In mammals, with a few notable exceptions, sex is determined by an XX female:XY male sex chromosome system in which the Y chromosome bears the male-dominant testis-determining gene SRY. The X chromosome of most placental mammals has a virtually identical gene content, whereas the degenerate Y chromosome contains overlapping subsets of only a few active genes (for review, see Graves 2006). Marsupial mammals have a smaller X and Y that represent an ancestral therian mammal sex pair and define the ancient region of the human sex chromosomes. The mammal X and Y evolved from a pair of autosomes after the proto-Y acquired a male-determining gene, X–Y recombination between male-advantage genes was suppressed, and the Y degenerated (Charlesworth et al. 2005; Fraser and Heitman 2005; Graves 2006). Mapping orthologs of mammal X-borne genes in other vertebrates will help our understanding of how the mammal XY and the sex-determining (SRY) gene evolved, and also how they function.
In other amniotes (reptiles and birds), sex is determined by a variety of different mechanisms that belong in two broad classes: genetic or environmental, although recent data show that some species combine both (Quinn et al. 2007). Genetic sex determination is the most widespread within lineages and commonly involves a pair of differentiated sex chromosomes (for review, see Ezaz et al. 2006). Sex chromosomes are named XY when the heterogametic sex is the male (female XX and male XY as in mammals) and ZW when heterogametic sex is the female (female ZW and male ZZ as in birds or snakes).
Amniote XY and ZW sex chromosome pairs are superficially similar. The paired chromosome (X or Z) in the homogametic sex is generally well conserved, large, and gene-rich, whereas the sex-specific Y or W is usually small, heterochromatic, and almost devoid of active genes. However, comparative gene mapping shows that the sex chromosomes of mammals (XY), birds (ZW), and snakes (ZW) are entirely nonhomologous, implying that they evolved from different pairs of autosomes (Fridolfsson et al. 1998; Nanda et al. 2000; Graves and Shetty 2001; Kohn et al. 2004; Matsubara et al. 2006). The sex-determining gene that initiates testis development is also different; for instance, there is no SRY in non-mammal vertebrates, and sex determination in birds seems to be largely controlled by dosage of a Z-borne gene (possibly DMRT1) (Raymond et al. 1999) rather than a male-dominant gene. The similarities shared by sex chromosomes are thus the results of convergent evolutionary forces in different vertebrate lineages. The acquisition of a sex-determining gene on one member of an autosome pair and the accumulation of sex-specific alleles nearby were followed by suppression of recombination between the new proto-sex chromosomes, which resulted in vulnerability to mutation and deletions of the sex-specific chromosome (Y or W) and, therefore, its degradation (Charlesworth and Charlesworth 2000; Charlesworth et al. 2005; Fraser and Heitman 2005; Steinemann and Steinemann 2005; Graves 2006).
The original autosome from which the mammal X and Y diverged can be deduced from the relationship of the X pair to other vertebrate sex chromosome systems. The short arm of chromosome 4 in the chicken (Gallus gallus [GGA]) and several autosomal regions in fish share genes with the mammal X, and thus represent this ancestral autosome. How this genomic region took over a sex-determining function from an ancestral temperature-determining system, or from nonhomologous ancestral sex chromosomes, demands exploration of the sex-determining system of the most basal mammal group, the monotremes. Monotremes diverged ∼165 million years ago (Mya) from therian mammals (placentals and marsupials) and therefore fall in the phylogenetic tree between birds/reptiles and therians (Van Rheede et al. 2006; Bininda-Emonds et al. 2007). Monotremes, represented only by the duck-billed platypus Ornithorhynchus anatinus and the echidnas (four species), display a fascinating mixture of mammalian and avian/reptilian morphological, physiological, and karyological features and have a unique reproductive system that combines egg-laying with lactation (for review, see Grützner and Graves 2004).
Monotremes have long been known to possess a complex male heterogametic system in which multiple X and Y chromosomes form a chain at male meiosis (Bick and Sharman 1975; Murtagh 1977; Wrigley and Graves 1988). Recently, individual X and Y chromosomes were identified by chromosome painting (Rens et al. 2004). The male was discovered to have 10 unpaired chromosomes that included five male-specific Y chromosomes and five X chromosomes; the female possesses two copies of the five Xs. In male meiosis, the 10 sex chromosomes form an alternating XY chain, X1Y1X2Y2X3Y3X4Y4X5Y5 (Grützner et al. 2004), unique in vertebrates (for review, see Grützner et al. 2006). These 10 chromosomes pair and recombine in pseudoautosomal regions at the termini of adjacent X and Y chromosomes.
Little is known about the gene content of the 10 platypus sex chromosomes, but the few available data are extremely striking. Early comparative mapping using radioactive in situ hybridization with heterologous probes suggested that X1, located at one end of the chain, shared homology with the ancient part of the mammalian X (Watson et al. 1990; Wilcox et al. 1996; Mitchell et al. 1998; but see also Waters et al. 2005). At the other end of the chain, fluorescent in situ hybridization (FISH) of a large insert BAC-clone showed that X5 contained the Z-borne putative bird sex-determination gene DMRT1 (Grützner et al. 2004; El-Mogharbel et al. 2007). This suggested that the monotreme meiotic chain has homology with the therian XY system at one end and to the bird ZW system at the other, and represents an evolutionary link between two systems that were previously thought to have evolved independently (Grützner et al. 2004; Ezaz et al. 2006). The identity of the platypus sex-determining gene remains mysterious. Many efforts to identify a platypus homolog of the therian testis-determining gene SRY proved fruitless (Grützner et al. 2004).
Progress in platypus genome sequencing and the availability of BAC clones whose gene content can be identified from the database now allow us to partially characterize the gene content of X specific regions and identify genes on the pairing regions between adjacent X and Y chromosomes. We have also tested the hypotheses that platypus sex chromosomes share homology with both the mammal XY and the bird ZW systems. In complete contradiction to early data, we find that the 10 sex chromosomes of platypus share no homology with the ancestral therian X chromosome, which is homologous to platypus chromosome 6. Instead, we find that regions homologous to the chicken Z are scattered throughout the chain, principally on X5 and X3. These results have major implications for our understanding of mammalian sex chromosome evolution.
