Abstract
MicroRNA (miRNA) and other types of small regulatory RNAs play a crucial role in the regulation of gene expression in eukaryotes. Several distinct classes of small regulatory RNAs have been discovered in recent years. To extend the repertoire of small regulatory RNAs characterized in chickens we used a deep sequencing approach developed by Solexa (now Illumina Inc.). We sequenced three small RNA libraries prepared from different developmental stages of the chicken embryo (days five, seven, and nine) to produce over 9.5 million short sequence reads. We developed a bioinformatics pipeline to distinguish authentic mature miRNA sequences from other classes of small RNAs and short RNA fragments represented in the sequencing data. Using this approach we detected almost all of the previously known chicken miRNAs and their respective miRNA* sequences. In addition we discovered 449 new chicken miRNAs including 88 miRNA candidates. Of these, 430 miRNAs appear to be specific to the avian lineage. Another six new miRNAs had evidence of evolutionary conservation in at least one vertebrate species outside of the bird lineage. The remaining 13 putative miRNAs appear to represent chicken orthologs of known vertebrate miRNAs. We discovered 39 additional putative miRNA candidates originating from miRNA generating intronic sequences known as mirtrons.
MicroRNAs (miRNAs) are small 21–23 nucleotide regulatory RNAs that modulate gene expression in animals and plants. In animals, regulation of gene expression by miRNAs is achieved by sequence-specific targeting of the 3′ untranslated regions of messenger RNAs by the RNA induced silencing complex (RISC). This results in translational repression of protein synthesis and, in some cases, destabilization of messenger RNA (He and Hannon 2004). The number of newly discovered miRNAs is growing rapidly (Griffiths-Jones et al. 2006). Moreover, several other classes of small regulatory RNAs, distinguished by their origin and biological function, have been identified in recent years (He and Hannon 2004; Chapman and Carrington 2007). These include small interfering RNAs (siRNAs), encompassing trans-acting siRNAs (tasiRNAs) and natural antisense transcript derived siRNAs (natsiRNAs), repeat-associated siRNAs (rasiRNAs, also referred to as PIWI-interacting RNAs or piRNAs) (for review see Chapman and Carrington 2007); and a recently identified class of small RNAs associated with gene promoters (PASRs) and 3′ termini (TASRs) (Kapranov et al. 2007).
Identification of comprehensive sets of miRNAs and other small regulatory RNAs in different organisms is a critical step to facilitate our understanding of genome organization, genome biology, and evolution. The chicken embryo continues to be a classic model of vertebrate developmental biology that has been used for many decades (Brown et al. 2003). A rapidly developing embryo is an excellent biological system to study the repertoire and expression dynamics of small regulatory RNAs. The developmental processes of the embryo are well defined (Hamburger and Hamilton 1992) and live embryos can be readily manipulated in ovo (Brown et al. 2003). Moreover, the recently sequenced chicken genome (red jungle fowl, Gallus gallus) is the first nonmammalian amniote genome that represents the Diapsida lineage of vertebrate evolution and provides a resource for comparative genomics studies that help our understanding of mammalian evolution (Supplemental Fig. S1) (Hillier et al. 2004). The latest release of the miRNA database (miRBase 10.1) contains 149 chicken miRNA genes that code for 121 distinct mature miRNAs and three sequences originating from the RNA hairpin arm opposite to the annotated mature miRNA containing arm, the so-called miRNA* (Griffiths-Jones et al. 2006). Many of these miRNAs have been identified based only on sequence similarity to known miRNA orthologs and have never been confirmed experimentally (Hillier et al. 2004; Griffiths-Jones et al. 2006). Furthermore, the total number of chicken miRNA genes is currently lower than that identified in mouse (442 miRNA genes), human (533 miRNA genes), or even zebrafish (337 miRNA genes). The latter suggests that there are still many undiscovered miRNAs in birds.
We aimed to extend the known repertoire of small regulatory RNAs expressed in the chicken embryo by using a deep sequencing approach developed by Solexa (now Illumina Inc.). We constructed three small RNA libraries, which were prepared from chicken embryos collected at day five, day seven, and day nine of incubation (hereafter refred to as CE5, CE7, and CE9, respectively). These represent the chicken embryonic developmental stages 25–27, 30–31, and 35, which cover major morphological changes (e.g., limb and craniofacial development) as well as several underlying developmental processes including vasculogenesis, myogenesis, and osteogenesis (Hamburger and Hamilton 1992). Each library was sequenced individually and generated more than three million short sequence reads resulting in a total of over 9.5 million sequence reads. We developed a bioinformatics pipeline to distinguish authentic mature miRNA sequences from other small RNAs and short RNA fragments represented in the sequencing data. The following manuscript presents a detailed analysis of this sequence data and its interpretation.
