Abstract
Small RNA pathways play evolutionarily conserved roles in gene regulation and defense from parasitic nucleic acids. The character and expression patterns of small RNAs show conservation throughout animal lineages, but specific animal clades also show variations on these recurring themes, including species-specific small RNAs. The monotremes, with only platypus and four species of echidna as extant members, represent the basal branch of the mammalian lineage. Here, we examine the small RNA pathways of monotremes by deep sequencing of six platypus and echidna tissues. We find that highly conserved microRNA species display their signature tissue-specific expression patterns. In addition, we find a large rapidly evolving cluster of microRNAs on platypus chromosome X1, which is unique to monotremes. Platypus and echidna testes contain a robust Piwi-interacting (piRNA) system, which appears to be participating in ongoing transposon defense.
Initially regarded as a scientific hoax by European biologists, the platypus (Ornithorhynchus anatinus), along with four species of echidna (family Tachyglossidae), represent the only remaining members of the basal mammalian clade of prototheria. These animals diverged from remaining mammalian lineages around 166 million years ago (Bininda-Emonds et al. 2007). Morphologically, monotremes display a mixture of mammalian, reptilian, and unique features. As mammals, monotremes have fur, but they share with reptiles a cloaca, the single external opening for both the gastrointestinal and urogenital systems. Monotremes lay eggs that are nourished in part through a placental structure before deposition, but they provide milk to their hatched young through mammary glands. Monotreme embryos show meroblastic rather than holoblastic cleavage, which is more typical of reptiles and fish than of mammals.
The platypus is a specialized semiaquatic carnivore that displays several unique features. Its leathery bill is equipped with specialized neurons, which serve as electrosensors, and male platypuses have spikes on their hind legs, which are used in an elaborate venom delivery system. The platypus karyotype is also very unusual, with small chromosomes reminiscent of reptilian microchromosomes and multiple sex chromosomes (5X, 5Y) that form a chain during meiosis (Grutzner et al. 2004; Rens et al. 2004). Remarkably, the unique mix of mammalian, reptilian, and specialized features of monotremes is also revealed by its genomic sequence. This together with its phylogenetic position makes it a uniquely placed comparative tool for studying mammalian evolution (Warren et al. 2007).
RNAi pathways are deeply conserved and can be traced at least as far back as the divergence of prokaryotes and eukaryotes, ∼2.7 billion years ago. Individual eukaryotic lineages have adapted these pathways in different ways, although most use RNAi generally as both a genome defense and a gene regulatory mechanism. The RNAi machinery uses small RNAs of between 18 and 33 nucleotides (nts) in length as specificity determinants to regulate RNA stability and gene expression, through both post-transcriptional and transcriptional modes.
Two endogenous RNAi pathways have been described in mammals, the microRNA (miRNA) pathway and the Piwi-interacting (piRNA) pathway. The miRNA pathway uses small RNAs, around 18–24 nt in length, to regulate genes post-transcriptionally (Bartel 2004). miRNA duplexes are liberated from genome-encoded hairpins by successive processing by RNase III enzymes RNASEN (also known as Drosha) and DICER1 (Bartel 2004). Incorporation into complexes containing members of the EIF2C (also known as Argonaute) family of RNaseH-like enzymes coincides with the release of the second, or star, strand, leaving the mature strand to direct sequence-specific silencing events. miRNAs act in diverse biological networks, with many having roles in reinforcing cell-type and tissue identity (Plasterk 2006). Some families of miRNA genes have deeply conserved expression patterns, extending throughout animal lineages, whereas others are more rapidly evolving and lineage specific (Plasterk 2006). piRNAs are a second class of small RNAs, slightly larger in length than miRNAs (∼25–33 nt), which interact with the Piwi clade of Argonaute-family proteins. piRNAs show restricted expression patterns, being prominent in gonad, with mammalian piRNAs thus far being restricted to the germline compartment (Aravin et al. 2007a). Genetic and sequence analyses of piRNAs in Drosophila have revealed a role for piRNAs in transposon control via a pathway that uses small RNA generating loci as transposon sensors (Brennecke et al. 2007; Gunawardane et al. 2007). In post-pubescent mammalian testis, piRNAs arise from dense strand-biased clusters whose biogenesis and functions remain largely unknown, although roles in transposon defense are supported for piRNAs that are expressed in germ cell precursors earlier in development (Aravin et al. 2007a).