Results
Identification and localization of BACs on platypus sex chromosomes
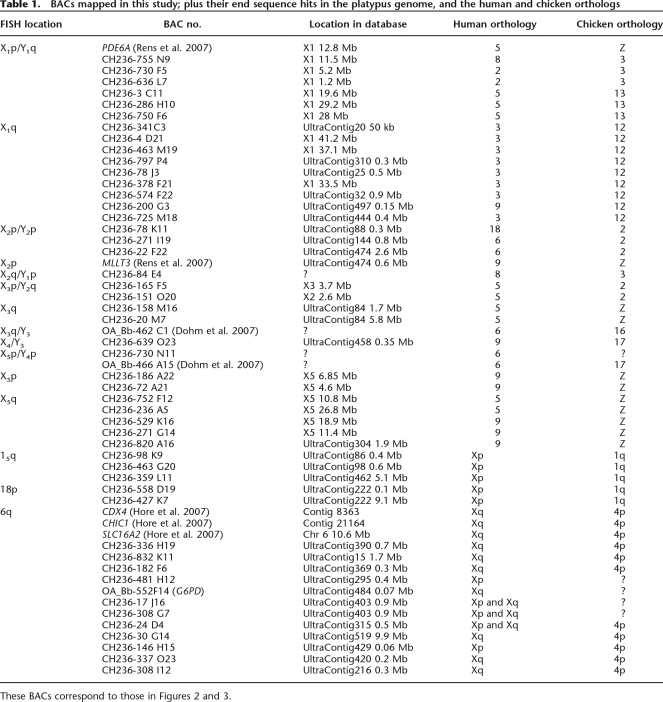
To aid assembly of the platypus genome sequence (Warren et al. 2008), the Graves laboratory contributed to the physical map by FISH mapping 150 randomly selected BACs (from the female platypus library CH-236; inserts ranging from 150 to 200 kb) to female metaphase spreads. Thirty-two of these BACs that apparently mapped to platypus X chromosomes were then examined further in the present study.
These 32 BAC clones were hybridized to male platypus mitotic metaphase chromosomes (Fig. 1). Eighteen mapped to a single X chromosome (nine on X1q, two on X3q, two on X5p, and five on X5q), whereas 14 gave signals, not only on an X chromosome, but also on a Y adjacent to it in the chain (six on X1p and Y1q, one on Y1p and X2q, three on X2p and Y2p, two on Y2q and X3p, one on X4q and Y3q, and one on Y4p and X5p). Locations of all clones are shown on the sex chromosome map (Fig. 2). In addition, four genes/BACs from recent mapping studies were included in our analysis (Dohm et al. 2007; Rens et al. 2007; see Table 1). This confirms the order of the chromosomes in the chain and provides new information on the XY pairing (pseudoautosomal) regions.
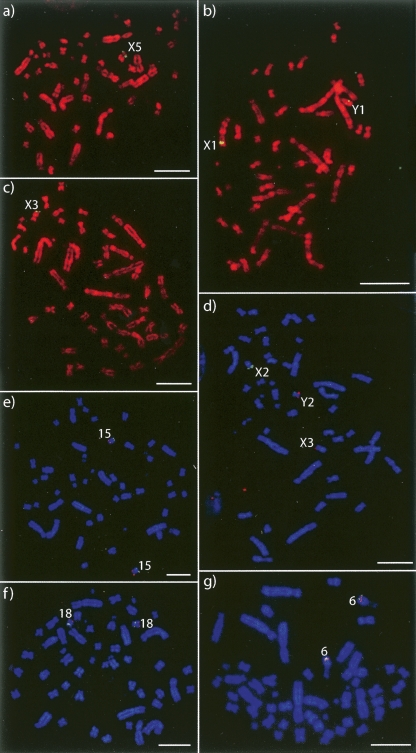
Figure 1.
BACs mapped by FISH to (a–d) the sex chromosomes and (e–g) autosomes orthologous to the human X on male platypus metaphase spreads: (a) CH236-72 A21; (b) CH236-3 C11; (c) CH236-20 M7; (d) (green) CH236-271 I19 and (red) CH236-165 F5; (e) (green) CH236-359 L11 (representing UltraContig462) and (red) CH236-97 I3 (chromosome 15 anchor BAC); (f) (green) CH236-427 K7 (representing UltraContig222) and (red) Ch236-330 L7 (chromosome 18 anchor BAC): (g) (green) CH236-337 O23 (representing UltraContig420) and (red) CH236-30 G14 (representing UltraContig519). Scale bars, 10 μm.
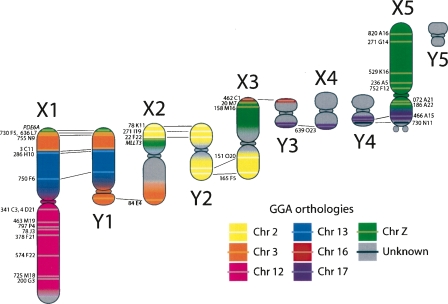
Figure 2.
The 10 sex chromosomes from male platypus with the location of the 32 BACs mapped in this study and four BACs/genes mapped in recent studies (colored bands). BAC numbers are indicated, and homology with chicken is represented by different colors. BACs that hybridized to both an X and a Y revealed pseudoautosomal regions and are connected by lines.
Table 1.
BACs mapped in this study; plus their end sequence hits in the platypus genome, and the human and chicken orthologs
Gene content of BACs on platypus sex chromosomes
To compare the position of orthologous genes across species, BAC-end sequences for 27 of the clones were obtained through the platypus NCBI BAC-end trace archive. The remaining five clones were BAC-end-sequenced using vector primers. BLAST searches of the sequences obtained for the 32 clones against the most recent platypus assembly (Ensembl Genome Browser; ornAna1 release March 2007, http://www.ensembl.org/Ornithorhynchus_anatinus/) established to which scaffolds and chromosomes the BACs were homologous (Table 1). This permitted homology between the meiotic chain and the human and chicken genome to be determined.
The 32 BACs represented scaffolds that contained genes with orthologs on human (HSA) chromosomes 2, 3, 4, 5, 6, 9, and 18; and that represent chicken (GGA) chromosomes 2, 3, 12, 13,16, 17, and Z (Fig. 2; Table 1). The majority of orthology with the chicken genome was to the Z (represented by regions of HSA 5 and 9), which was represented by 11 BACs distributed along X5 and the long arm of X3. Recent studies (Dohm et al. 2007; Rens et al. 2007) have also identified small regions of homology with the chicken Z on X2p and distal X1p.
Surprisingly, not a single BAC identified on the platypus sex chromosomes represented scaffolds that shared homology with the human X chromosome. The contradiction with early gene mapping of several human X orthologs to platypus X1 led us to further investigate the location of human X orthologs in the platypus genome.