Results
An overview of the sequencing results
To simplify the sequencing data, all identical sequence reads in each small RNA library were grouped and converted into sequence tags—unique sequences with associated counts of the individual sequence reads. Although the total numbers of sequence reads in all three RNA libraries were approximately the same (∼3.2 million; Supplemental Table S2), the numbers and fractions of the unique sequence tags were substantially different between the libraries (Supplemental Table S3). The CE5 library had the highest number of unique sequence tags; this was followed by the CE7 and then the CE9 libraries. The apparent decrease in the diversity of the small RNA populations from day five to day nine of chick embryo development is also evident in the relative increase in the fraction of sequence tags that were sequenced multiple times in a CE7 and CE9 sample (Supplemental Table S4). These data highlight differences in the overall complexity of the steady-state small RNA pools between the different developmental stages.
To gain insight into possible mechanisms underlying observed changes in the sequence diversity of the small RNA libraries we analyzed sequence tags associated with known RNA classes by mapping sequence tags onto the chicken genome assembly using BLAT (Kent 2002). We found that known chicken miRNAs accounted for ∼60% of all sequence reads in all three embryonic libraries (Supplemental Table S2). This result indicates that our small RNA libraries were highly enriched in mature miRNA sequences. However, when we analyzed numbers of the unique sequence tags, we discovered that these 60% of sequence reads derived from known miRNAs represented only a relatively small fraction (4%–12%) of the total numbers of the unique sequence tags. The highest fraction of the unique sequence tags (52%–73%) was attributed to unclassified small RNAs (Supplemental Table S3). We conclude that while known miRNAs might be the most abundant class of small regulatory RNAs in the chicken embryo, there is a less abundant but much more diverse pool of small RNAs that may represent new miRNAs and other classes of regulatory RNA.
Known miRNAs
To assess the efficiency of the deep sequencing procedure for miRNA detection we analyzed the numbers and distribution of the sequence tags associated with known chicken miRNA genes. We found that out of 121 known chicken miRNAs 118 were expressed in at least two of our three embryonic small RNA libraries, and 117 miRNAs were detected in all three RNA libraries (Supplemental Table S1). These corresponded to 144 and 145 miRNA genes respectively. These results illustrate that a rapidly developing chicken embryo employs almost the entire repertoire of the known miRNAs. The high sensitivity of the deep sequencing approach was clearly evident when we looked at the distributions and numbers of sequence tags originating from different arms of the miRNA precursors (pre-miRNAs). We identified sequence tags representing miRNA* sequences for 136 out of 145 expressed miRNA genes, a large increase over the three reported previously (Supplemental Table S9). As expected, in most of the cases total sequence read counts were heavily skewed toward the RNA hairpin arm containing the annotated miRNA. This observation is consistent with the current knowledge of miRNA biogenesis and strand selection. The dsRNA-specific endonuclease known as DICER excises a 21–23 bp small RNA duplex from a hairpin-like miRNA precursor, producing an equimolar ratio of miRNA/miRNA*. Subsequently, the strand with lower thermodynamic stability in its 5′ end is preferentially incorporated into the RISC complex and, unlike the other strand, is protected from rapid degradation (He and Hannon 2004). In many cases, miRNA* cannot be detected by conventional methods due to their rapid turnover; however, the deep sequencing procedure allows many of them to be identified.
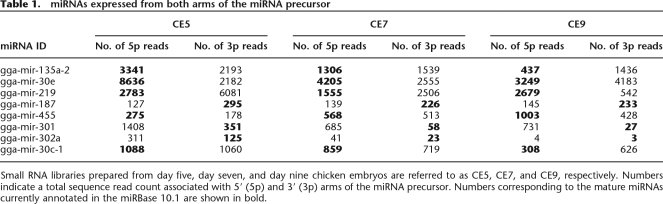
Although the majority of miRNA genes display strand bias, some of them give rise to miRNA and miRNA* with similar 5′ end stability that leads to equal incorporation of either strand into the RISC and their protection from degradation. These miRNA genes are usually characterized by similar expression and/or cloning recovery rates of miRNA and miRNA* (He and Hannon 2004). A few such miRNA genes have been predicted and validated in different species (Aravin et al. 2003; Schwarz et al. 2003). Our analysis of the sequence tags originating from the known miRNA genes identified eight miRNA genes that appeared to encode miRNAs on both arms of the pre-miRNA (Table 1). These genes demonstrated a nearly equal number of sequence reads originating from the 5′ and 3′ arms of the miRNA hairpin precursor (5p and 3p, respectively). Moreover, six of the eight genes had a higher number of sequence reads originating from the arm of the pre-miRNA hairpin opposite to the annotated mature miRNA containing arm in at least one of the RNA libraries (Fig. 1A; Table 1). Furthermore, four of these miRNAs (miR-135a-2, miR-30e, miR-219, and miR-30c) showed a reversal in the ratios of the 5p- and 3p-derived sequence tags across the three RNA libraries (Table 1). The latter cases may indicate developmental switching in preferential use of mature miRNAs originating from different arms of the pre-miRNA precursor and suggest additional levels of complexity in miRNA processing which remain to be uncovered.
