected fragment), and 25 cycles of 95°C for 15 s, 58°C for 30 s, 68°C for 1 to 5 min starting from 100 ng of DNA. In the first 10 cycles, a touchdown approach was followed starting from 65°C, and every cycle the temperature was decreased with 0.5°C. For the next 25 cycles, the extension time was extended with 20 sec per cycle. PCR fragments were either directly sequenced or first cloned into pGEM-T-Easy vector (Promega) and subsequently sequenced with the T7 and SP6 vector primers. For mapping the exact position and extent of the second duplications in K9228 and T88, qPCR and ELT-PCR was performed as described above. For this, 29 additional primer sets were developed and sequences are also provided in the Supplemental Table S1.
Inverse PCR
Inverse PCR (iPCR) allows identifying neighboring unknown DNA sequence starting from known sequence (Willis et al. 1997). iPCR was successfully performed for the patients T88 and K9228. Ten micrograms of genomic DNA of control and patient were digested overnight according to the manufacturer’s instructions (New England Biolabs). DNA from T88 was digested with EaeI (YGGCCR) and K9228 DNA with BtsCI (GGATG). Digested DNA was purified with PCR purification kit (QIAGEN) and eluted in 50 μL of water. All purified DNA was used in a 1-mL ligation reaction supplied with 50 units T4 DNA ligase (Promega), 10× One-Phor-All buffer (100 mM tris-acetate at pH 7.5, 100 mM Mg-acetate, 500 mM K-acetate), and 500 μM adenosine tris-phosphate (GE Healthcare). The ligation mixture was incubated for 48 h at 4°C, purified with the PCR purification kit, and eluted in 50 μL of water. ELT-PCR was performed on 10 μL of this ligated DNA. Distinct PCR products were cloned into pGEM-T-Easy and subsequently sequenced with the T7 and SP6 primers.
In silico analysis of breakpoint regions
To determine the genomic architecture of the Xq28 region, we used several online bioinformatics tools. Repetitive DNA was masked with RepeatMasker. Tandem repeats were screened for via ETANDEM (http://bioweb.pasteur.fr/seqanal/interfaces/etandem.html). Fractional GC content of DNA sequences was calculated using GEECEE (http://bioweb.pasteur.fr/seqanal/interfaces/geecee.html) and RepeatMasker. Alternative tools used for the search of repetitive structures or similar blocks of sequences were REPuter (http://woody.embl-heidelberg.de/repeatmask/), Censor (http://www.girinst.org/censor/help.html), PipMaker (http://pipmaker.bx.psu.edu/cgi-bin/pipmaker?basic), and TOUCAN (http://homes.esat.kuleuven.be/~saerts/software/toucan.php). The Genamics Expression Software (http://genamics.com/expression/index.htm) enabled us to identify well-defined DNA patterns (Immunoglobulin heavy chain: GAGCT, GGGCT, GGGGT, TGGGG, TGAGC; translin target sites: ATGCAG, GCCCWSSW; heptamer recombination motif: CACAGTG; nonamer recombination motif: ACAAAAACC; alternating purine-pyrimidine tract; Chi sequence: GCTGGTGG; or the pentanucleotide CCAGC present in the Alu core sequence). M-fold (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html) was used to predict secondary DNA structures.
Additionally, a 1-Mb region surrounding MECP2 (152.55–153.55 Mb) was in silico divided in 100-kb fragments, and each fragment was then compared with itself and with each of the other fragments by BLAST 2 sequences (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi) using default parameters. This 1-Mb region was also checked for the presence of segmental duplications in the online available Human Genome Segmental Duplication Database (http://projects.tcag.ca/humandup/), and the Self chain tool of the UCSC genome browser (http://genome.ucsc.edu) was used to find highly similar DNA stretches elsewhere in the genome.
RepeatMasker was used to search for the involvement of any type of IR in the MECP2 duplication rearrangement. Every breakpoint region was therefore subjected to RepeatMasker. Because we reasoned that the enrichment in IRs could extend beyond the breakpoint region itself, up to 10 kb of neighboring DNA sequence was added and again analyzed with RepeatMasker. Regions with more than 75% masking were defined as enriched IR regions (LIR1-9; see Table 1).
