Abstract
Studies of microsatellites evolution based on marker data almost inherently suffer from an ascertainment bias because there is selection for the most mutable and polymorphic loci during marker development. To circumvent this bias we took advantage of whole-genome shotgun sequence data from three unrelated chicken individuals that, when aligned to the genome reference sequence, give sequence information on two chromosomes from about one-fourth (375,000) of all microsatellite loci containing di- through pentanucleotide repeat motifs in the chicken genome. Polymorphism is seen at loci with as few as five repeat units, and the proportion of dimorphic loci then increases to 50% for sequences with ∼10 repeat units, to reach a maximum of 75%–80% for sequences with 15 or more repeat units. For any given repeat length, polymorphism increases with decreasing GC content of repeat motifs for dinucleotides, nonhairpin-forming trinucleotides, and tetranucleotides. For trinucleotide repeats which are likely to form hairpin structures, polymorphism increases with increasing GC content, indicating that the relative stability of hairpins affects the rate of replication slippage. For any given repeat length, polymorphism is significantly lower for imperfect compared to perfect repeats and repeat interruptions occur in >15% of loci. However, interruptions are not randomly distributed within repeat arrays but are preferentially located toward the ends. There is negative correlation between microsatellite abundance and single nucleotide polymorphism (SNP) density, providing large-scale genomic support for the hypothesis that equilibrium microsatellite distributions are governed by a balance between rate of replication slippage and rate of point mutation.
Empirical data on microsatellite mutability and polymorphism almost always come with the limitation of suffering from an ascertainment bias. For instance, direct observations of de novo mutation events in pedigrees are essentially confined to loci with very high mutation rates, which are not necessarily representative for the majority of microsatellite loci in the genome when it comes to rate and pattern of evolution (Weber and Wong 1993; Ellegren 2000; Huang et al. 2002). The same applies to observations on microsatellite allele frequency distributions at loci genotyped in population samples (Estoup et al. 1995). Such data tend to be biased toward highly polymorphic loci because there is a selection for polymorphism at various stages of the process of marker development; short repeat tracts are avoided for marker design, monomorphic markers or markers with limited polymorphism are typically discarded at an early screening stage, and the most polymorphic loci would find most widespread use in subsequent studies. Using unusually mutable loci will lead to overestimates of genetic diversity and will give a biased picture of the microsatellite mutation process. Another example, and which is perhaps the most well-known aspect of microsatellite ascertainment bias, is the comparison of repeat lengths of orthologous loci in two related species. Everything else being equal, this will tend to give a pattern of longer repeats in the species from which markers were developed (the focal species), an inevitable consequence of the selection for long and polymorphic loci as described above (Ellegren et al. 1995, 1997; Webster et al. 2002; Vowles and Amos 2006). Again, this will lead to incorrect interpretations of microsatellite mutation and evolution.
Whole-genome sequence surveys for microsatellite occurrence avoid this ascertainment bias. Such analyses give a snapshot of the distribution of repeat lengths across the genome, which can be compared to expectations of theoretical models (Dieringer and Schlötterer 2003). However, in the absence of polymorphism data, they do not capture on-going evolutionary processes. For a few species genome sequencing has been augmented with large-scale initiatives toward obtaining sequence information from multiple individuals, like re-sequencing of targeted regions in the human HapMap (International HapMap Consortium 2005) or sparse shotgun sequencing made in different dog (Canis familiaris) breeds (Lindblad-Toh et al. 2005). One of the most extensive efforts of this kind is the light shotgun sequencing of three different domestic chicken (Gallus gallus domesticus) (International Chicken Polymorphism Map Consortium 2004), made in addition to the assembly of the chicken genome sequence, which was based on sequencing of a red jungle fowl (G. g. gallus, the wild ancestor to domestic chicken) (International Chicken Genome Sequencing Consortium 2004). This generated sequence data for another chromosome (than the reference sequence) from the chicken population for about half the genome, uncovering a total of 2.8 million single nucleotide polymorphisms (SNPs) (International Chicken Genome Sequencing Consortium 2004) and more than 270,000 length polymorphisms (Brandström and Ellegren 2007). Here, we use these data to obtain an unbiased picture of microsatellite variability in a vertebrate genome and to address several general questions pertinent to microsatellite evolution. Importantly, due to the more or less random nature of shotgun sequencing, this approach gives diversity data for one of the most polymorphic sequence categories in eukaryotic genomes without being confined by an ascertainment bias.
Results
The length dependence of microsatellite polymorphism
A survey of the chicken genome assembly identifies 1,615,000 loci with perfect di- through pentanucleotide microsatellites with a length of three repeat units or longer. Quality filtered data from 0.25× shotgun sequencing of three unrelated chicken individuals, from different breeds, provide information for ∼375,000 microsatellite loci (23% of the genomic total). In this draw of two chromosomes from the chicken population 7300 (1.8%) of the loci are polymorphic.