Table 1.
miRNAs expressed from both arms of the miRNA precursor
Small RNA libraries prepared from day five, day seven, and day nine chicken embryos are referred to as CE5, CE7, and CE9, respectively. Numbers indicate a total sequence read count associated with 5′ (5p) and 3′ (3p) arms of the miRNA precursor. Numbers corresponding to the mature miRNAs currently annotated in the miRBase 10.1 are shown in bold.
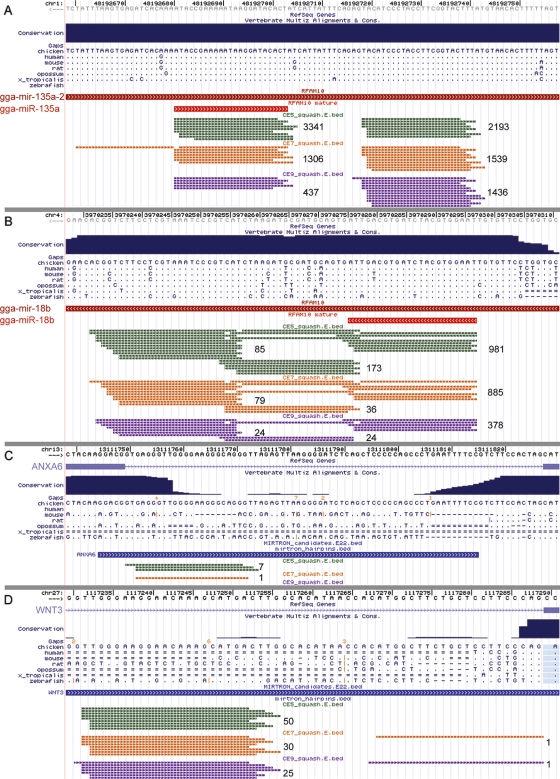
Figure 1.
The figure shows UCSC genome browser screens displaying chicken miRNAs gga-mir-135a-2 (A), gga-mir-18b (B), a mirtron within the ANXA6 gene (C), and an atypical mirtron within the WNT3 gene (D). Sequence tags originating from these loci are shown as thin lines of three different colors representing each of the small RNA libraries: CE5, green; CE7, orange; and CE9, magenta. Numbers to the right of the sequence tag clusters indicate total number of reads originating from this cluster. Arrow directions indicate positive or negative strand of genomic DNA. Identical bases in the multiple sequence alignment are shown as dots.
Similar to other deep sequencing studies, we observed heterogeneity at the 5′ and 3′ ends of the sequenced tags (Fig. 1A) (Ruby et al. 2006, 2007b; Landgraf et al. 2007; Morin et al. 2008). In some cases sequence read counts for these miRNA isoforms were higher than the corresponding sequence read counts for known miRNA sequences reported in the miRBase 10.1. We suggest that these most frequently sequenced miRNA isoforms should be utilized to refine miRBase annotations of chicken miRNAs (Supplemental Table S10).
In 15 cases we observed sequence tags originating from the terminal loop region of the pre-miRNA precursor (Fig. 1B; Supplemental Figs. S3, S4). For 12 of them the total numbers of small RNA sequence reads derived from the terminal hairpin loop were very small as compared to numbers of sequence reads derived from mature miRNAs. Examples like these have been reported before (Ruby et al. 2006), and may be explained by the larger volume of sequence data occasionally detecting pre-miRNA processing intermediates. However, in the three remaining cases (mir-18b, mir-451, and mir-218-1) we found that the numbers of sequence reads derived from the terminal hairpin loop were relatively high (Fig. 1B; Supplemental Figs. S3, S4). The discrete clustering of sequence tags along the miRNA hairpin precursor (Fig. 1B) suggests that the detected sequence tags are genuine products of pre-miRNA processing rather than random degradation products of the unprocessed pre-miRNA. These data indicate a complex post-transcriptional processing in these three miRNA genes.
As reflected by the total counts of miRNA-derived sequence reads, known miRNAs had a very broad range of expression which varied from hundreds of thousands of sequence reads for the most abundant miRNAs to zero for the three previously known chicken miRNAs that have not been detected (Supplemental Table S1). Similarly to the observations made by Takada and colleagues in the mouse embryo (Takada et al. 2006), we found that 10 of the most abundant known miRNAs account for approximately half of all miRNA-derived sequence tags in all three libraries (Supplemental Table S1).