Localization of platypus UltraContigs containing orthologs of human X genes
To discover which platypus chromosomes were orthologous to the human X, 128 markers distributed at ∼1–2 Mb along the human X chromosome were BLASTed to the most recent assembly of the platypus genome (Warren et al. 2008). Results of the BLAST searches were manually inspected to exclude false hits. Regions of the platypus genome that were orthologous to the human X chromosome were noted. Of the 155 Mb of the human X, only 14–15 Mb had been anchored to platypus chromosomes, all to chromosome 6.
The bulk of the human X was orthologous to unanchored scaffolds. BACs to anchor these scaffolds were obtained by BLASTing part of each scaffold to the platypus BAC end trace archive. Best hits to the trace archive were examined and clone numbers recorded. These clones were subsequently obtained from BACPAC resources CHORI (BAC library CH-263) for FISH mapping. In total, 16 BACs were identified and obtained to anchor 14 unanchored scaffolds orthologous to the human X (Table 1). Another BAC was obtained from CUGI (BAC library OA_Bb, clone 522F14) that represented a scaffold containing G6PD.
Five BACs represented four scaffolds that contained genes orthologous to the short arm of the human X distal from Xp11.23, a region (XAR) previously shown to have been added relatively recently (100–150 Mya) to the eutherian X (Graves 1995). These BACs were mapped by FISH to chromosomes 15q and 18p (Fig. 1e,f) in contradiction to previous assignments to chromosomes 1 and 2 by radioactive in situ hybridization (RISH) (Watson et al. 1991).
Twelve BACs represented 10 UltraContigs that contained genes orthologous to genes on the conserved region (XCR) of the human X (Xq and proximal Xp). All these BACs were mapped to platypus chromosome 6 (Figs. 1g, 3), in contradiction to early radioactive in situ localizations to platypus X1 (Watson et al. 1990). The anchored scaffolds contained six genes (ALAS2, F8, F9, PLP1, GLA, and G6PD) that were previously mapped to X1 by radioactive in situ hybridization. The present results indicate that no region of the human X chromosome shares homology with any of the monotreme X chromosomes (Fig. 3).
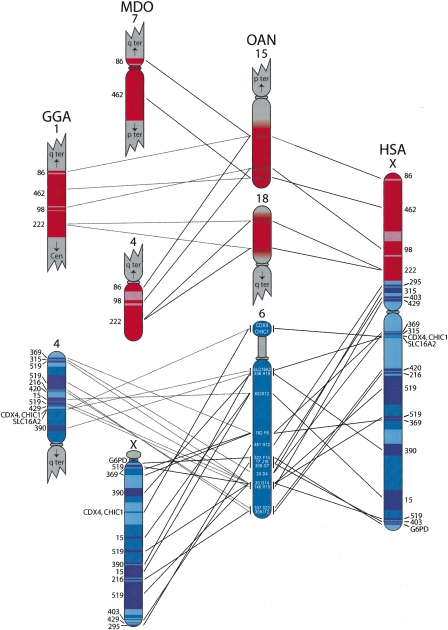
Figure 3.
The human (HSA) X chromosome compared to orthologous platypus (OAN), opossum (MDO), and chicken (GGA) chromosomes. All shades of blue are orthologous to the X conserved region (XCR) of the placental mammal X chromosome. Royal blue on the human X, opossum X, and chicken 4p were anchored to platypus chromosome 6 prior to this study; navy blue regions on these chromosomes are orthologous to unanchored UltraContigs in the platypus assembly; whereas pale blue is predicted orthology with platypus chromosome 6. Red is orthologous to X added region (XAR), and pink is the predicted orthology to platypus chromosomes 15 and 18. None of the X added orthologous regions were anchored in platypus before this study. The numbers of all unanchored UltraContigs (see Table 1 for list of anchoring BACs) are listed left of the chicken and opossum chromosomes, and right of the human X. FISH locations of BACs on platypus chromosomes 6, 15, and 18 are shown, and lines indicate orthology with the human, opossum, and chicken genomes. Some of the platypus UltraContigs have been broken up in human, opossum, and/or chicken. Vertical lines next to platypus chromosome 6 indicate that FISH could not achieve resolution between BACs.
These results confirm and extend the homology between platypus chromosome 6 and the human X that was previously established for seven XCR genes (Waters et al. 2005; Hore et al. 2007). However, we show here that the conserved region of the therian X is represented entirely, not partially, by platypus chromosome 6, which is rearranged with respect to the human X, opossum X, and chicken chromosome 4p (Fig. 3).
Discussion
The basal position of monotremes in the mammalian tree places them in a unique phylogenetic position for comparison of their genomes with those of therian mammals, birds, and fish. These comparisons help to unravel the functional and evolutionary histories of mammalian genomes (e.g., Grützner and Graves 2004; Margulies et al. 2005; Wakefield and Graves 2005). Our comparison of monotreme sex chromosomes to therian and bird sex chromosomes provides surprising new insights into the evolution of the monotreme multiple sex chromosome system and, more importantly, the evolution of therian sex chromosomes from those of an amniote ancestor.
Pseudoautosomal regions and the evolution of the meiotic chain in monotremes
The bizarre sex chromosome system of the platypus raises many questions of function and evolution. Of great significance for our understanding of how the multiple sex chromosome system functions is our definition of sequences shared between X and Y chromosomes that are adjacent in the meiotic chain (Fig. 2), and that are held together at meiotic metaphase by recombination within X–Y-shared pseudoautosomal regions (PARs). PARs were previously inferred from the positions of the chromosomes in the chain at meiosis, and most (particularly the whole arm homology between X1–Y1) could be detected using chromosome paints derived from sorted platypus chromosomes (Rens et al. 2004).
Of the 32 BACs mapped on the X chromosomes, 14 give a signal on one of the adjacent Y chromosomes: seven on Y1, five on Y2, one on Y3, and one on Y4 (Fig. 2). Only four BACs had been mapped to platypus sex chromosome in previous studies (one to X1/Y1, one to X2p, one to X3q/Y3, and one to X5p/Y4) (see Table 1; Fig. 2). We here confirmed homology between the whole arms of X1p–Y1q, and between the terminal regions of Y1p–X2q, X2p–Y2p, and Y2q–X3p, which is in agreement with homology identified previously by chromosome painting, and we also report homology between Y4p–X5p (Fig. 2). The identification of genes in these regions shows that the PARs of these multiple X and Y chromosomes contain active genes, as do the human and mouse PARs. RNA-FISH has shown that both X- and Y-borne alleles of four PAR genes are expressed from both X and Y alleles (J.E. Deakin, T.A. Hore, E. Koina, and J.A.M. Graves, in prep.).