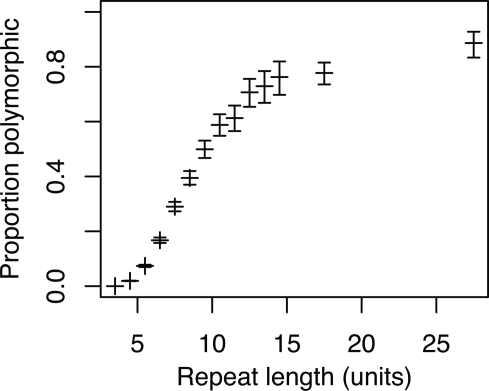
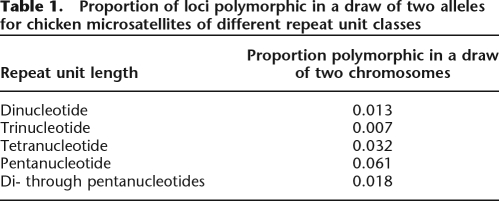
With two chromosomes sampled, per locus, sequencing might have revealed either two identical alleles or two different alleles. Subsequently we combine all loci of any given repeat length to obtain the proportion of polymorphic loci for that size class (using the arithmetic mean for loci with two alleles). An analysis of the relationship between microsatellite length and degree of polymorphism shows how the proportion of dimorphic loci increases with repeat length (Fig. 1). Although rare, intraspecific length variation does occur at repeats with as few as five repeat units. Fifty percent of all loci are dimorphic for sequences with ∼10 repeat units, and polymorphism then increases asymptotically to reach a plateau of 75%–80% of loci being polymorphic for repeat tracts with >15–20 repeat units. Logistic regression models of the dependence of microsatellite length on polymorphism level show a more uniform relationship for di-, tri-, tetra-, and pentanucleotide repeats when considering the effect of the number of repeat units than when considering the length of the repeat tract in base pairs (Supplemental Fig. 1). Yet, there is significant variation among di-, tri-, tetra-, and pentanucleotide repeats in the length dependence given by the number of repeat units on polymorphism level (logistic regression, P < 10−15). Table 1 summarizes the proportion of loci found to be polymorphic for different repeat length classes. A larger fraction of tetra- and, in particular, pentanucleotide repeats are variable, compared to di- and trinucleotide repeats. A breakdown on individual repeat motifs is presented in Supplemental Table 1.
Figure 1.
Proportion of dimorphic loci in relation to repeat length for all microsatellites. Whiskers indicate the 95% confidence interval. Because of small sample size, results from the 15–20 and 20–35 repeat unit intervals have been pooled.
Table 1.
Proportion of loci polymorphic in a draw of two alleles for chicken microsatellites of different repeat unit classes
Microsatellite polymorphism and repeat motifs
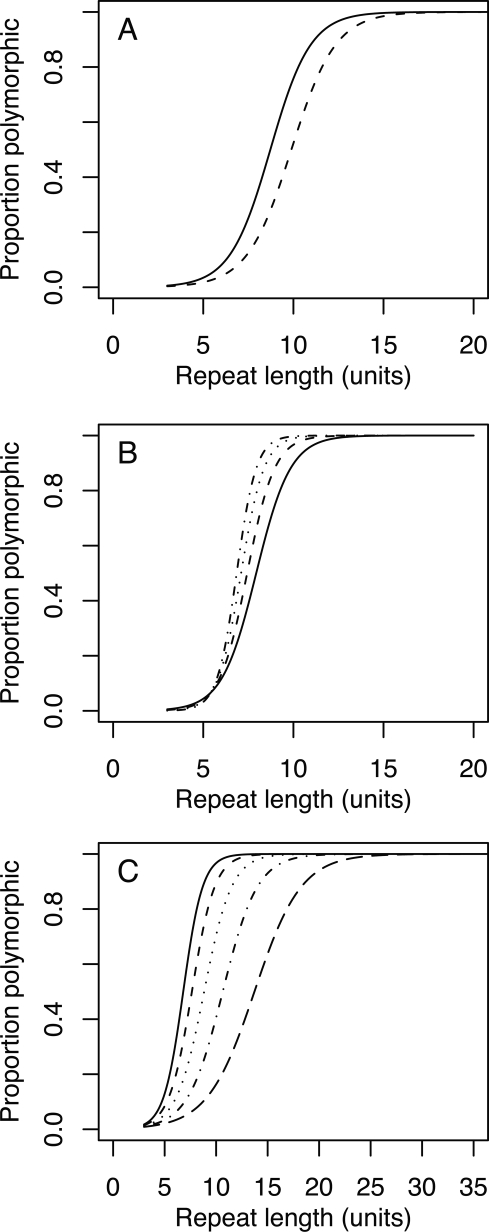
A closer examination of polymorphism levels for different repeat motifs reveals distinct differences among motifs (for details, see Supplemental Figs. 2, 3). For dinucleotide repeats and at any given repeat length (Fig. 2A), (AT)n shows higher variability than (AC)n and (AG)n, and this is the case throughout the whole spectrum of repeat lengths. As this could potentially be related to base composition (GC-poor motifs being less variable), we analyzed polymorphism levels of different tri- and tetranucleotide repeat motifs with respect to their GC content. Figure 2C shows a very clear pattern for tetranucleotide repeats, with the highest polymorphism seen at motifs with 0% GC and the lowest at 100% GC. This mimics the trend for dinucleotide repeats. However, for trinucleotide repeats the opposite relationship is observed: For any given repeat length, motifs with 100% GC show the highest polymorphism and those with 0% the lowest (Fig. 2B). While unusual secondary structures may be attained by several types of microsatellites, some trinucleotide repeats may be more prone to form stabilizing hairpin structures during strand dissociation than other repeat classes, increasing the rate of replication slippage (Mitas 1997). To test if this could potentially explain the deviating pattern for trinucleotide repeats, we divided them in motifs that have two adjacent self-complementary nucleotides (like ACT and AGC) and that would have a tendency for hairpin formation, and those that have not (like AAC and AGG). Repeats with less hairpin-forming potential show the pattern observed for other repeat classes with the highest variability for GC-poor motifs and lowest for GC-rich motifs (Supplemental Fig. 4). In contrast, motifs which are more likely to form hairpin structure are less variable when GC-poor and more variable when GC-rich.