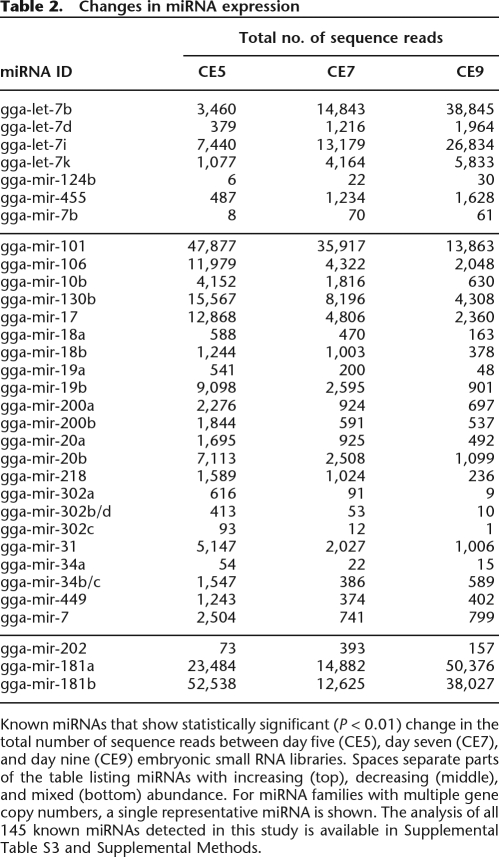
Although our experiment was not designed for direct comparison of miRNA abundance between the three different developmental stages, we applied a statistical evaluation procedure enabling us to identify potentially significant changes in relative miRNA abundance between the three embryonic small RNA libraries (see Supplemental Methods). Table 2 lists the known chicken miRNAs that show statistically significant (P < 0.01) changes in their relative abundance between samples from embryonic stages at days five, seven, and nine. (The full list of the miRNAs and the detailed analysis is available in Supplemental Table S1.) Seven out of 32 miRNAs listed in the Table 2 appeared to be up-regulated in the order of CE5 < CE7 < CE9, while 22 miRNAs seemed to be down-regulated in the same order (CE5 > CE7 > CE9).
Table 2.
Changes in miRNA expression
Known miRNAs that show statistically significant (P < 0.01) change in the total number of sequence reads between day five (CE5), day seven (CE7), and day nine (CE9) embryonic small RNA libraries. Spaces separate parts of the table listing miRNAs with increasing (top), decreasing (middle), and mixed (bottom) abundance. For miRNA families with multiple gene copy numbers, a single representative miRNA is shown. The analysis of all 145 known miRNAs detected in this study is available in Supplemental Table S3 and Supplemental Methods.
Newly identified miRNAs
To identify novel miRNAs in the sequencing data from the three small RNA libraries, we used the following criteria: (1) genomic loci annotated as known chicken miRNAs or as other classes of noncoding RNA were excluded; (2) to be considered for further analysis an individual locus had to be supported by at least two independent sequence reads originating from at least two small RNA libraries; (3) the loci lacking hairpin-like RNA secondary structures including the positions of the small RNA tags were eliminated. The resulting set of sequences and their respective RNA structures were analyzed further to distinguish genuine miRNA precursors from other RNAs that contain similar RNA structures (e.g., tRNA-derived repeat elements; Supplemental Methods).
The resulting data set was composed of 361 unique sequences identified as novel chicken miRNAs (Supplemental Table S7; http://www.livestockgenomics.csiro.au/courses/chicken/). In addition we identified another 88 miRNA candidates that met all of our inclusion criteria except that these were detected in only one miRNA library (Supplemental Table S13). Sequence comparisons between the new chicken miRNA candidates and other vertebrate miRNAs present in miRBase (miRBase 10.1) revealed that 13 of 361 of these new chicken miRNAs were orthologous to miRNAs identified in other vertebrate species (Supplemental File S1; Supplemental Table S5). To investigate evolutionary conservation of the remaining 348 chicken miRNAs, we searched for highly similar DNA sequences in the human, dog, opossum, zebrafish, xenopus, and lizard genome assemblies. Sequences returned by sequence similarity searches were then confirmed as orthologous miRNA candidates by analysis of their predicted RNA structures. We found six new chicken miRNA genes that were conserved in at least one of the analyzed vertebrate genomes (Supplemental File S1; Supplemental Table S5). To identify potential avian-specific miRNAs we used a similar approach to search the NCBI archive for sequence traces of the zebra finch (Taeniopygia guttata). Traces with high sequence identity scores were analyzed for the presence of evolutionarily conserved hairpin-like structures. We identified 22 chicken miRNA sequences that are also present in zebra finch. This number is likely to be an underestimate of the avian-specific miRNAs, as our searches were limited to zebra finch sequences available in the NCBI trace archive at the time of this study. It is not yet possible to give an accurate estimate of what portion of the zebra finch genome has been covered by the NCBI sequence entries.
When we looked at the relative abundance of the new miRNAs (as reflected by the total counts of the most frequently sequenced reads), we found that evolutionarily conserved miRNAs were often among the most abundant ones (Supplemental Tables S5, S6). This result is consistent with many other studies demonstrating some correlation between evolutionary conservation of miRNAs and their expression levels (for the most recent studies see Berezikov et al. 2006a; Landgraf et al. 2007; Ruby et al. 2007b). Interestingly several new miRNAs identified in our study that appeared to be chicken specific had expression levels comparable to some evolutionarily conserved miRNAs (Supplemental Tables S5, S6).