The sizes and pairing relationships of the sex chromosomes provide clues to the genesis of the bizarre multiple sex chromosome system of monotremes. It has been proposed that a translocation chain was formed by recurrent translocations between sex chromosomes and autosomes, as in spiders and the evening primrose (for review, see Grützner et al. 2006). Once autosomes were captured in the sex chromosome chain, degradation of the neo-Y chromosomes occurred, reducing the homology between chromosomes, as has been proposed for the Y and W chromosomes of standard XY and ZW systems. Platypus Y chromosomes are smaller than the X chromosomes with which they pair, as might be expected from a progressive degeneration of male-specific sequence. Hence, the continued presence of Y-borne copies is likely to reflect the timing of when chromosomes were incorporated into the meiotic chain (i.e., the oldest Y sharing less homology with its paired Xs than the youngest). This supports the scenario that chain-building began with a sex chromosome–autosome fusion at the X5 end (only two BACs hybridized on Y4) and progressed with serial translocations toward the X1 end (12 of our BACs hybridized on Y1 or Y2).
Platypus Y chromosomes and platypus sex determination
Paradoxically, the first data on the gene content of the platypus Y chromosomes came from the sequencing of the female platypus genome. The genes that we have identified on the Y chromosomes, using X-specific probes, may offer us clues to the identity of the monotreme sex-determining gene.
There is no platypus ortholog of the therian testis-determining gene SRY, which is not surprising because SOX3 (from which SRY evolved) maps with other human X-borne genes on platypus chromosome 6. The autosomal location of SOX3 is consistent with the hypothesis that SRY did not acquire its sex-determining function until the part of the genome corresponding to platypus pair 6 became a proto-XY in therian mammals 165–150 Mya (Wallis et al. 2007).
The platypus sex-determining gene must lie on one of the platypus sex chromosomes; either a dominant male-determining gene on a Y or a dosage-dependent gene on an X. The assignment of genes to four Y chromosomes at least identifies the genomic regions that evolved into Ys, although none of the characterized Y regions are likely to bear the unknown sex-determining gene because they represent PARs and thus are identical between male and female. However, genes in regions that are adjacent on the human and chicken chromosomes may prove to contain interesting candidate genes. For instance, the unpaired part of X5 contains the DMRT1 gene that is a candidate for sex determination in birds and for sex reversal in humans; however, it is present in two doses in females and one in males (which is the opposite situation from birds), and therefore is unlikely to play the role of sex-determining switch in platypus (El-Mogharbel et al. 2007; Wallis et al. 2007). Grafodatskaya et al. (2007) demonstrated that GATA4, a gene that interacts with human and mouse male gonad development, lies on the platypus sex chromosomes, but on the Y1/X2 pairing region, and is therefore a poor sex-determining candidate. Other putative sex-determining genes (genes know to play a role in vertebrate sex determination) lie on autosomes (Grafodatskaya et al. 2007); thus the hunt for a putative platypus sex-determining switch continues.
Gene content and evolution of the platypus sex chromosomes
The physical mapping of 32 BACs on all but the smallest platypus sex chromosome (Y5) along with consideration of genes close to these BACs in the assembly give us partial information about the gene contents of all five Xs and the corresponding Ys (excluding Y5).
Our striking finding is that orthology of the 10 sex chromosomes with the chicken Z is now extended from the DMRT gene cluster on X5 (El-Mogharbel et al. 2007), to most of the X5 chromosome and the long arm of X3. In addition, Rens et al. (2007) recently found more chicken Z homologs on X1/Y1, X2, and also on autosome pairs, but some of those genes were PCR-mapped, and the position on these chromosomes was not determined. The chicken Z is homologous to regions of human chromosomes 9 and 5. The mixture of HSA 9 and HSA 5 genes on platypus X5 and X3 implies that a region corresponding to the chicken Z was intact in an ancestor of monotremes and birds 310 Mya.
In addition, on the short arm of X3 lie orthologs of GGA 2 genes, some of which lie adjacent to the chicken Z genes on human chromosome 5. The large X1 also contains some genes whose human orthologs (on GGA 13) map near the GGA Z genes on human chromosome 5 (Fig. 2). This implies that fusion of GGA Z, GGA 2, and GGA 13 regions occurred in the mammal lineage before monotremes and therians diverged, or alternatively that an ancestral superchromosome gave rise to the Z by fission in the chicken lineage (see also Rens et al. 2007).
Platypus sex chromosomes are more bird-like than mammal-like
None of the BACs that we mapped to platypus X1 contained orthologs of human X-borne genes. This is such an important but indicative finding that we decided to localize homologs of human X genes in the platypus genome. We assigned several large UltraContigs containing orthologs of human X genes to platypus chromosomes 6, 15q, and 18p (Fig. 3). These localizations are inconsistent with early mapping using radioactive in situ hybridization with human cDNA probes (Watson et al. 1990; Wilcox et al. 1996; Mitchell et al. 1998) but consistent with later studies using FISH mapping of homologous lambda phage clones (Waters et al. 2005). These contradictions can be attributed to previous misidentification of Giemsa-stained pair 6 and X1, and to the unreliability of radioactive in situ hybridization of small heterologous human clones onto divergent platypus chromosomes.
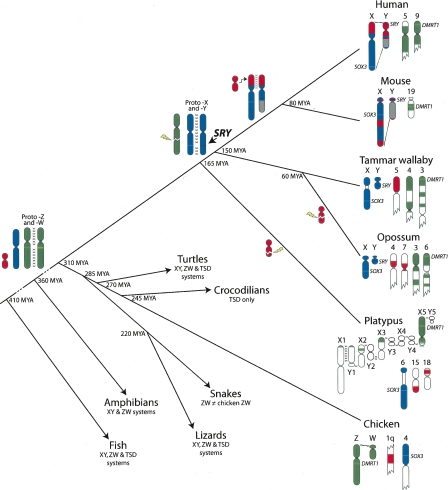
Thus, the platypus meiotic chain shares no gene homology with the therian X chromosome, whereas bird Z orthologs are scattered throughout it. The platypus sex chromosome complex is, therefore, not a link between the bird and mammal systems as previously proposed (Grützner et al. 2004) but is, in fact, derived from a chromosome with homology with a region on the bird Z and human associated regions (Fig. 4).
Figure 4.
Phylogeny displaying the different sex chromosome systems of major vertebrate groups. Homology between genomes, and with ancestral autosomes, is indicated by the same color. Key events are marked on the tree. (Green) A chicken-like ZW system appears to be ancestral to all amniotes, which was retained in birds and caught up in the meiotic chain of monotremes. There was a switch from ZW to XY systems independently in both therian and monotreme mammals. (Red) The region added to the sex chromosomes of placental mammals was broken independently in opossum and monotremes. (Green) The autosome orthologous to the bird Z chromosome was broken in the therian ancestor and subsequently scrambled onto several autosomes in mouse; therefore only the chromosome carrying Dmrt1 is displayed in that species.