Figure 2.
Fitted logistic regression models of proportion polymorphic microsatellites as a function of microsatellite length for dinucleotide repeats (A), trinucleotide repeats (B), and tetranucleotide repeats (C). (A) (solid line) (AT)n; (dashed line) (AC)n and (AG)n. (B) (Solid line) Motifs with 0% GC; (dashed line) 33% GC; (dotted line) 67% GC; (dotted-dashed line) 100% GC. (C) (Solid line) 0% GC motifs; (short dashed line) 25% GC; (dotted line) 50%; (dotted-dashed line) 75%; (long dashed line) 100% GC.
The reduction of microsatellite polymorphism from repeat interruptions
It has been recognized that sequence variation within repeat tracts can generate microsatellite alleles identical in size but different in sequence (Estoup et al. 1995; Garza and Freimer 1996). We were able to quantify the genome-wide occurrence of this form of microsatellite heterogeneity by observing that 6% of all loci show one perfect and one imperfect (interrupting nucleotide) allelic repeat array. For microsatellites longer than 15 repeat units, this proportion is as high as 16%. Interruptions within perfect repeat arrays reduce the likelihood for a microsatellite locus being polymorphic. By including the presence of imperfections in one of the two sequenced alleles as an independent variable in logistic regressions of polymorphism on length, we found that, for any given length, imperfect microsatellites are significantly less variable compared to perfect repeat loci (P < 10−15, Supplemental Fig. 5).
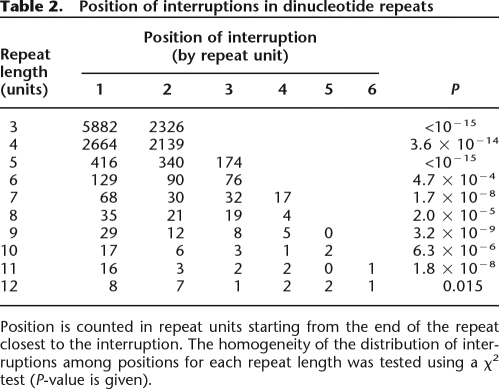
Interestingly, the position in repeat sequences at which interruptions occur is clearly nonrandom. For all dinucleotide repeat lengths from three to 12 units, which is the size interval we have sufficient data on interruptions for, there is a highly significant tendency for interruptions to be biased toward the end of repeat regions (Table 2). The very first position is particularly prone to point mutation throughout all repeat lengths. A similar trend is found for trinucleotide repeats (Supplemental Table 2).
Table 2.
Position of interruptions in dinucleotide repeats
Position is counted in repeat units starting from the end of the repeat closest to the interruption. The homogeneity of the distribution of interruptions among positions for each repeat length was tested using a χ2 test (P-value is given).
Microsatellite abundance
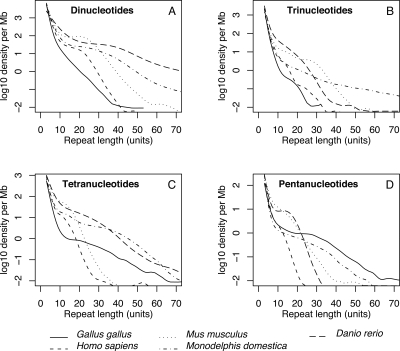
Using the same search criteria as applied to chicken for determining the genomic occurrence of microsatellites, we also surveyed the human, mouse, opossum, and zebrafish genomes for microsatellite abundance. Microsatellites account for a smaller proportion of the chicken genome than they do for other vertebrate genomes (Supplemental Table 3). The microsatellite frequency is 30%–80% higher in mammals than in chicken and, given the larger genome size of the three investigated mammals, the total number of microsatellites in these species is thus three to five times higher than in chicken.
There is a well-known negative relationship between microsatellite density and microsatellite length (Toth et al. 2000; Dieringer and Schlötterer 2003), which is also seen in the chicken genome as well as in human, mouse, opossum, and zebra fish (Fig. 3). However, the character of this relationship varies between species and repeat types. For di- and trinucleotide repeats, microsatellite density in chicken is lower than in the other vertebrates over the whole range of repeat lengths analyzed. On the other hand, long tetra- and, in particular, pentanucleotide repeats tend to be more common in chicken relative to the other species. Coupled with the positive correlation between length and polymorphism, this is consistent with the observation of a higher proportion of chicken tetra- and pentanucleotide repeats being polymorphic, compared to di- and trinucleotide repeats (Table 1).
Figure 3.
Genomic occurrence of di- through pentanucleotide microsatellites in five vertebrates in relation to repeat length. (A) Dinucleotides; (B) trinucleotides; (C) tetranucleotides; (D) pentanucleotides.