Mirtrons
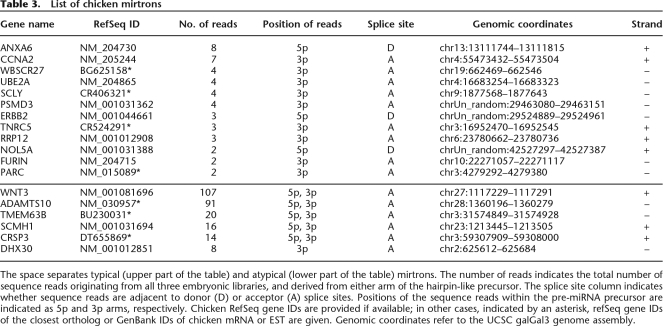
Three recent studies have described an alternative miRNA processing pathway that uses intron splicing machinery instead of the DROSHA (RNASEN) endonuclease to generate miRNA precursors from short intronic sequences (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007a). A distinct feature of such miRNA-generating introns is that the miRNA hairpin-like precursor is directly adjacent to the splice sites such that mature miRNA sequences often start directly at the 5′ terminus of the intron and/or end at its 3′ terminus. The few mirtrons identified to date originate from the diverse evolutionarily lineages of insect (Drosophila sp.), worms (Caenorhabditis elegans), and mammals (mostly represented by primates) (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007a). A bioinformatics approach similar to the one described by Berezikov et al. (2007) was utilized to investigate the presence of mirtrons in the chicken genome. We found 12 mirtrons that were supported by multiple sequence reads (minimum two) in at least two embryonic libraries (Fig. 1C; Table 3; Supplemental Figs. S2a–S2l). None of these mirtrons were identified in previous studies. Having identified 12 chicken mirtrons we extended our search to larger (>200 nt) introns, looking for intronic sequence tags directly adjacent to splice sites. This search yielded another six mirtron candidates (Fig. 1D; Table 3; Supplemental Figs. S2m–S2r). Although we cannot call these candidates “typical” mirtrons, as the predicted pre-miRNA precursor is anchored by only one of the two splice sites, these miRNAs otherwise have typical characteristics of the known mirtrons reported in literature (Berezikov et al. 2007). Sequence analysis of the 3′ and 5′ ends of the representative sequence tags uncovered sequence motifs similar to ones reported by Berezikov and colleagues for mammalian mirtrons (Supplemental Fig. S5). Surprisingly, we found more sequence reads supporting expression of the “atypical” mirtrons originating from the large introns than we did for the “typical” mirtrons derived from small introns (Table 3). Additional analyses of chicken introns identified a further 21 mirtron candidates that had some typical mirtron characteristics but which did not meet our filtering criteria and therefore are considered as mirtron candidates (Supplemental Table S8). Analysis of the evolutionary conservation of the newly identified mirtrons revealed that only one mirtron, located within the neurexin 1 gene, was evolutionarily conserved between mammals and birds. Another mirtron candidate was found within the chicken ortholog of the human DEAH-box RNA helicase gene DHX30, which has already been reported to contain a mirtron (Berezikov et al. 2007). However, examination of the microsynteny between chicken and human orthologs revealed that the chicken mirtron is located in a different DHX30 intron compared to human. We did not find any small sequence reads supporting expression of the human mirtron ortholog in chicken.
Table 3.
List of chicken mirtrons
The space separates typical (upper part of the table) and atypical (lower part of the table) mirtrons. The number of reads indicates the total number of sequence reads originating from all three embryonic libraries, and derived from either arm of the hairpin-like precursor. The splice site column indicates whether sequence reads are adjacent to donor (D) or acceptor (A) splice sites. Positions of the sequence reads within the pre-miRNA precursor are indicated as 5p and 3p arms, respectively. Chicken RefSeq gene IDs are provided if available; in other cases, indicated by an asterisk, refSeq gene IDs of the closest ortholog or GenBank IDs of chicken mRNA or EST are given. Genomic coordinates refer to the UCSC galGal3 genome assembly.
Discussion
The results presented here provide experimental and bioinformatic evidence supporting the discovery of 361 new miRNAs, 88 new miRNA candidates, 18 mirtrons (including six novel atypical mirtron candidates), and 21 mirtron candidates that are expressed in the chicken embryo. Together with the existing set of 121 known chicken miRNAs, this brings the total number of miRNAs in the chicken to 609. Analysis of the evolutionary conservation of the newly identified putative miRNAs revealed that only six of them are conserved in non-avian vertebrates, with the majority of the remaining miRNAs likely to be specific to the bird and/or chicken lineages. The small number of the newly identified miRNAs conserved in vertebrates is not surprising, given the results of other recent studies. Berezikov et al. (2006a) identified 447 new miRNA genes, most of which appeared to be specific to primates; with only 11 of 447 miRNA genes identified in other groups of vertebrates. It appears that while most of the miRNAs discovered in early studies were found to be highly conserved in evolution, more and more of the newly identified miRNAs are present in only a small group of organisms and in some cases in a single species (Bentwich et al. 2005; Berezikov et al. 2006a, b; Ruby et al. 2007b). This phenomenon can be explained in part by the frequently observed correlation between the level of evolutionary conservation of a given miRNA and its expression level, and hence a technical capacity for it to be detected. It is noteworthy that the oldest known miRNA, let-7 (Pasquinelli et al. 2000), and its family members, were sequenced more than one million times in our three embryonic libraries (Supplemental Table S1). Historically, this correlation between evolutionary conservation and expression level led to a widespread notion that all miRNAs are highly conserved. However, this notion was formulated before the abundant lineage-specific miRNAs were identified and hence might be misleading. Although, the functional significance of these evolutionarily divergent miRNAs has not been established experimentally, it has been hypothesized that these miRNAs might play a role in establishing and maintaining phenotypic diversity between different groups of organisms (Plasterk 2006; Sempere et al. 2006). It is plausible that the conserved miRNAs are responsible for control of the basic cellular and developmental pathways common to most eukaryotes (e.g., cell cycle) whereas the nonconserved miRNAs are involved in regulation of the lineage-specific pathways and functions.