Our findings mean that the egg-laying monotreme mammals, representing the basal mammalian group, have an XY sex determination system quite unlike that of the therian mammal XY, but one derived from an ancestral bird-like ZW system.
Evolution of amniote sex chromosomes
These results demand reconsideration of the theories of sex chromosome evolution in vertebrates. Previous demonstrations that the mammal XY and bird ZW chromosomes share no homology were interpreted as independent evolution from different autosomes in a common ancestor that probably had a temperature-dependent sex determination (Nanda et al. 2000; Graves and Shetty 2001; Matsubara et al. 2006). However, our mapping results suggest that the amniote ancestor may have had a bird-like ZZ male: ZW female sex chromosome system, from which all other unrelated ZW and XY systems evolved (Fig. 4). Such transitions from one genetic sex determination system to another involving different sex chromosomes have already been recorded in many taxa, in particular fish (for review, see Ezaz et al. 2006).
The homology of bird Z and monotreme Xs implies a switch of sex heterogamety involving the same sex chromosomes, which was previously thought to be impossible, or at the very least unlikely. However, just such a system has been described in the frog Rana rugosa, where an XY-to-ZW transition has occurred via a hybridization event that created a male sex bias (Ogata et al. 2007). A dominant female-determining gene was subsequently selected for, creating a ZW sex chromosome system. The new Z is homologous to the original Y in these frog populations. If an analogous event occurred in the mammalian ancestor, the monotreme X chromosomes represent an undifferentiated bird W chromosome. Hence, the switch of heterogamety occurred before the Z and W were significantly diverged from each other; thus, a switch of complicated dosage compensation systems was not required. Therefore, the considerably leaky and region-specific bird dosage compensation (Itoh et al. 2007), the partial monotreme dosage compensation (Deakin et al. submitted), and therian X inactivation systems probably all evolved independently.
The transition from a female heterogametic ZW to a male heterogametic XY-based system could have been independent in the therian and monotreme lineages, since monotreme and therian sex chromosomes are nonhomologous. Under this scenario, in monotremes there must have been transition from a bird-like ZW to an XY system that was subsequently modified by recurrent translocations with autosomes, one of which may have donated, or acquired a dominant male-determining gene on a Y, or a dosage-sensitive gene on the X (Wallis et al. 2007). In the therian lineage, one member of an autosome pair (represented by platypus chromosome 6q and chicken 4p) acquired the dominant testis-determining gene SRY, perhaps by modification of the X-borne SOX3 gene (Graves 1998) to assume a controlling function and supplant the ancestral chicken-like ZW system. Alternatively, if the ZW-to-XY transition occurred before mammalian radiation, in therians a dominant SRY gene must have supplanted an ancestral mammalian (monotreme-like) XY system.
The differentiation of the therian X and Y therefore must have occurred after the divergence of the monotremes (∼166 Mya), but before the divergence of marsupial and placental mammals (∼148 Mya). This estimate is inconsistent with the division of the human X chromosome into four evolutionary strata by comparing synonymous nucleotide changes between X and Y shared protein-coding genes (Lahn and Page 1999). The oldest of these strata was thought to have arisen by an inversion on the Y that isolated it from the X soon after the divergence of birds from the mammalian lineage 310 Mya (Lahn and Page 1999). However, our results demonstrate that the upper age limit of this oldest stratum must be less than 166 million years (Myr). The mammal X and Y are, therefore, more than 145 Myr younger than previously deduced. Hence, the degradation of the human Y has been much more rapid than previously proposed, and brings forward the date of its proposed extinction (Graves 2006) to 7 Myr.
Conclusions
Our findings have major implications for our understanding of the evolution of the great variety of vertebrate sex determination systems. The proposal that the ancestral amniote had a genetic system mediated by a bird-like ZZ male: ZW female system has the corollary that other sex chromosome systems such as those of therian mammals and snakes (the latter having an unrelated ZZ:ZW system) (Matsubara et al. 2006), and even temperature-sensitive systems such as that in alligators and marine turtles, all derived from this chromosomal system. This makes it very important to understand the interaction of temperature and genetic systems in species that exhibit both (Quinn et al. 2007), and the evolutionary forces that affect the frequency of W and Z chromosomes in systems that have XX males or ZZ females. Finally, our results demonstrate that the therian sex chromosomes evolved <165 Mya (and not 310 Mya as previously thought), which increases dramatically the rates of sex-linked gene evolution and Y degeneration.
The imminent accessibility of the complete platypus genome, the ever-growing genome data in a wide range of taxa, and future comparative gene mapping, notably involving reptiles, will test the significance of the monotreme sex determination system and will provide further insights into sex chromosome evolution in vertebrates.
Methods
A male platypus fibroblast cell line was established from skin biopsy following standard procedures and in accordance with the Australian National University Animal Experimentation Ethics Committee permit no. R.CG.07.03. and Environment ACT permit no. LI 2002 270. Chromosomes were prepared as described previously (Alsop et al. 2005).
Fluorescence in situ hybridization (FISH)
BAC DNA was labeled by nick-translation with Biotin-16-dUTP, Digoxygenin-11-dUTP (Roche Diagnostics). Hybridization and detection were carried out following the procedure described by Alsop et al. (2005). Images were captured and enhanced using IPLab software (Scanalytics Inc.). Hybridization signals were assigned to specific chromosomal regions identified by DAPI staining.
Genomic sequence analyses
For all BACs that were mapped to the platypus sex chromosomes, BAC-end sequences were retrieved from the platypus NCBI trace archive and BLASTed against the platypus assembly (Ensembl Genome Browser; ornAna1 release March 2007; http://www.ensembl.org/Ornithorhynchus_anatinus/) to determine which contig they were assigned to. Orthology with the human and chicken genomes was established by locating the genes contained in these contigs in human and chicken.
To identify platypus BACs to anchor large unanchored scaffolds orthologous to the human X, 128 cDNAs at 1–2-Mb intervals on the human X chromosome were BLASTed against the platypus assembly. Results were sorted through manually, and hits to UltraContigs were noted. These UltraContigs were BLASTed back to the platypus BAC-end trace archive, and appropriate BACs were chosen to map by FISH, thus anchoring the contigs of interest.
BAC library and end sequencing
BAC clones from a female platypus library (CH236) were obtained through BACPAC resources CHORI (http://bacpac.chori.org/).