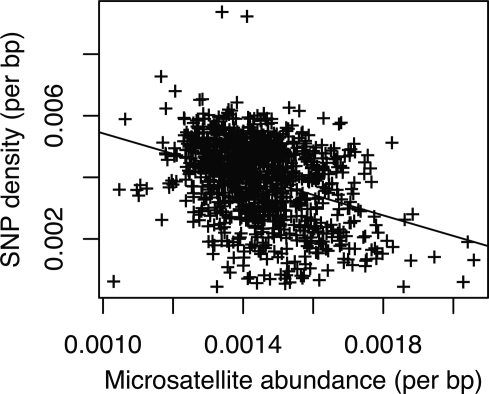
Using the same shotgun sequence data from the three chickens we analyzed the relationship between the density of SNPs and microsatellite abundance. There is a negative correlation between SNP density and microsatellite abundance (P < 10−10; Fig. 4). There is a significant heterogeneity in microsatellite density both within and among chromosomes when analyzed in nonoverlapping 1 Mb windows (ANOVA, P < 10−10). For example, the Z chromosome tends to have more frequent, and longer, tetra- and pentanucleotide repeats compared to the autosomes (data not shown). This is also reflected in that, overall, microsatellites on the Z chromosome are significantly longer than loci on the autosomes (t-test, P < 10−6). Base composition is an important factor explaining microsatellite density, although the effect varies among motifs. There is a negative correlation between GC content and the density of AT-rich motifs, while the opposite is seen for GC-rich motifs (Supplemental Fig. 6).
Figure 4.
The relationship between SNP density and microsatetellite abundance in nonoverlapping 1 Mb windows (R2 = −0.35, P = 10−15).
Discussion
It has been clearly shown that microsatellites used as genetic markers differ in several respects to a genomic sample of microsatellite loci, including in length, structure, and base composition (Pardi et al. 2005). The most important aspect of this study is therefore that it seeks to circumvent the general problem of ascertainment bias in the analysis of microsatellite polymorphism, something that has been an issue in basically all previous studies of microsatellite evolution and mutation using population data.
We found significant heterogeneity in polymorphism levels among microsatellites of different repeat motifs and this was evident even after controlling for variation in repeat length. For both di- and tetranucleotide repeats there were very clear trends in the direction of polymorphism increasing with decreasing GC content. The most straightforward and intuitive explanation for this is that the weaker hydrogen bonds between the two strands of AT-rich repeats result in more frequent strand dissociation and, subsequently, replication slippage-induced length mutation. Moreover, since AT-rich repeats are preferentially located in AT-rich genomic regions, the effect might be augmented by increased instability in immediately flanking regions. This is consistent with in vitro experiments with synthetic oligonucleotide which revealed a negative correlation between GC content and slippage rate (Schlötterer and Tautz 1992).
A different relationship between GC content and polymorphism level was seen for trinucleotide repeats. This class of tandem repeats is well-known for their unusual helical properties in the formation of DNA structures (for review, see Pearson and Sinden 1998). One important observation from biophysical work is that trinucleotide repeats are more flexible and curved molecules compared to other repeats (Bacolla et al. 1997; Chastain and Sinden 1998). Most notably, many of them attain hairpin structures in the leading daughter strand synthesized during DNA replication. In addition, other structures such as hairpins on both strands, cruciforms, triplexes, and quadruplexes are known to occur (Pearson and Sinden 1998). Hairpin structures stabilize slipped strand intermediates and thereby increase the rate of slippage-generated length mutations (Gellibolian et al. 1997); this is thought to be an important mechanism behind neurodegenerative diseases caused by trinucleotide expansion.
As not all trinucleotide repeat motifs are likely to form hairpin structures, we separately analyzed motifs with two adjacent self-complementary nucleotides (potential hairpin formation) and motifs without; this classification broadly corresponds to high and low instability of the different motifs seen in in vitro experiments (Mitas 1997) as well as in in vivo studies of Escherichia coli and Saccharomyces cerevisiae (Lenzmeier and Freudenreich 2003). For motifs less likely to form hairpins the same trend as for di- and tetranucleotide repeats was observed, with AT-rich repeats showing the highest variability. In contrast, for trinucleotide repeats with hairpin-forming potential, genetic diversity was highest for GC-rich repeats. This leads to a model in which polymorphism in di- and tetranucleotide repeats, as well as in nonhairpin-forming trinucleotide repeats, is governed primarily by the instability of the double helix over the repeat tract as determined by base composition. For hairpin-forming trinucleotide repeats, the model suggests that it is the stability of within-strand secondary structures, as determined by GC content, that plays an overall role in governing polymorphism levels.
It has been suggested that species-specific rates of point mutation determine the genomic equilibrium length distribution of microsatellites (Bell and Jurka 1997; Kruglyak et al. 1998, 2000). According to this model, microsatellite growth is promoted by replication slippage while point mutations act in the opposite direction, introducing interruptions within repeat arrays that hinder further expansion by lowering the slippage rate (see below). There is significant variation in point mutation rates also within genomes (Ellegren et al. 2003), including in chicken (Webster et al. 2006), and this could potentially affect the length of individual microsatellite loci. Specifically, it predicts that, in regions with high point mutation rates, the frequent occurrence of microsatellite imperfections from point mutation will impede the evolution of long repeat arrays, essentially making microsatellites rarer. Preliminary support for this hypothesis was provided by Santibáñez-Koref et al. (2001) who, for a set of rodent microsatellite markers, found a negative correlation between flanking sequence divergence and repeat lengths at (CA)n loci. Nucleotide diversity is at least in part determined by variation in the underlying mutation rate, and SNP density can thus be used as a measure of the local rate of point mutation. The negative correlation seen between SNP density and microsatellite abundance in chicken, therefore, provides genome-wide support that the local rate of point mutation is a general governor of microsatellite evolution at the level of individual loci.