Control DNA sequences used to compare for enrichment in IRs and GC content were randomly chosen along the X chromosome (sequences available upon request).
DXS8087 and SNP marker analysis
DXS8087 marker analysis was performed on 50 ng of DNA of male patients and their mothers and grandmothers (when available), injected together with the ROX-labeled genotyping marker 100-500 (Applied Biosystems) on an ABI3130xl system, and analyzed with Genemapper software (Applied Biosystems).
For SNP analysis, we searched two polymorphic SNPs in the CEPH population within the MECP2 region using the HapMap SNP tool (http://www.hapmap.org/cgi-perl/gbrowse/hapmap_B35/). Primers were designed to amplify rs4898460 at 152.83 Mb (allele frequency of A:60 and G:40), and rs5987215 at 153.05 Mb (allele frequency of T:62 and G:38). PCR was performed on 50 ng of DNA with Platinum Taq (Invitrogen), and 50 ng of PCR product was directly sequenced with the forward PCR primer using the BigDye v3.1 reagent (Applied Biosystems). Sequencing samples were analyzed on an ABI3130xl apparatus, and the nucleotide present at each SNP position was scored for each sample. Primer sequences can be found in Supplemental Table S1.
Acknowledgments
We thank all the patients and their families for their long-time cooperation. This work was supported by a grant from NIH (HD26202) to C.E.S. and in part by a grant from the South Carolina Department of Disabilities and Special Needs. M.B. is supported by the IWT (Instituut voor Innovatie door Technologie en Wetenschap), Flanders, Belgium. H.V.E. is a postdoctoral researcher of the Fund for Scientific Research–Flanders (FWO-Vlaanderen), Belgium. This paper is dedicated to the memory of Ethan Francis Schwartz (1996–1998).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075903.107.
References
- Aradhya S., Woffendin H., Bonnen P., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Woffendin H., Bonnen P., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Bonnen P., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Poustka A., D'Urso M., Kenwrick S., D'Urso M., Kenwrick S., Kenwrick S., et al. Physical and genetic characterization reveals a pseudogene, an evolutionary junction, and unstable loci in distal Xq28. Genomics. 2002;79:31–40. doi: 10.1006/geno.2001.6680. [DOI] [PubMed] [Google Scholar]
- Bagnall R.D., Ayres K.L., Green P.M., Giannelli F., Ayres K.L., Green P.M., Giannelli F., Green P.M., Giannelli F., Giannelli F. Gene conversion and evolution of Xq28 duplicons involved in recurring inversions causing severe hemophilia A. Genome Res. 2005;15:214–223. doi: 10.1101/gr.2946205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J., Feng N., Iyadurai K., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Feng N., Iyadurai K., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Iyadurai K., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Staaf J., Oseth L., Czapansky-Beilman D., Oseth L., Czapansky-Beilman D., Czapansky-Beilman D., et al. Recurrent 10q22-q23 deletions: A genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am. J. Hum. Genet. 2007;80:938–947. doi: 10.1086/513607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters M., Van Esch H., Marynen P., Froyen G., Van Esch H., Marynen P., Froyen G., Marynen P., Froyen G., Froyen G. X chromosome array-CGH for the identification of novel X-linked mental retardation genes. Eur. J. Med. Genet. 2005;48:263–275. doi: 10.1016/j.ejmg.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Deeb S.S., Kohl S., Kohl S. Genetics of color vision deficiencies. Dev. Ophthalmol. 2003;37:170–187. doi: 10.1159/000072046. [DOI] [PubMed] [Google Scholar]