BAC-end sequences were not available in the database for all BACs, so several were obtained using the vector primers. BAC DNA was extracted from bacterial cultures with the Promega Wizard Plus SV Minipreps DNA Purification System following the manufacturer’s instructions. PCR amplifications were carried out in 10-μL reaction volumes and included 400 ng of template DNA, 4 μL of BigDye (BDTv3.1), 2 μL of 5× buffer, and 15 pmol of primer (SP6 and T7). The cycling conditions employed were an initial denaturation of 5 min at 95°C, followed by 99 cycles of 10 sec at 96°C/10 sec at 50°C/4 min at 60°C; reactions were then held at 4°C.
Acknowledgments
We thank Rosalind Attenborough for her expert technical help. F.V. was supported by a Lavoisier fellowship from the Ministère Français des Affaires Etrangères. The Cambridge Resource Centre is supported by a grant from the Wellcome Trust. Most support was provided by the Research School of Biological Sciences, ANU.
Footnotes
In therian mammals (placentals and marsupials), sex is determined by an XX female: XY male system, in which a gene (SRY) on the Y affects male determination. There is no equivalent in other amniotes, although some taxa (notably birds and snakes) have differentiated sex chromosomes. Birds have a ZW female: ZZ male system with no homology with mammal sex chromosomes, in which dosage of a Z-borne gene (possibly DMRT1) affects male determination. As the most basal mammal group, the egg-laying monotremes are ideal for determining how the therian XY system evolved. The platypus has an extraordinary sex chromosome complex, in which five X and five Y chromosomes pair in a translocation chain of alternating X and Y chromosomes. We used physical mapping to identify genes on the pairing regions between adjacent X and Y chromosomes. Most significantly, comparative mapping shows that, contrary to earlier reports, there is no homology between the platypus and therian X chromosomes. Orthologs of genes in the conserved region of the human X (including SOX3, the gene from which SRY evolved) all map to platypus chromosome 6, which therefore represents the ancestral autosome from which the therian X and Y pair derived. Rather, the platypus X chromosomes have substantial homology with the bird Z chromosome (including DMRT1) and to segments syntenic with this region in the human genome. Thus, platypus sex chromosomes have strong homology with bird, but not to therian sex chromosomes, implying that the therian X and Y chromosomes (and the SRY gene) evolved from an autosomal pair after the divergence of monotremes only 166 million years ago. Therefore, the therian X and Y are more than 145 million years younger than previously thought.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7101908.
References
- Alsop A.E., Miethke P., Rofe R., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Miethke P., Rofe R., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Rofe R., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Koina E., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Sankovic N., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Deakin J.E., Haines H., Rapkins R.W., Graves J.A.M., Haines H., Rapkins R.W., Graves J.A.M., Rapkins R.W., Graves J.A.M., Graves J.A.M. Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby) Chromosome Res. 2005;13:627–636. doi: 10.1007/s10577-005-0989-2. [DOI] [PubMed] [Google Scholar]
- Bick Y., Sharman G., Sharman G. The chromosomes of the platypus (Ornithorhynchus): Monotremata. Cytobios. 1975;14:17–28. [Google Scholar]
- Bininda-Emonds O.R.P., Cardillo M., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Cardillo M., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Vos R.A., Gittleman J.L., Purvis A., Gittleman J.L., Purvis A., Purvis A. The delayed rise of present-day mammals. Nature. 2007;466:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., Charlesworth B., Marais G., Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Dohm J.C., Tsend-Ayush E., Reinhardt R., Grutzner F., Himmelbauer H., Tsend-Ayush E., Reinhardt R., Grutzner F., Himmelbauer H., Reinhardt R., Grutzner F., Himmelbauer H., Grutzner F., Himmelbauer H., Himmelbauer H. Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes. Genome Biol. 2007;8:R175. doi: 10.1186/gb-2007-8-8-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mogharbel N., Wakefield M., Deakin J.E., Tsend-Ayush E., Grützner F., Alsop A., Ezaz T., Graves J.A.M., Wakefield M., Deakin J.E., Tsend-Ayush E., Grützner F., Alsop A., Ezaz T., Graves J.A.M., Deakin J.E., Tsend-Ayush E., Grützner F., Alsop A., Ezaz T., Graves J.A.M., Tsend-Ayush E., Grützner F., Alsop A., Ezaz T., Graves J.A.M., Grützner F., Alsop A., Ezaz T., Graves J.A.M., Alsop A., Ezaz T., Graves J.A.M., Ezaz T., Graves J.A.M., Graves J.A.M. DMRT gene cluster analysis in the platypus: New insights into genomic organization and regulatory regions. Genomics. 2007;89:10–21. doi: 10.1016/j.ygeno.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Ezaz T., Stiglec R., Veyrunes F., Graves J.A.M., Stiglec R., Veyrunes F., Graves J.A.M., Veyrunes F., Graves J.A.M., Graves J.A.M. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 2006;16:736–743. doi: 10.1016/j.cub.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Fraser J.A., Heitman J., Heitman J. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 2005;15:645–651. doi: 10.1016/j.gde.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Fridolfsson A.K., Cheng H., Copeland N.G., Jenkins N.A., Liu H.C., Raudsepp T., Woodage T., Chowdhary B., Halverson J., Ellegren H., Cheng H., Copeland N.G., Jenkins N.A., Liu H.C., Raudsepp T., Woodage T., Chowdhary B., Halverson J., Ellegren H., Copeland N.G., Jenkins N.A., Liu H.C., Raudsepp T., Woodage T., Chowdhary B., Halverson J., Ellegren H., Jenkins N.A., Liu H.C., Raudsepp T., Woodage T., Chowdhary B., Halverson J., Ellegren H., Liu H.C., Raudsepp T., Woodage T., Chowdhary B., Halverson J., Ellegren H., Raudsepp T., Woodage T., Chowdhary B., Halverson J., Ellegren H., Woodage T., Chowdhary B., Halverson J., Ellegren H., Chowdhary B., Halverson J., Ellegren H., Halverson J., Ellegren H., Ellegren H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl. Acad. Sci. 1998;95:8147–8152. doi: 10.1073/pnas.95.14.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D., Rens W., Wallis M.C., Trifonov V., O’Brien P.C.M., Clarke O., Graves J.A.M., Ferguson-Smith M.A., Rens W., Wallis M.C., Trifonov V., O’Brien P.C.M., Clarke O., Graves J.A.M., Ferguson-Smith M.A., Wallis M.C., Trifonov V., O’Brien P.C.M., Clarke O., Graves J.A.M., Ferguson-Smith M.A., Trifonov V., O’Brien P.C.M., Clarke O., Graves J.A.M., Ferguson-Smith M.A., O’Brien P.C.M., Clarke O., Graves J.A.M., Ferguson-Smith M.A., Clarke O., Graves J.A.M., Ferguson-Smith M.A., Graves J.A.M., Ferguson-Smith M.A., Ferguson-Smith M.A. Search for the sex-determining switch in monotremes: Mapping WT1, SF1, LHX1, LHX2, FGF9, WNT4, RSPO1 and GATA4 in platypus. Chromosome Res. 2007;15:777–785. doi: 10.1007/s10577-007-1161-y. [DOI] [PubMed] [Google Scholar]