Our study confirms the well-known relationship between microsatellite length and polymorphism (Weber 1990). In addition, it is able to determine the character of this relationship over the full spectrum of repeat lengths. Comparisons of orthologous regions in human and chimpanzee genomes have revealed that, over evolutionary time scale, mutations leading to interspecific length variation do occur even for short repeats, albeit at a low rate (Webster et al. 2002). We show that intraspecific length polymorphism is present at chicken microsatellite loci with as few as five repeat units. Assuming that replication slippage is the main mechanism of microsatellite mutation, length mutation thus occurs at a sufficiently high rate in genomic regions with only a limited number of repeat units, to generate polymorphism in a population sample (cf. Zhu et al. 2000; Nishizawa and Nishizawa 2002). This is in line with recent work on short insertions and deletions (indels) in the chicken genome showing that tandem duplication are highly overrepresented at indel sites (Brandström and Ellegren 2007).
Selkoe and Toonen (2006) concluded that detectable microsatellite homoplasy, the presence of two or more alleles identical by state but not by descent (i.e., when an interrupted allele has the same length as a perfect repeat array), appears “to affect only a fraction of genotypes at a fraction of loci.” However, our data indicates that interruptions in microsatellite sequences are more common than previously thought, making this statement invalid. Sixteen percent of microsatellites with >15 repeat units showed one allele with a perfect and one with an imperfect repeat array. Given the multiallelic nature of long microsatellite loci, it seems evident that an even larger proportion of loci, perhaps the majority, would have shown interrupted alleles had more than two chromosomes been sequenced per locus. This is not unexpected since nucleotide diversity in the chicken genome is high. For example, Sundström et al. (2004) found one segregating site every 39 bp of autosomal sequence in a population sample of 25 chickens. A species with lower levels of single nucleotide polymorphisms should be expected to have less microsatellite interruptions.
Clearly, for any given total length of a microsatellite locus, interruptions have the consequence of lowering variability, most likely due to the stabilizing effects of unique sequence within tandem array that prevent replication slippage (Petes et al. 1997; Rolfsmeier and Lahue 2000; Sibly et al. 2003). This may contribute to the variance in genetic diversity often seen among microsatellites of similar length (cf. Fig. 1). Homoplasy can affect population genetic analyses like inflating estimates of gene flow and genetic differentiation (Adams et al. 2004; Curtu et al. 2004).
Brohede and Ellegren (1999) analyzed a number of sheep and ovine microsatellite orthologs and found a tendency for point mutations to be enriched in microsatellite ends and in the immediate microsatellite flanking regions. A larger data set of human–chimpanzee orthologs was analyzed for flanking sequence divergence by Vowles and Amos (2006), who also found a higher substitution rate in the flanking positions closest to repeat regions. In our genome-wide set of chicken microsatellites there is a very clear trend of interruptions being more common in the very end of repeat regions. There are several possible explanations to this observation. Obviously, the point mutation rate may be higher in these regions, for example, because of structural alterations when DNA goes from unique to repetitive sequence or because of a propensity for loop formation during strand slippage in end regions coupled with a relative mutational fragility of looped regions. Another possibility is that point mutations occur more randomly within repeat arrays but somehow “migrate” toward the ends during subsequent slippage mutations or gene conversion-like processes.
The correlation between microsatellite length and variability has implications for the relative polymorphism content of different classes of repeats in the chicken genome. This can be concluded from the observations of a comparatively high proportion of tetra- and pentanucleotide repeats being represented by long arrays and the higher proportion of polymorphic loci among tetra- and pentanucleotide than di- and trinucleotide repeats. Long and highly polymorphic tetra- and pentanucleotide repeats have been found in several different bird species (e.g., Primmer et al. 1998). Genomic surveys show that the length distributions of tetra- and pentanucleotides differ between birds and mammals, birds having much longer repeats. Such difference in length distributions adds a further dimension on microsatellite heterogeneity to previous observations of differences in the relative occurrence of repeat motifs in eukaryotic genomes (Toth et al. 2000; International Human Genome Sequencing Consortium 2001; Katti et al. 2001; Morgante et al. 2002; Dieringer and Schlötterer 2003). Elucidating the mechanisms behind such differences shall be an important topic for further research. As shown here and elsewhere (Dieringer and Schlötterer 2003), base composition correlates with the relative abundance of different repeat motifs within genomes and may therefore also be a factor explaining differences among genomes. However, overall, microsatellite abundance is lower in birds than in mammals (cf. Primmer et al. 1997), which is also the case when it comes to interspersed repeats (International Chicken Genome Sequencing Consortium 2004). Compared to the common ancestor of amniotes (Shedlock et al. 2007), there thus seems to have been a general loss of repeat sequences in the lineage, leading to the minimalist avian genome.
Conclusions
This genome-wide study in chicken has attempted to provide an unbiased picture of microsatellite evolution by circumventing the ascertainment bias associated with inferring evolutionary processes in microsatellite sequences using data from genetic markers. We confirmed the well-known relationship between microsatellite length and polymorphism level and were able to quantify this relationship from lengths of just a few repeat units up to several tens. We show for the first time how polymorphism is dependent on base composition, with the degree of diversity being positively correlated with GC for di- and tetranucleotide repeats but negatively correlated for trinucleotide repeats. We show that repeat interruptions (imperfect repeats) occur at a significant fraction of all loci, more often than previously thought, and that such interruptions reduce polymorphism levels. Related to the latter, we provide genome-wide evidence that a high local rate of point mutation lowers microsatellite abundance, supporting the hypothesis that the occurrence of microsatellite at equilibrium is a balance between point mutation and replication slippage rates. Altogether, the approach of using genomic sequence data from multiple individuals for inferring microsatellite evolution offers a new and important means for an increased understanding of the dynamics of this abundant class of repeat sequences.