As compared to the numbers of primate-specific miRNAs (Berezikov et al. 2006a), the number of potential avian-specific miRNAs identified in our study appears relatively low; only 22 of the newly identified miRNAs were found to have homologs in the available genomic sequence of zebra finch. However, as suggested earlier, this number is likely to be an underestimate, and poorly reflects the true extent of the evolutionary conservation of miRNAs among birds. There are at least two reasons to suggest this. Firstly, the availability of the zebra finch genome sequences is currently limited, and the full analysis of the miRNA conservation can only be performed after the whole-genome sequencing and assembly are complete for the zebra finch. Secondly, the estimated divergence time between the two bird genomes analyzed in this study is much higher than that of human and chimp genomes: ∼105 million years in birds as compared to five million years in primates. In fact the estimated divergence time between the two bird lineages is comparable to the divergence of the human and dog lineages (Supplemental Fig. S1). Therefore, it is conceivable that many new species-specific miRNAs have evolved since the divergence of the two bird lineages.
The large number of sequence tags originating from known chicken miRNAs in all three embryonic RNA libraries enabled us to analyze changes in miRNA expression and observe less common events in the miRNA biogenesis that have not been previously reported. Data on sequence reads originating from the terminal loop of the miRNA precursor, exemplified by chicken mir18b (Fig. 1B), and apparent shifts in strand preference in some miRNA genes (Table 1) highlight the complexity of the post-transcriptional regulation of miRNA processing. Evidence for the regulated and/or alternative processing of pre-miRNAs is just beginning to emerge. The processing of a human miRNA from the mir-18 family (hsa-mir-18a) was recently demonstrated to be dependent on the splicing repressor protein HNRNPA1 (Guil and Caceres 2007). In addition, a new sub-class of intronic miRNAs named mirtrons was reported to bypass the DROSHA-dependent step of miRNA processing by utilizing the splicing machinery to excise RNA hairpin precursors (Okamura et al. 2007; Ruby et al. 2007a). In this study we have reported 39 new chicken mirtrons including 21 mirtron candidates. The identification of six atypical mirtrons provides further evidence for the complex relationship between the splicing machinery and pre-miRNA processing. Analysis of the orthologous sequences for newly identified mirtrons revealed that none of these have been reported in other vertebrates. These results may indicate either rapid divergence and/or independent evolution of mirtrons in different evolutionary lineages.
Given that our small RNA libraries were prepared from whole-embryo extracts at relatively advanced stages of embryonic development, it is difficult to discuss observed changes in abundance of different miRNAs in the context of specific biological processes. However, certain trends appear to be clear. For example, an apparent decrease in the abundance of 22 miRNAs presented in Table 2 and potentially of some others (Supplemental Table S1) is likely to reflect progressive restriction of the expression of these miRNAs from broad embryonic domains such as endoderm, mesoderm, and ectoderm to specific regions, organs, and potentially cell types, as embryo development progresses to maturity. A similar trend was observed in zebrafish and chicken embryonic development using in situ hybridization and miRNA microarray techniques (Wienholds et al. 2005; Darnell et al. 2006).
Although the examples described above by no means represent all of the molecular events in miRNA biogenesis and expression, they illustrate the high value of the deep sequencing data for qualitative and potentially quantitative studies of small regulatory RNAs. The substantial proportion of unclassified small RNAs identified in this study suggests that there may be other classes of small regulatory RNA in chicken embryos that were not covered by our analyses. Some of these may also include rare miRNAs that were represented by single sequence reads and thus did not pass our filtering criteria. These may be confirmed in future studies applying the deep sequencing approach to specialized tissues or cells.