Using the platypus genome sequence, we have performed a comparative analysis of small RNA pathways in platypus, echidna, chicken, and eutherian mammals and have found deep conservation of many small RNA species. We also note miRNAs with mammalian or chicken/monotreme lineage-specific conservation as well as a surprisingly extensive set of monotreme-specific and platypus-specific small RNAs. Strikingly, our analysis implicates small RNA functions in monotreme reproduction, as we find large clusters of germline-expressed fast-evolving miRNA loci on one of the X chromosomes in platypus and evidence for ongoing transposon defense by piRNAs in the adult platypus testis.
Results
Comparative analysis of RNAi components in platypus
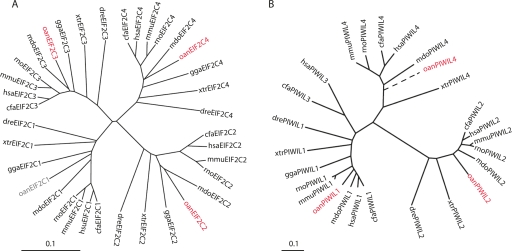
The completion of the platypus genome sequence offers an opportunity to examine the conservation of small RNA pathways in prototherian mammals (Warren et al. 2007). Indeed, homologs of key RNAi components DICER1, RNASEN, and EIF2C/Argonaute family members 1 through 4 could be readily identified in platypus (Fig. 1A; Supplemental Table 1). Moreover, synteny of the EIF2C/Argonaute gene family was conserved between platypus and eutherian mammals, with platypus EIF2C1, EIF2C3, and EIF2C4 closely linked on Ultracontig 472, and EIF2C2 on chromosome 4. Platypus also had obvious orthologs for PIWI family members PIWIL1, PIWIL2, and PIWIL4 but appeared to be missing PIWIL3 (Fig. 1B; Supplemental Tables 1 and 2). As PIWIL3 is only found in a subset of eutherian mammals and could not be found in a marsupial genome (Monodelphis domestica), it is likely that this gene emerged after the divergence of the eutherian and prototherian lineages. Interestingly, the DDH motif, necessary for the endoribonuclease activity of EIF2C/Argonaute enzymes, was conserved in platypus EIF2C2 and all three PIWI proteins, as well as in EIF2C3, even though mouse and human orthologs of EIF2C3 appear to be noncatalytic (Liu et al. 2004; Meister et al. 2004). The conservation of this motif in the EIF2C/Argonaute clade throughout mammalian evolution is mysterious, as the biological significance of Argonaute-mediated RNA cleavage in mammals is unclear (Yekta et al. 2004; Davis et al. 2005; Yu et al. 2005; O'Carroll et al. 2007). Overall, our findings suggest that the platypus maintains a fully functional set of RNAi enzymes, the evolution of which reconstructs the phylogeny of the vertebrate lineage.
Figure 1.
RNA interference genes in platypus. Phylogenetic trees of EIF2C (A) and PIWI (B) families in multiple vertebrate species. The platypus PIWIL4 (oanPIWIL4) sequence was incomplete, and a partial coding sequence was used to place this gene on the tree (broken line) (Supplemental Table 2). Platypus genes are highlighted in red. Scale: 0.1 = 0.1 base substitutions expected. (oan) Ornithorhynchus anatinus; (cfa) Canis familiaris; (hsa) Homo sapiens; (mmu) Mus musculus; (rno) Rattus norvegicus; (mdo) Monodelphis domesctica; (gga) Gallus gallus; (xtr) Xenopus tropicalis; (dre) Danio rerio.
Identification of conserved and novel miRNAs in monotremes
To determine the extent of conservation of miRNA pathways in monotremes, we cloned and deep-sequenced the 18- to 24-nt small RNA content of six tissues (brain, kidney, heart, lung, liver, and testis) from adult male platypus (Ornithorhynchus anatinus) and adult male short-beaked echidna (Tachyglossus aculeatus). A total of 2,982,604 unique small RNAs were identified from these twelve libraries that mapped to the platypus genome: 1,409,428 small RNAs from platypus and 1,532,352 small RNAs from echidna. We devised a miRNA discovery pipeline combining a computational heuristic with manual curation of miRNAs (see Methods). This approach yielded 332 miRNA candidates, 149 orthologs of known miRNAs, and 183 novel miRNAs (Supplemental Table 3). Three of the novel miRNAs (miR-1329, miR-1397, and miR-1416) were also identified in chicken (Gallus gallus), whereas 180 miRNAs were apparently unique to monotremes.