Methods
Sequence data
We downloaded version 2.1 (galGal3) of the chicken genome assembly as well as the complete genomes of human (Hg18), mouse (Mm8), opossum (MonDom4), and zebra fish (DanRer4) from the University of California at Santa Cruz genome browser (http://genome.ucsc.edu). In conjunction to the sequencing of the chicken (red jungle fowl) genome, one individual of each of three domestic chicken breeds (Layer, Broiler, Silke) has also been sequenced to a low coverage, with approximately one million reads per breed (International Chicken Polymorphism Map Consortium 2004). Alignments of these reads to version 2.1 of the chicken genome assembly were kindly provided by G.K.-S. Wong (Beijing Institute of Genomics of the Chinese Academy of Sciences). To extract high-quality alignments we used an approach similar to Mills et al. (2006), filtering the alignments to only contain the longest region with a sequence quality of >Q25 over at least 100 bp. The alignments were also filtered for overlaps within each breed, to ensure that only two chromosomes were compared in each pairwise (breed to reference sequence) comparison. After quality filtering the coverage was roughly 10% from each of the three breeds.
Microsatellite detection
We used a modified version of the program sputnik (C. Abajian, unpubl.; Morgante et al. 2002) to search whole genome sequence data for microsatellites. For all species we used the same settings to extract all perfect microsatellites 6 bp or longer, where the repeat unit was 5 bp or shorter (equivalent to the flags -R 0 -v 1 -u 5 -s 4 -L 4 -l 0). The output from sputnik was then filtered to only include microsatellites of three repeat units or longer of di- through pentanucleotide repeats. The inclusion of simple repeats containing as few as three repeat units was motivated by the fact that tandem repeat length mutations do occur in tandem repeats of such short lengths (Zhu et al. 2000; Brandström and Ellegren 2007). Mononucleotide repeats were excluded from this analysis as they tend to be more sensitive to sequencing errors (International Chicken Polymorphism Map Consortium 2004). All microsatellites were grouped into their canonical motifs by sputnik. Note that (GC)n does not exist in the chicken genome in sufficient numbers to allow a meaningful comparison to other dinucleotide motifs.
Polymorphism data
We extracted polymorphism data for microsatellites, defined using the sputnik searches described above, with the same parameters, from the alignments of chicken breed sequences to the genome reference sequence. In order to find cases of one perfect and one imperfect repeat both sequences of the alignment were searched for microsatellites. Alignment gaps in microsatellite regions were interpreted as length polymorphisms and we also recorded all other forms of sequence differences between the two alleles within microsatellite regions. Cases of compound microsatellites (i.e., when one microsatellite is followed directly by another microsatellite motif) were discarded (17,672 loci). Data on single nucleotide polymorphisms (SNPs) have previously been extracted from the same alignments (Brandström and Ellegren 2007) and were used herein for comparative purposes. When assessing the relationship between microsatellite length and proportion of dimorphic loci, or genomic parameters, we used the arithmetic mean of the length of the two alleles seen at dimorphic repeat loci.
Throughout the paper we refer to variability in pooled samples of loci as the proportion of loci dimorphic or polymorphic. In essence, this is the mean heterozygosity although we have the somewhat unusual situation of only having two chromosomes sampled per locus. For each locus, the observed heterozygosity is either 0 (two identical alleles sequenced) or 1 (two different alleles). While variance will obviously be larger with data from just a few chromosomes sampled per locus, mean heterozygosity is not affected by the number of chromosomes sampled per locus.
Statistical models
All statistical tests and models were done using the R statistics environment (R Development Core Team 2007). Models with microsatellite density and proportion polymorphic microsatellites were fitted using ordinary linear models. Models of microsatellite mutability were fitted using logistic regression. The binary value of whether a microsatellite had a length polymorphism or not was used as response variable. Logistic regression models were evaluated based on their Akaike Information Criterion (AIC) to find the best fitting models (Venables and Ripley 2002).
Acknowledgments
Financial support was obtained from the Swedish Research Council. We thank Gane Wong for the alignments. The comments of two anonymous reviewers helped to improve an earlier version of this manuscript.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075242.107.