- del Gaudio D., Fang P., Scaglia F., Ward P.A., Craigen W.J., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., Fang P., Scaglia F., Ward P.A., Craigen W.J., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., Scaglia F., Ward P.A., Craigen W.J., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., Ward P.A., Craigen W.J., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., Craigen W.J., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., Neul J.L., Patel A., Lee J.A., Irons M., Patel A., Lee J.A., Irons M., Lee J.A., Irons M., Irons M., et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet. Med. 2006;8:784–792. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- Friez M.J., Jones J.R., Clarkson K., Lubs H., Abuelo D., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., Jones J.R., Clarkson K., Lubs H., Abuelo D., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., Clarkson K., Lubs H., Abuelo D., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., Lubs H., Abuelo D., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., Abuelo D., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., Pai S., Simensen R., Williams C., Giampietro P.F., Simensen R., Williams C., Giampietro P.F., Williams C., Giampietro P.F., Giampietro P.F., et al. Recurrent infections, hypotonia, and mental retardation caused by duplication of MECP2 and adjacent region in Xq28. Pediatrics. 2006;118:e1687–e1695. doi: 10.1542/peds.2006-0395. [DOI] [PubMed] [Google Scholar]
- Froyen G., Van Esch H., Bauters M., Hollanders K., Frints S.G., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P., Van Esch H., Bauters M., Hollanders K., Frints S.G., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P., Bauters M., Hollanders K., Frints S.G., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P., Hollanders K., Frints S.G., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P., Frints S.G., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P., Vermeesch J.R., Devriendt K., Fryns J.P., Marynen P., Devriendt K., Fryns J.P., Marynen P., Fryns J.P., Marynen P., Marynen P. Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: Important role for increased gene dosage of XLMR genes. Hum. Mutat. 2007;28:1034–1042. doi: 10.1002/humu.20564. [DOI] [PubMed] [Google Scholar]
- Gotter A.L., Nimmakayalu M.A., Jalali G.R., Hacker A.M., Vorstman J., Conforto D.D., Medne L., Emanuel B.S., Nimmakayalu M.A., Jalali G.R., Hacker A.M., Vorstman J., Conforto D.D., Medne L., Emanuel B.S., Jalali G.R., Hacker A.M., Vorstman J., Conforto D.D., Medne L., Emanuel B.S., Hacker A.M., Vorstman J., Conforto D.D., Medne L., Emanuel B.S., Vorstman J., Conforto D.D., Medne L., Emanuel B.S., Conforto D.D., Medne L., Emanuel B.S., Medne L., Emanuel B.S., Emanuel B.S. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X.Y., Ray P.N., Worton R.G., Ray P.N., Worton R.G., Worton R.G. Mechanisms of tandem duplication in the Duchenne muscular dystrophy gene include both homologous and nonhomologous intrachromosomal recombination. EMBO J. 1991;10:2471–2477. doi: 10.1002/j.1460-2075.1991.tb07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Osaka H., Thurston V.C., Clarke J.T., Yoneyama A., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Osaka H., Thurston V.C., Clarke J.T., Yoneyama A., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Thurston V.C., Clarke J.T., Yoneyama A., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Clarke J.T., Yoneyama A., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Yoneyama A., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R., Hodes M.E., Shaffer L.G., Lupski J.R., Shaffer L.G., Lupski J.R., Lupski J.R. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am. J. Hum. Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomietz E., Meyn M.S., Pandita A., Squire J.A., Meyn M.S., Pandita A., Squire J.A., Pandita A., Squire J.A., Squire J.A. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- Koszul R., Caburet S., Dujon B., Fischer G., Caburet S., Dujon B., Fischer G., Dujon B., Fischer G., Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus E., Leung W.Y., Haber J.E., Leung W.Y., Haber J.E., Haber J.E. Break-induced replication: A review and an example in budding yeast. Proc. Natl. Acad. Sci. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsche K., Ressler B., Katzera H.G., Orth U., Gillessen-Kaesbach G., Morlot S., Schwinger E., Gal A., Ressler B., Katzera H.G., Orth U., Gillessen-Kaesbach G., Morlot S., Schwinger E., Gal A., Katzera H.G., Orth U., Gillessen-Kaesbach G., Morlot S., Schwinger E., Gal A., Orth U., Gillessen-Kaesbach G., Morlot S., Schwinger E., Gal A., Gillessen-Kaesbach G., Morlot S., Schwinger E., Gal A., Morlot S., Schwinger E., Gal A., Schwinger E., Gal A., Gal A. Characterization of breakpoint sequences of five rearrangements in L1CAM and ABCD1 (ALD) genes. Hum. Mutat. 