The platypus genome sequence has revealed commonalities with both therian mammals and nonmammalian vertebrates (Warren et al. 2007). Monotremes share at least 10 miRNA genes with eutherian mammals (mouse/human) but not chicken, 4 with chicken but not mouse or human, and one with fish (Dania rerio, Fugu rubripes, and Tetraodon nigroviridis) but not mouse, human, or chicken (Warren et al. 2007) (Fig. 2; Supplemental Table 3). Although it is possible that some miRNAs that have apparently been lost during vertebrate evolution are missing because of incomplete miRNA identification or genome sequence, it is likely that some miRNA gain and loss reflects genuine evolutionary adaptation.
Figure 2.
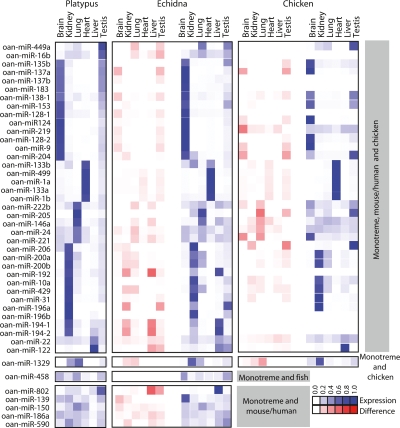
Conservation of microRNA expression patterns in monotremes. Comparison of platypus/echidna and platypus/chicken miRNA tissue expression profiles. miRNA orthology was determined by perfect match (platypus and echidna) or at least 16-nt identity in the mature miRNA (mouse, human, chicken, and fish). The expression score represented by blue squares is a ratio of the number of normalized reads in that tissue versus all tissues in that organism, and the difference score represented by red squares is the difference between the confidence intervals of the expression ratios for each platypus/echidna or platypus/chicken pair of miRNAs. A subset of miRNAs shared between monotremes, mouse/human, and chicken; monotreme and chicken; monotreme and fish; and monotreme and mouse/human are shown. miRNAs with the most distinct tissue-specific expression patterns are shown here clustered by tissue expression; the complete data sets can be found in Supplemental Figs. 1–4.
miRNA expression patterns have been extensively characterized in several vertebrates (Chen et al. 2005; Wienholds et al. 2005; Ason et al. 2006; Darnell et al. 2006; Kloosterman et al. 2006a, b; Takada et al. 2006; Xu et al. 2006; Landgraf et al. 2007). Using miRNA cloning frequency data, we determined miRNA signatures for platypus and echidna brain, heart, lung, kidney, liver, and testis (Fig. 2; Supplemental Fig. 1). Although expression patterns of miRNAs tend to be conserved across vertebrates, variation has been observed in some cases (Ason et al. 2006). We found strong conservation of expression domains and expression levels of orthologous miRNAs among monotremes (platypus and echidna), among mammals (platypus and mouse/human), and between platypus and chicken (Fig. 2; Supplemental Figs. 1–4). Interestingly, the tissue with the greatest differences in miRNA expression level between the aquatic platypus and the terrestrial, predominantly ant-eating echidna was liver, perhaps reflecting the unique dietary adaptations of these two monotremes (Fig. 2; Supplemental Fig. 1).