References
- Adams R.I., Brown K.M., Hamilton M.B., Brown K.M., Hamilton M.B., Hamilton M.B. The impact of microsatellite electromorph size homoplasy on multilocus population structure estimates in a tropical tree (Corythophora alta) and an anadromous fish (Morone saxatilis) Mol. Ecol. 2004;13:2579–2588. doi: 10.1111/j.1365-294X.2004.02256.x. [DOI] [PubMed] [Google Scholar]
- Bacolla A., Gellibolian R., Shimizu M., Amirhaeri S., Kang S., Ohshima K., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Gellibolian R., Shimizu M., Amirhaeri S., Kang S., Ohshima K., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Shimizu M., Amirhaeri S., Kang S., Ohshima K., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Amirhaeri S., Kang S., Ohshima K., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Kang S., Ohshima K., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Ohshima K., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Larson J.E., Harvey S.C., Stollar B.D., Wells R.D., Harvey S.C., Stollar B.D., Wells R.D., Stollar B.D., Wells R.D., Wells R.D. Flexible DNA: Genetically unstable CTG.CAG and CGG.CCG from human hereditary neuromuscular disease genes. J. Biol. Chem. 1997;272:16783–16792. doi: 10.1074/jbc.272.27.16783. [DOI] [PubMed] [Google Scholar]
- Bell G.I., Jurka J., Jurka J. The length distribution of perfect dimer repetitive DNA is consistent with its evolution by an unbiased single-step mutation process. J. Mol. Evol. 1997;44:414–421. doi: 10.1007/pl00006161. [DOI] [PubMed] [Google Scholar]
- Brandström M., Ellegren H., Ellegren H. The genomic landscape of short insertion and deletion polymorphisms in the chicken (Gallus gallus) genome: A high frequency of deletions in tandem duplicates. Genetics. 2007;176:1691–1701. doi: 10.1534/genetics.107.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohede J., Ellegren H., Ellegren H. Microsatellite evolution: Polarity of substitutions within repeats and neutrality of flanking sequences. Proc. R. Soc. Lond. B. Biol. Sci. 1999;266:825–833. doi: 10.1098/rspb.1999.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain P.D., Sinden R.R., Sinden R.R. CTG repeats associated with human genetic disease are inherently flexible. J. Mol. Biol. 1998;275:405–411. doi: 10.1006/jmbi.1997.1502. [DOI] [PubMed] [Google Scholar]
- Curtu A.L., Finkeldey R., Gailing O., Finkeldey R., Gailing O., Gailing O. Comparative sequencing of a microsatellite locus reveals size homoplasy within and between European oak species (Quercus spp.) Plant Mol. Biol. Rep. 2004;22:339–346. [Google Scholar]
- Dieringer D., Schlötterer C., Schlötterer C. Two distinct modes of microsatellite mutation processes: Evidence from the complete genomic sequences of nine species. Genome Res. 2003;13:2242–2251. doi: 10.1101/gr.1416703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 2000;24:400–402. doi: 10.1038/74249. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Primmer C.R., Sheldon B.C., Primmer C.R., Sheldon B.C., Sheldon B.C. Microsatellite 'evolution': Directionality or bias? Nat. Genet. 1995;11:360–362. doi: 10.1038/ng1295-360. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Moore S., Robinson N., Byrne K., Ward W., Sheldon B.C., Moore S., Robinson N., Byrne K., Ward W., Sheldon B.C., Robinson N., Byrne K., Ward W., Sheldon B.C., Byrne K., Ward W., Sheldon B.C., Ward W., Sheldon B.C., Sheldon B.C. Microsatellite evolution—A reciprocal study of repeat lengths at homologous loci in cattle and sheep. Mol. Biol. Evol. 1997;14:854–860. doi: 10.1093/oxfordjournals.molbev.a025826. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Smith N.G., Webster M.T., Smith N.G., Webster M.T., Webster M.T. Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 2003;13:562–568. doi: 10.1016/j.gde.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Estoup A., Garnery L., Solignac M., Cornuet J.M., Garnery L., Solignac M., Cornuet J.M., Solignac M., Cornuet J.M., Cornuet J.M. Microsatellite variation in honey bee (Apis mellifera L.) populations: Hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics. 1995;140:679–695. doi: 10.1093/genetics/140.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza J.C., Freimer N.B., Freimer N.B. Homoplasy for size at microsatellite loci in humans and chimpanzees. Genome Res. 1996;6:211–217. doi: 10.1101/gr.6.3.211. [DOI] [PubMed] [Google Scholar]
- Gellibolian R., Bacolla A., Wells R.D., Bacolla A., Wells R.D., Wells R.D. Triplet repeat instability and DNA topology: An expansion model based on statistical mechanics. J. Biol. Chem. 1997;272:16793–16797. doi: 10.1074/jbc.272.27.16793. [DOI] [PubMed] [Google Scholar]
- Huang Q.Y., Xu F.H., Shen H., Deng H.Y., Liu Y.J., Liu Y.Z., Li J.L., Recker R.R., Deng H.W., Xu F.H., Shen H., Deng H.Y., Liu Y.J., Liu Y.Z., Li J.L., Recker R.R., Deng H.W., Shen H., Deng H.Y., Liu Y.J., Liu Y.Z., Li J.L., Recker R.R., Deng H.W., Deng H.Y., Liu Y.J., Liu Y.Z., Li J.L., Recker R.R., Deng H.W., Liu Y.J., Liu Y.Z., Li J.L., Recker R.R., Deng H.W., Liu Y.Z., Li J.L., Recker R.R., Deng H.W., Li J.L., Recker R.R., Deng H.W., Recker R.R., Deng H.W., Deng H.W. Mutation patterns at dinucleotide microsatellite loci in humans. Am. J. Hum. Genet. 2002;70:625–634. doi: 10.1086/338997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Chicken Polymorphism Map Consortium A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Katti M.V., Ranjekar P.K., Gupta V.S., Ranjekar P.K., Gupta V.S., Gupta V.S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- Kruglyak S., Durrett R.T., Schug M.D., Aquadro C.F., Durrett R.T., Schug M.D., Aquadro C.F., Schug M.D., Aquadro C.F., Aquadro C.F. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl. Acad. Sci. 1998;95:10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S., Durrett R., Schug M.D., Aquadro C.F., Durrett R., Schug M.D., Aquadro C.F., Schug M.D., Aquadro C.F., Aquadro C.F. Distribution and abundance of microsatellites in the yeast genome can be explained by a balance between slippage events and point mutations. Mol. Biol. Evol. 2000;17:1210–1219. doi: 10.1093/oxfordjournals.molbev.a026404. [DOI] [PubMed] [Google Scholar]