- Graves J.A.M. The origin and function of the mammalian Y chromosome and y-borne genes—An evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves J.A.M. Interactions between SRY and SOX genes in mammalian sex determination. Bioessays. 1998;20:264–269. doi: 10.1002/(SICI)1521-1878(199803)20:3<264::AID-BIES10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Graves J.A.M. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Graves J.A.M., Shetty S., Shetty S. Sex from W to Z—Evolution of vertebrate sex chromosomes and sex determining genes. J. Exp. Zool. 2001;290:449–462. doi: 10.1002/jez.1088. [DOI] [PubMed] [Google Scholar]
- Grützner F., Graves J.A.M., Graves J.A.M. A platypus’ eye view of the mammalian genome. Curr. Opin. Genet. Dev. 2004;14:642–649. doi: 10.1016/j.gde.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Grützner F., Rens W., Tsend-Ayush E., El-Mogharbel N., O’Brien P.C.M., Jones R.C., Ferguson-Smith M.A., Graves J.A.M., Rens W., Tsend-Ayush E., El-Mogharbel N., O’Brien P.C.M., Jones R.C., Ferguson-Smith M.A., Graves J.A.M., Tsend-Ayush E., El-Mogharbel N., O’Brien P.C.M., Jones R.C., Ferguson-Smith M.A., Graves J.A.M., El-Mogharbel N., O’Brien P.C.M., Jones R.C., Ferguson-Smith M.A., Graves J.A.M., O’Brien P.C.M., Jones R.C., Ferguson-Smith M.A., Graves J.A.M., Jones R.C., Ferguson-Smith M.A., Graves J.A.M., Ferguson-Smith M.A., Graves J.A.M., Graves J.A.M. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004;432:913–917. doi: 10.1038/nature03021. [DOI] [PubMed] [Google Scholar]
- Grützner F., Ashley T., Rowell D.M., Graves J.A.M., Ashley T., Rowell D.M., Graves J.A.M., Rowell D.M., Graves J.A.M., Graves J.A.M. How did the platypus get its sex chromosome chain? A comparison of meiotic multiples and sex chromosomes in plants and animals. Chromosoma. 2006;115:75–88. doi: 10.1007/s00412-005-0034-4. [DOI] [PubMed] [Google Scholar]
- Hore T.A., Koina E., Wakefield M.J., Graves J.A.M., Koina E., Wakefield M.J., Graves J.A.M., Wakefield M.J., Graves J.A.M., Graves J.A.M. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007;15:147–161. doi: 10.1007/s10577-007-1119-0. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Melamed E., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Melamed E., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Van Nas A., Replogle K., Band M.R., Clayton D.F., Replogle K., Band M.R., Clayton D.F., Band M.R., Clayton D.F., Clayton D.F., et al. Dosage compensation is less effective in birds than in mammals. J. Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M., Kehrer-Sawatzki H., Vogel W., Graves J.A.M., Hameister H., Kehrer-Sawatzki H., Vogel W., Graves J.A.M., Hameister H., Vogel W., Graves J.A.M., Hameister H., Graves J.A.M., Hameister H., Hameister H. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 2004;20:598–603. doi: 10.1016/j.tig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C., Page D.C. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Maduro V.V.B., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., NISC Comparative Sequencing Program. Maduro V.V.B., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., Tomkins J.P., Amemiya C.T., Luo M., Green E.D., Amemiya C.T., Luo M., Green E.D., Luo M., Green E.D., Green E.D. Comparative sequencing provides insights about the structure and conservation of marsupial and monotreme genomes. Proc. Natl. Acad. Sci. 2005;102:3354–3359. doi: 10.1073/pnas.0408539102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Tarui H., Toriba M., Yamada K., Nishida-Umehara C., Agata K., Matsuda Y., Tarui H., Toriba M., Yamada K., Nishida-Umehara C., Agata K., Matsuda Y., Toriba M., Yamada K., Nishida-Umehara C., Agata K., Matsuda Y., Yamada K., Nishida-Umehara C., Agata K., Matsuda Y., Nishida-Umehara C., Agata K., Matsuda Y., Agata K., Matsuda Y., Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.M., Wilcox S.A., Watson J.M., Lerner J.L., Woods D.R., Schefler J., Hearn J.P., Bishop C.E., Graves J.A.M., Wilcox S.A., Watson J.M., Lerner J.L., Woods D.R., Schefler J., Hearn J.P., Bishop C.E., Graves J.A.M., Watson J.M., Lerner J.L., Woods D.R., Schefler J., Hearn J.P., Bishop C.E., Graves J.A.M., Lerner J.L., Woods D.R., Schefler J., Hearn J.P., Bishop C.E., Graves J.A.M., Woods D.R., Schefler J., Hearn J.P., Bishop C.E., Graves J.A.M., Schefler J., Hearn J.P., Bishop C.E., Graves J.A.M., Hearn J.P., Bishop C.E., Graves J.A.M., Bishop C.E., Graves J.A.M., Graves J.A.M. The origin and loss of the ubiquitin activating enzyme gene on the mammalian Y chromosome. Hum. Mol. Genet. 1998;7:429–434. doi: 10.1093/hmg/7.3.429. [DOI] [PubMed] [Google Scholar]
- Murtagh C. A unique cytogenetics system in monotremes. Chromosoma. 1977;65:37–57. [Google Scholar]
- Nanda I., Zend-Ajusch E., Shan Z., Grützner F., Schartl M., Burt D.W., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Zend-Ajusch E., Shan Z., Grützner F., Schartl M., Burt D.W., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Shan Z., Grützner F., Schartl M., Burt D.W., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Grützner F., Schartl M., Burt D.W., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Schartl M., Burt D.W., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Burt D.W., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Koelher M., Fowler V.M., Goodwin G., Schneider W.J., Fowler V.M., Goodwin G., Schneider W.J., Goodwin G., Schneider W.J., Schneider W.J., et al. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: A comparative (re)view on avian sex determination. Cytogenet. Cell Genet. 2000;89:67–78. doi: 10.1159/000015567. [DOI] [PubMed] [Google Scholar]