Methods
Sources of sequences and assemblies
Draft genome assembly of the green lizard (Anolis carolinensis) was produced by the Broad Institute at MIT and Harvard (http://www.broad.mit.edu/). Draft genome assembly of the frog (Xenopus tropicalis) was produced by the DOE Joint Genome Institute (JGI) (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html/). Draft genome assembly of the zebrafish (Danio rerio) was produced by The Wellcome Trust Sanger Institute in collaboration with the Max Planck Institute for Developmental Biology in Tuebingen, and the Netherlands Institute for Developmental Biology (http://www.sanger.ac.uk/). Zebra finch (Taeniopygia guttata) sequence data were produced by the Genome Sequencing Center at Washington University School of Medicine in St. Louis (http://genome.wustl.edu/genome_group_index.cgi). The NCBI Trace Archive accession nos. for zebra finch (Taeniopygia guttata) sequences used in this study are provided in supplemental data. The NCBI Taxonomy ID numbers for chicken (Gallus gallus) and zebra finch (Taeniopygia guttata) are 9031 and 59729, respectively. Human, chicken, and opossum genomes were produced by their respective genome sequencing consortiums (Lander et al. 2001; Hillier et al. 2004; Mikkelsen et al. 2007). Unless specified otherwise, sequences of miRNA precursors and mature miRNAs were obtained from the latest release of miRNA database (miRBase 10.1, December 2007, http://microrna.sanger.ac.uk/registry/) (Griffiths-Jones et al. 2006).
Chicken embryo collection and RNA isolation
Fertilized Ross-308 eggs were obtained from Barter Enterprises. The eggs were incubated in a Multiquip incubator (Multiquip) at 37.5°C with rotation every 6 h. Chick embryos were collected at days five, seven, and nine of incubation and selected to represent the embryonic developmental stages 25–27, 30–31, and 35, respectively (Hamburger and Hamilton 1992). After removal of the amnion, embryos were rinsed in 1× PBS, and immediately processed for RNA isolation.
Low molecular weight RNA was extracted using mirVana miRNA Isolation Kit (Ambion) following the manufacturer’s protocol. In brief, embryos were dissected and homogenized in the lysis buffer supplied with the kit. The RNA concentration and purity were determined photometrically by measuring absorbance at 260 nm and A260/A280 ratio using the NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies). RNA samples were stored at –80°C until further use.
Small RNA library construction and sequencing
For small RNA library construction and deep sequencing, RNA samples were prepared as follows: for each developmental stage equal quantities (7 μg) of small RNA isolated from three individual embryos were pooled. Approximately 20 μg of small RNA representing each developmental stage were submitted to Solexa (now Illumina Inc.) for sequencing.
In brief, the sequencing was performed as follows: RNA was purified by polyacrylamide gel electrophoresis (PAGE), to enrich for molecules in the range of 18–30 nt, and ligated with proprietary adapters to the 5′ and 3′ termini of the RNA. The samples were used as templates for cDNA synthesis. The cDNA was amplified with 18 PCR cycles to produce sequencing libraries that were subjected to Solexa’s proprietary sequencing-by-synthesis method.
Analysis of sequencing data
Individual sequence reads with the base quality scores were produced by Illumina/Solexa. All identical sequences were counted and eliminated from the initial data set. The resulting set of the unique sequences with associated read counts is referred to as sequence tags. A mirror of the UCSC genome browser and database was created with the Gallus gallus v.2.1 genome sequence and annotations (galGal3, May 2006) (Kent et al. 2002; Karolchik et al. 2003). After trimming the 3′ adaptor sequence, sequence tags were mapped onto the chicken genome assembly using BLAT software (Kent 2002). To identify sequence tags originating from coding exons, repeats, rRNA, tRNA, snRNA, and snoRNA, we used UCSC RefGene, RepeatMasker, and NCBI RefSeq data (Pruitt and Maglott 2001; Karolchik et al. 2003; Kuhn et al. 2007), as well as our own sets of ncRNA annotations compiled from the NCBI GenBank data (http://www.ncbi.nlm.nih.gov/). To identify novel miRNA genes we identified all hairpin-like RNA structures encompassing small RNA sequence tags using RNAfold (Hofacker 2003); then we analyzed sequence and structural features of the predicted hairpin-like structures to distinguish genuine miRNA precursors from other RNA classes that may contain similar RNA structures (e.g., tRNA-derived repeat elements; see Supplemental Methods for details). All sequences identified as new miRNA precursors and the most frequently sequenced tags for each mature miRNA were submitted to the microRNA database (miRBase at the Sanger center http://microrna.sanger.ac.uk/registry/).
Acknowledgments
This work was supported by CSIRO Emerging Sciences Initiatives in Epigenetics and Cellular Reprogramming. We thank the members of the zebra finch, the lizard, and the zebrafish genome sequencing consortiums for making their data available in advance of formal publications. We also thank Toni Reverter for the assistance with the statistical analysis of the data, and Fai Wong and Volker Haring for critical reading of the manuscript and discussion.
Footnotes
[Supplemental material is available online at www.genome.org. The raw sequence data from this study have been submitted to Gene Expression Omnibus under accession no. GSE10686.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.074740.107.