2002;19:526–535. doi: 10.1002/humu.10072. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Lupski J.R., Lupski J.R. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Inoue K., Cheung S.W., Shaw C.A., Stankiewicz P., Lupski J.R., Inoue K., Cheung S.W., Shaw C.A., Stankiewicz P., Lupski J.R., Cheung S.W., Shaw C.A., Stankiewicz P., Lupski J.R., Shaw C.A., Stankiewicz P., Lupski J.R., Stankiewicz P., Lupski J.R., Lupski J.R. Role of genomic architecture in PLP1 duplication causing Pelizaeus-Merzbacher disease. Hum. Mol. Genet. 2006;15:2250–2265. doi: 10.1093/hmg/ddl150. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Carvalho C.M., Lupski J.R., Carvalho C.M., Lupski J.R., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Albertson D.G., Pinkel D., Altshuler D.M., Pinkel D., Altshuler D.M., Altshuler D.M., et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood W.W., Chari R., Chi B., Lam W.L., Chari R., Chi B., Lam W.L., Chi B., Lam W.L., Lam W.L. Recent advances in array comparative genomic hybridization technologies and their applications in human genetics. Eur. J. Hum. Genet. 2006;14:139–148. doi: 10.1038/sj.ejhg.5201531. [DOI] [PubMed] [Google Scholar]
- Lu X., Shaw C.A., Patel A., Li J., Cooper M.L., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Shaw C.A., Patel A., Li J., Cooper M.L., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Patel A., Li J., Cooper M.L., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Li J., Cooper M.L., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Cooper M.L., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., Sahoo T., Yatsenko S.A., Bacino C.A., Yatsenko S.A., Bacino C.A., Bacino C.A., et al. Clinical implementation of chromosomal microarray analysis: Summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs H., Abidi F., Bier J.A., Abuelo D., Ouzts L., Voeller K., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Abidi F., Bier J.A., Abuelo D., Ouzts L., Voeller K., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Bier J.A., Abuelo D., Ouzts L., Voeller K., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Abuelo D., Ouzts L., Voeller K., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Ouzts L., Voeller K., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Voeller K., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Fennell E., Stevenson R.E., Schwartz C.E., Arena F., Stevenson R.E., Schwartz C.E., Arena F., Schwartz C.E., Arena F., Arena F. XLMR syndrome characterized by multiple respiratory infections, hypertelorism, severe CNS deterioration and early death localizes to distal Xq28. Am. J. Med. Genet. 1999;85:243–248. doi: 10.1002/(sici)1096-8628(19990730)85:3<243::aid-ajmg11>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Lupski J.R. Genome structural variation and sporadic disease traits. Nat. Genet. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- Madrigal I., Rodriguez-Revenga L., Armengol L., Gonzalez E., Rodriguez B., Badenas C., Sanchez A., Martinez F., Guitart M., Fernandez I., Rodriguez-Revenga L., Armengol L., Gonzalez E., Rodriguez B., Badenas C., Sanchez A., Martinez F., Guitart M., Fernandez I., Armengol L., Gonzalez E., Rodriguez B., Badenas C., Sanchez A., Martinez F., Guitart M., Fernandez I., Gonzalez E., Rodriguez B., Badenas C., Sanchez A., Martinez F., Guitart M., Fernandez I., Rodriguez B., Badenas C., Sanchez A., Martinez F., Guitart M., Fernandez I., Badenas C., Sanchez A., Martinez F., Guitart M., Fernandez I., Sanchez A., Martinez F., Guitart M., Fernandez I., Martinez F., Guitart M., Fernandez I., Guitart M., Fernandez I., Fernandez I., et al. X-chromosome tiling path array detection of copy number variants in patients with chromosome X-linked mental retardation. BMC Genomics. 