- Lenzmeier B.A., Freudenreich C.H., Freudenreich C.H. Trinucleotide repeat instability: A hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Mills R.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E., Tsui C., Pittard W.S., Devine S.E., Pittard W.S., Devine S.E., Devine S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2254. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Hanafey M., Powell W., Hanafey M., Powell W., Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Nishizawa K., Nishizawa K. A DNA sequence evolution analysis generalized by simulation and the markov chain monte carlo method implicates strand slippage in a majority of insertions and deletions. J. Mol. Evol. 2002;55:706–717. doi: 10.1007/s00239-002-2366-5. [DOI] [PubMed] [Google Scholar]
- Pardi F., Sibly R.M., Wilkinson M.J., Whittaker J.C., Sibly R.M., Wilkinson M.J., Whittaker J.C., Wilkinson M.J., Whittaker J.C., Whittaker J.C. On the structural differences between markers and genomic AC microsatellites. J. Mol. Biol. 2005;60:688–693. doi: 10.1007/s00239-004-0274-6. [DOI] [PubMed] [Google Scholar]
- Pearson C.E., Sinden R.R., Sinden R.R. Trinucleotide repeat DNA structures: Dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol. 1998;8:321–330. doi: 10.1016/s0959-440x(98)80065-1. [DOI] [PubMed] [Google Scholar]
- Petes T.D., Greenwell P.W., Dominska M., Greenwell P.W., Dominska M., Dominska M. Stabilization of microsatellite sequences by variant repeats in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:491–498. doi: 10.1093/genetics/146.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer C.R., Raudsepp T., Chowdhary B., Ellegren H., Raudsepp T., Chowdhary B., Ellegren H., Chowdhary B., Ellegren H., Ellegren H. Low frequency of microsatellites in the avian genome. Genome Res. 1997;7:471–482. doi: 10.1101/gr.7.5.471. [DOI] [PubMed] [Google Scholar]
- Primmer C.R., Saino N., Møller A.P., Ellegren H., Saino N., Møller A.P., Ellegren H., Møller A.P., Ellegren H., Ellegren H. Unravelling the process of microsatellite evolution through analysis of germline mutations in barn swallows. Mol. Biol. Evol. 1998;15:1047–1054. [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2007. [Google Scholar]
- Rolfsmeier M.L., Lahue R.S., Lahue R.S. Stabilizing effects of interruptions on trinucleotide repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:173–180. doi: 10.1128/mcb.20.1.173-180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santibáñez-Koref M.F., Gangeswaran R., Hancock J.M., Gangeswaran R., Hancock J.M., Hancock J.M. A relationship between lengths of microsatellites and nearby substitution rates in mammalian genomes. Mol. Biol. Evol. 2001;18:2119–2123. doi: 10.1093/oxfordjournals.molbev.a003753. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Tautz D., Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992;20:211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe K.A., Toonen R.J., Toonen R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- Shedlock A.M., Botka C.W., Zhao S., Shetty J., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V., Botka C.W., Zhao S., Shetty J., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V., Zhao S., Shetty J., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V., Shetty J., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V., Zhang T., Liu J.S., Deschavanne P.J., Edwards S.V., Liu J.S., Deschavanne P.J., Edwards S.V., Deschavanne P.J., Edwards S.V., Edwards S.V. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc. Natl. Acad. Sci. 2007;104:2767–2772. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibly R.M., Meade A., Boxall N., Wilkinson M.J., Corne D.W., Whittaker J.C., Meade A., Boxall N., Wilkinson M.J., Corne D.W., Whittaker J.C., Boxall N., Wilkinson M.J., Corne D.W., Whittaker J.C., Wilkinson M.J., Corne D.W., Whittaker J.C., Corne D.W., Whittaker J.C., Whittaker J.C. The structure of interrupted human AC microsatellites. Mol. Biol. Evol. 2003;20:453–459. doi: 10.1093/molbev/msg056. [DOI] [PubMed] [Google Scholar]
- Sundström H., Webster M.T., Ellegren H., Webster M.T., Ellegren H., Ellegren H. Reduced variation on the chicken Z chromosome. Genetics. 2004;167:377–385. doi: 10.1534/genetics.167.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth G., Gaspari Z., Jurka J., Gaspari Z., Jurka J., Jurka J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D., Ripley B.D. Modern applied statistics with S. Springer; New York: 2002. [Google Scholar]
- Vowles E.J., Amos W., Amos W. Quantifying ascertainment bias and species-specific length differences in human and chimpanzee microsatellites using genome sequences. Mol. Biol. Evol. 2006;23:598–607. doi: 10.1093/molbev/msj065. [DOI] [PubMed] [Google Scholar]
- Weber J.L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weber J.L., Wong C., Wong C. Mutation of human short tandem repeats. Hum. Mol. Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Webster M.T., Smith N.G.C., Ellegren H., Smith N.G.C., Ellegren H., Ellegren H. Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc. Natl. Acad. Sci. 2002;99:8748–8753. doi: 10.1073/pnas.122067599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.T., Axelson E., Ellegren H., Axelson E., Ellegren H., Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Mol. Biol. Evol. 2006;23:1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Strassmann J.E., Queller D.C., Strassmann J.E., Queller D.C., Queller D.C. Insertions, substitutions, and the origin of microsatellites. Genet. Res. 2000;76:227–236. doi: 10.1017/s001667230000478x. [DOI] [PubMed] [Google Scholar]