References
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Snyder B., Gaasterland T., Meyer J., Tuschl T., Gaasterland T., Meyer J., Tuschl T., Meyer J., Tuschl T., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H.A., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H.A., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H.A., Kondova I., Bontrop R., Cuppen E., Plasterk R.H.A., Bontrop R., Cuppen E., Plasterk R.H.A., Cuppen E., Plasterk R.H.A., Plasterk R.H.A. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006a;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Berezikov E., van Tetering G., Verheul M., de van Belt J., van Laake L., Vos J., Verloop R., de van Wetering M., Guryev V., Takada S., van Tetering G., Verheul M., de van Belt J., van Laake L., Vos J., Verloop R., de van Wetering M., Guryev V., Takada S., Verheul M., de van Belt J., van Laake L., Vos J., Verloop R., de van Wetering M., Guryev V., Takada S., de van Belt J., van Laake L., Vos J., Verloop R., de van Wetering M., Guryev V., Takada S., van Laake L., Vos J., Verloop R., de van Wetering M., Guryev V., Takada S., Vos J., Verloop R., de van Wetering M., Guryev V., Takada S., Verloop R., de van Wetering M., Guryev V., Takada S., de van Wetering M., Guryev V., Takada S., Guryev V., Takada S., Takada S., et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006b;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Chung W.J., Willis J., Cuppen E., Lai E.C., Chung W.J., Willis J., Cuppen E., Lai E.C., Willis J., Cuppen E., Lai E.C., Cuppen E., Lai E.C., Lai E.C. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W.R., Hubbard S.J., Tickle C., Wilson S.A., Hubbard S.J., Tickle C., Wilson S.A., Tickle C., Wilson S.A., Wilson S.A. The chicken as a model for large-scale analysis of vertebrate gene function. Nat. Rev. Genet. 2003;4:87–98. doi: 10.1038/nrg998. [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C., Carrington J.C. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Darnell D.K., Kaur S., Stanislaw S., Konieczka J.H., Yatskievych T.A., Antin P.B., Kaur S., Stanislaw S., Konieczka J.H., Yatskievych T.A., Antin P.B., Stanislaw S., Konieczka J.H., Yatskievych T.A., Antin P.B., Konieczka J.H., Yatskievych T.A., Antin P.B., Yatskievych T.A., Antin P.B., Antin P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J., Grocock R.J., van Dongen S., Bateman A., Enright A.J., van Dongen S., Bateman A., Enright A.J., Bateman A., Enright A.J., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Caceres J.F., Caceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H.L., Hamilton H.L. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- He L., Hannon G.J., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Ponting C.P., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Bork P., Burt D.W., Groenen M.A.M., Delany M.E., Burt D.W., Groenen M.A.M., Delany M.E., Groenen M.A.M., Delany M.E., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Hertel J., Hackermuller J., Hofacker I.L., Hackermuller J., Hofacker I.L., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Stanke M., Smith K.E., Siepel A., Smith K.E., Siepel A., Siepel A., et al. The UCSC genome browser database: Update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rice A., Kamphorst A.O., Landthaler M., Kamphorst A.O., Landthaler M., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Garber M., Gentles A.J., Goodstadt L., Heger A., Gentles A.J., Goodstadt L., Heger A., Goodstadt L., Heger A., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Zhao Y., McDonald H., Zeng T., Hirst M., McDonald H., Zeng T., Hirst M., Zeng T., Hirst M., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C., Hagen J.W., Duan H., Tyler D.M., Lai E.C., Duan H., Tyler D.M., Lai E.C., Tyler D.M., Lai E.C., Lai E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., Hayward D.C., Ball E.E., Degnan B., Muller P., Ball E.E., Degnan B., Muller P., Degnan B., Muller P., Muller P., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Plasterk R.H.A. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Maglott D.R., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Lee W., Nusbaum C., Ge H., Bartel D.P., Nusbaum C., Ge H., Bartel D.P., Ge H., Bartel D.P., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Jan C.H., Bartel D.P., Jan C.H., Bartel D.P., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C., Johnston W.K., Kellis M., Bartel D.P., Lai E.C., Kellis M., Bartel D.P., Lai E.C., Bartel D.P., Lai E.C., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Du T., Xu Z., Aronin N., Zamore P.D., Xu Z., Aronin N., Zamore P.D., Aronin N., Zamore P.D., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Cole C.N., McPeek M.A., Peterson K.J., Cole C.N., McPeek M.A., Peterson K.J., McPeek M.A., Peterson K.J., Peterson K.J. The phylogenetic distribution of metazoan microRNAs: Insights into evolutionary complexity and constraint. J. Exp. Zoolog. Part B. Mol. Dev. Evol. 2006;306B:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- Takada S., Berezikov E., Yamashita Y., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Berezikov E., Yamashita Y., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Yamashita Y., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Fujiwara S., Watanabe H., Soda M., Watanabe H., Soda M., Soda M., et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Kauppinen S., Plasterk R.H.A., Plasterk R.H.A. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]