2007;8:443. doi: 10.1186/1471-2164-8-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J., Haber J.E., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- Menten B., Maas N., Thienpont B., Buysse K., Vandesompele J., Melotte C., De Ravel T., Van Vooren S., Balikova I., Backx L., Maas N., Thienpont B., Buysse K., Vandesompele J., Melotte C., De Ravel T., Van Vooren S., Balikova I., Backx L., Thienpont B., Buysse K., Vandesompele J., Melotte C., De Ravel T., Van Vooren S., Balikova I., Backx L., Buysse K., Vandesompele J., Melotte C., De Ravel T., Van Vooren S., Balikova I., Backx L., Vandesompele J., Melotte C., De Ravel T., Van Vooren S., Balikova I., Backx L., Melotte C., De Ravel T., Van Vooren S., Balikova I., Backx L., De Ravel T., Van Vooren S., Balikova I., Backx L., Van Vooren S., Balikova I., Backx L., Balikova I., Backx L., Backx L., et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: A new series of 140 patients and review of published reports. J. Med. Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimault C., Giraud G., Courtois V., Cailloux F., Boire J.Y., Dastugue B., Boespflug-Tanguy O., Giraud G., Courtois V., Cailloux F., Boire J.Y., Dastugue B., Boespflug-Tanguy O., Courtois V., Cailloux F., Boire J.Y., Dastugue B., Boespflug-Tanguy O., Cailloux F., Boire J.Y., Dastugue B., Boespflug-Tanguy O., Boire J.Y., Dastugue B., Boespflug-Tanguy O., Dastugue B., Boespflug-Tanguy O., Boespflug-Tanguy O. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher Disease: Duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. The Clinical European Network on Brain Dysmyelinating Disease. Am. J. Hum. Genet. 1999;65:360–369. doi: 10.1086/302483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Albertson D.G., Albertson D.G. Comparative genomic hybridization. Annu. Rev. Genomics Hum. Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- Plenge R.M., Stevenson R.A., Lubs H.A., Schwartz C.E., Willard H.F., Stevenson R.A., Lubs H.A., Schwartz C.E., Willard H.F., Lubs H.A., Schwartz C.E., Willard H.F., Schwartz C.E., Willard H.F., Willard H.F. Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am. J. Hum. Genet. 2002;71:168–173. doi: 10.1086/341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., Glaze D., Krull K., Lee J.A., Lewis R.A., Krull K., Lee J.A., Lewis R.A., Lee J.A., Lewis R.A., Lewis R.A., et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M., Dujon B., Fairhead C., Dujon B., Fairhead C., Fairhead C. Distance from the chromosome end determines the efficiency of double strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 2003;328:847–862. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- Richardson C., Jasin M., Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooms L., Reyniers E., Kooy R.F., Reyniers E., Kooy R.F., Kooy R.F. Diverse chromosome breakage mechanisms underlie subtelomeric rearrangements, a common cause of mental retardation. Hum. Mutat. 2007;28:177–182. doi: 10.1002/humu.20421. [DOI] [PubMed] [Google Scholar]
- Rosenberg C., Knijnenburg J., Bakker E., Vianna-Morgante A.M., Sloos W., Otto P.A., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Knijnenburg J., Bakker E., Vianna-Morgante A.M., Sloos W., Otto P.A., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Bakker E., Vianna-Morgante A.M., Sloos W., Otto P.A., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Vianna-Morgante A.M., Sloos W., Otto P.A., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Sloos W., Otto P.A., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Otto P.A., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Kriek M., Hansson K., Krepischi-Santos A.C., Fiegler H., Hansson K., Krepischi-Santos A.C., Fiegler H., Krepischi-Santos A.C., Fiegler H., Fiegler H., et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: Clinical significance of imbalances present both in affected children and normal parents. J. Med. Genet. 2006;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlaville D., Prieur M., de Blois M.C., Genevieve D., Lapierre J.M., Ozilou C., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., Prieur M., de Blois M.C., Genevieve D., Lapierre J.M., Ozilou C., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., de Blois M.C., Genevieve D., Lapierre J.M., Ozilou C., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., Genevieve D., Lapierre J.M., Ozilou C., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., Lapierre J.M., Ozilou C., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., Ozilou C., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., Picq M., Gosset P., Morichon-Delvallez N., Munnich A., Gosset P., Morichon-Delvallez N., Munnich A., Morichon-Delvallez N., Munnich A., Munnich A., et al. Functional disomy of the Xq28 chromosome region. Eur. J. Hum. Genet. 2005;13:579–585. doi: 10.1038/sj.ejhg.5201384. [DOI] [PubMed] [Google Scholar]