- Ogata M., Hasegawa Y., Ohtani H., Mineyama M., Miura I., Hasegawa Y., Ohtani H., Mineyama M., Miura I., Ohtani H., Mineyama M., Miura I., Mineyama M., Miura I., Miura I. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity. 2007;100:92–99. doi: 10.1038/sj.hdy.6801068. [DOI] [PubMed] [Google Scholar]
- Quinn A.E., Georges A., Sarre S.D., Guarino F., Ezaz T., Graves J.A.M., Georges A., Sarre S.D., Guarino F., Ezaz T., Graves J.A.M., Sarre S.D., Guarino F., Ezaz T., Graves J.A.M., Guarino F., Ezaz T., Graves J.A.M., Ezaz T., Graves J.A.M., Graves J.A.M. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Raymond C.S., Kettlewell J.R., Hirsch B., Bardwell V.J., Zarkower D., Kettlewell J.R., Hirsch B., Bardwell V.J., Zarkower D., Hirsch B., Bardwell V.J., Zarkower D., Bardwell V.J., Zarkower D., Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev. Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- Rens W., Grützner F., O’Brien P.C.M., Fairclough H., Graves J.A.M., Ferguson-Smith M.A., Grützner F., O’Brien P.C.M., Fairclough H., Graves J.A.M., Ferguson-Smith M.A., O’Brien P.C.M., Fairclough H., Graves J.A.M., Ferguson-Smith M.A., Fairclough H., Graves J.A.M., Ferguson-Smith M.A., Graves J.A.M., Ferguson-Smith M.A., Ferguson-Smith M.A. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc. Natl. Acad. Sci. 2004;101:16257–16261. doi: 10.1073/pnas.0405702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W., O’Brien P.C.M., Grützner F., Clarke O., Grafodatskaya D., Tsend-Ayush E., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., O’Brien P.C.M., Grützner F., Clarke O., Grafodatskaya D., Tsend-Ayush E., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., Grützner F., Clarke O., Grafodatskaya D., Tsend-Ayush E., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., Clarke O., Grafodatskaya D., Tsend-Ayush E., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., Grafodatskaya D., Tsend-Ayush E., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., Tsend-Ayush E., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., Trifonov V.A., Skelton H., Wallis M.C., Johnston S., Skelton H., Wallis M.C., Johnston S., Wallis M.C., Johnston S., Johnston S., et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 2007;8:R243. doi: 10.1186/gb-2007-8-11-r243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann S., Steinemann M., Steinemann M. Y chromosomes: Born to be destroyed. Bioessays. 2005;27:1076–1083. doi: 10.1002/bies.20288. [DOI] [PubMed] [Google Scholar]
- Van Rheede T., Bastiaans T., Boone D.N., Blair Hedges S., de Jong W.W., Madsen O., Bastiaans T., Boone D.N., Blair Hedges S., de Jong W.W., Madsen O., Boone D.N., Blair Hedges S., de Jong W.W., Madsen O., Blair Hedges S., de Jong W.W., Madsen O., de Jong W.W., Madsen O., Madsen O. The platypus is in its place: Nuclear genes and indels confirm the sister group relation of monotremes and therians. Mol. Biol. Evol. 2006;23:587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- Wakefield M.J., Graves J.A.M., Graves J.A.M. Marsupials and monotremes sort genome treasures from junk. Genome Biol. 2005;6:218. doi: 10.1186/gb-2005-6-5-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis M.C., Delbridge M.L., Pask A.J., Alsop A.E., Grützner F., O’Brien P.C.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., Delbridge M.L., Pask A.J., Alsop A.E., Grützner F., O’Brien P.C.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., Pask A.J., Alsop A.E., Grützner F., O’Brien P.C.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., Alsop A.E., Grützner F., O’Brien P.C.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., Grützner F., O’Brien P.C.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., O’Brien P.C.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., Rens W., Ferguson-Smith M.A., Graves J.A.M., Ferguson-Smith M.A., Graves J.A.M., Graves J.A.M. Mapping platypus SOX genes; autosomal location of SOX9 excludes it from sex determining role. Cytogenet. Genome Res. 2007;116:2332–2334. doi: 10.1159/000098192. [DOI] [PubMed] [Google Scholar]
- Warren W.C., Hillier L.W., Graves J.A.M., Birney E., Pontig C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Hillier L.W., Graves J.A.M., Birney E., Pontig C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Graves J.A.M., Birney E., Pontig C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Birney E., Pontig C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Pontig C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Belov K., Miller W., Clarke L., Chinwalla A.T., Miller W., Clarke L., Chinwalla A.T., Clarke L., Chinwalla A.T., Chinwalla A.T., et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters P.D., Deldridge M.L., Deakin J.E., El-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Deldridge M.L., Deakin J.E., El-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Deakin J.E., El-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., El-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Carvalho-Silva D.R., Graves J.A.M., Graves J.A.M. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus): Implications for mammalian sex chromosome evolution. Chromosome Res. 2005;13:401–410. doi: 10.1007/s10577-005-0978-5. [DOI] [PubMed] [Google Scholar]
- Watson J.M., Spencer J.A., Riggs A.D., Graves J.A.M., Spencer J.A., Riggs A.D., Graves J.A.M., Riggs A.D., Graves J.A.M., Graves J.A.M. The X chromosome of monotremes shares a highly conserved region with the eutherian and marsupial X chromosomes despite the absence of X chromosome inactivation. Proc. Natl. Acad. Sci. 1990;87:7125–7129. doi: 10.1073/pnas.87.18.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.M., Spencer J.A., Riggs A.D., Graves J.A.M., Spencer J.A., Riggs A.D., Graves J.A.M., Riggs A.D., Graves J.A.M., Graves J.A.M. Sex chromosome evolution: Platypus gene mapping suggests that part of the human X chromosome was originally autosomal. Proc. Natl. Acad. Sci. 1991;88:11256–11260. doi: 10.1073/pnas.88.24.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox S.A., Watson J.M., Spencer J.A., Graves J.A.M., Watson J.M., Spencer J.A., Graves J.A.M., Spencer J.A., Graves J.A.M., Graves J.A.M. Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics. 1996;35:66–70. doi: 10.1006/geno.1996.0323. [DOI] [PubMed] [Google Scholar]
- Wrigley J.M., Graves J.A.M., Graves J.A.M. Karyotypic conservation in the mammalian order Monotremata (subclass Prototheria) Chromosoma. 1988;96:231–247. doi: 10.1007/BF00302363. [DOI] [PubMed] [Google Scholar]