- Sen S.K., Han K., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Han K., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Cordaux R., Liang P., Batzer M.A., Liang P., Batzer M.A., Batzer M.A. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Sheen C.R., Jewell U.R., Morris C.M., Brennan S.O., Ferec C., George P.M., Smith M.P., Chen J.M., Jewell U.R., Morris C.M., Brennan S.O., Ferec C., George P.M., Smith M.P., Chen J.M., Morris C.M., Brennan S.O., Ferec C., George P.M., Smith M.P., Chen J.M., Brennan S.O., Ferec C., George P.M., Smith M.P., Chen J.M., Ferec C., George P.M., Smith M.P., Chen J.M., George P.M., Smith M.P., Chen J.M., Smith M.P., Chen J.M., Chen J.M. Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication. Hum. Mutat. 2007;28:1198–1206. doi: 10.1002/humu.20591. [DOI] [PubMed] [Google Scholar]
- Small K., Warren S.T., Warren S.T. Emerin deletions occurring on both Xq28 inversion backgrounds. Hum. Mol. Genet. 1998;7:135–139. doi: 10.1093/hmg/7.1.135. [DOI] [PubMed] [Google Scholar]
- Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Stallings R.L., Yoon K., Kwek S., Ko D., Yoon K., Kwek S., Ko D., Kwek S., Ko D., Ko D. The origin of chromosome imbalances in neuroblastoma. Cancer Genet. Cytogenet. 2007;176:28–34. doi: 10.1016/j.cancergencyto.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P., Bauer-Sardina I., Saccone N.L., Putzel J., Laitinen T., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y., Bauer-Sardina I., Saccone N.L., Putzel J., Laitinen T., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y., Saccone N.L., Putzel J., Laitinen T., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y., Putzel J., Laitinen T., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y., Laitinen T., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y., Kere J., Pilia G., Rice J.P., Kwok P.Y., Pilia G., Rice J.P., Kwok P.Y., Rice J.P., Kwok P.Y., Kwok P.Y. Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat. Genet. 2000;25:324–328. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- Van Esch H., Bauters M., Ignatius J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Bauters M., Ignatius J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Ignatius J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., Bienvenu T., Jensen L.R., Gecz J., Jensen L.R., Gecz J., Gecz J., et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers L.E., Veltman J.A., van Kessel A.G., Brunner H.G., Veltman J.A., van Kessel A.G., Brunner H.G., van Kessel A.G., Brunner H.G., Brunner H.G. Identification of disease genes by whole genome CGH arrays. Hum. Mol. Genet. 2005;14:R215–R223. doi: 10.1093/hmg/ddi268. [DOI] [PubMed] [Google Scholar]
- Willis T.G., Jadayel D.M., Coignet L.J.A., Abdul-Rauf M., Treleaven J.G., Catovsky D., Dyer M.J., Jadayel D.M., Coignet L.J.A., Abdul-Rauf M., Treleaven J.G., Catovsky D., Dyer M.J., Coignet L.J.A., Abdul-Rauf M., Treleaven J.G., Catovsky D., Dyer M.J., Abdul-Rauf M., Treleaven J.G., Catovsky D., Dyer M.J., Treleaven J.G., Catovsky D., Dyer M.J., Catovsky D., Dyer M.J., Dyer M.J. Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood. 1997;90:2456–2464. [PubMed] [Google Scholar]
- Woodward K.J., Cundall M., Sperle K., Sistermans E.A., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Cundall M., Sperle K., Sistermans E.A., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Sperle K., Sistermans E.A., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Sistermans E.A., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., Burford D.C., Carter N.P., Hobson D.L., Carter N.P., Hobson D.L., Hobson D.L., et al. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am. J. Hum. Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]