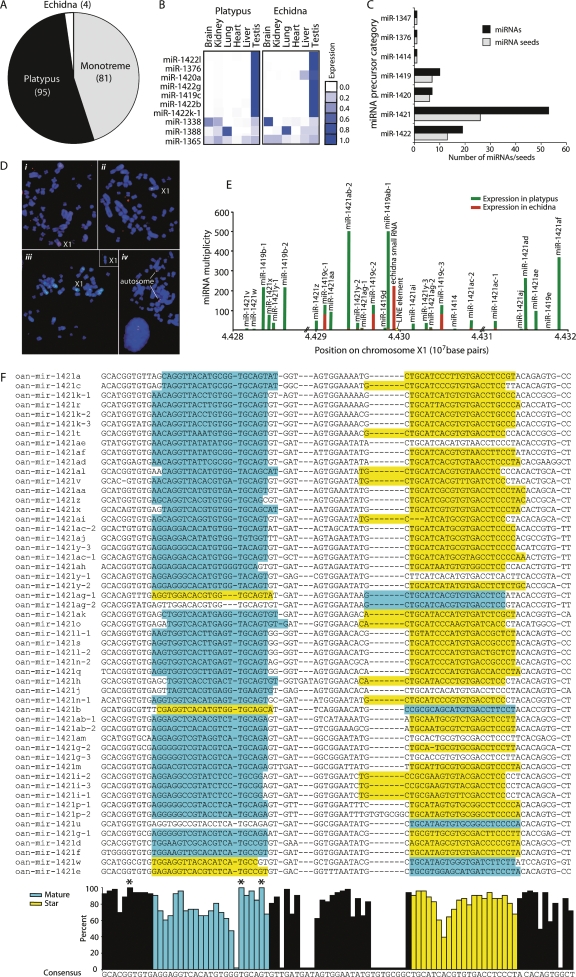
The platypus and echidna lineages are thought to have diverged ∼16.5 million years ago, although recent re-examination of fossil evidence suggest the possibility of an earlier divergence (Warren et al. 2007; Rowe et al. 2008). Of the 180 novel monotreme miRNA candidates identified through our screening approach, 95 were found only in platypus, 4 only in echidna (although their mature sequence was perfectly conserved in the platypus genome), and 81 from both platypus and echidna (Fig. 3A; Supplemental Table 3). Although we used stringent criteria for determining platypus and echidna orthology (perfect identity) and likely missed a number of echidna orthologs, it is striking that such a large number of miRNAs were found only in platypus. This suggests either that platypus and echidna indeed diverged earlier than previously estimated (Rowe et al. 2008) or that a subset of platypus miRNAs have evolved rapidly.
Figure 3.
Novel microRNAs in monotremes. (A) Pie graph of monotreme species distribution of 180 novel miRNAs. Eighty-one miRNAs were sequenced in both platypus and echidna small (gray); 95 were only in platypus (black); and four were cloned only in echidna (white), although their sequences were present in the platypus genome. (B) Novel miRNA tissue expression profiles. The expression score represented by blue squares is a ratio of the number of normalized reads in that tissue versus all tissues in that organism. Expression data for 10 of the most highly expressed novel miRNAs is shown. (C) Testis miRNA classification. Ninety-two novel miRNAs were highly expressed in testis and were derived from one of seven related hairpin precursors. The number of miRNAs and miRNA seed sequences derived from each precursor category is shown. (D) Five out of nine contigs containing testis-expressed novel miRNAs map to X1q. Fluorescence in situ hybridization with BAC and Fosmid clones containing miRNA clusters on male platypus metaphase chromosomes. (i) BAC 175h10 from contig 8388 (green) and BAC 273L19 from contig 7359 (red) on the long arm of X1. (ii) Colocalization of BAC 175h10 from contig 8388 (green) and BAC 272K3 from contig 22,847 (red) on X1q. (iii) Localization of Fosmid 0879F15 from contig 7160 (green) on X1q. (Insert) Localization of Fosmid 1002L21 (contig 11,344) on X1q. (iv) BAC clones 686C10 (green) and 802P20 (red) from contig 1754 map to a metacentric autosome. (E) Novel testis-expressed chromosome X1 miRNA cluster. Only small RNAs that map to the genome three times or less are shown. The cloning frequency divided by the number of genome matches for each miRNA is represented by a bar. Platypus miRNAs are plotted in green, echidna in red; platypus and echidna bars are independent and not additive. Gaps in the genome assembly are shown by //. A LINE element insertion that disrupted an echidna small RNA precursor is represented in yellow. (F) Orientation and sequence conservation of 53 miRNAs derived from the miR-1421 group of precursors. Multiple sequence alignment of 53 miR-1421 precursors with the sequence of the mature miRNA highlighted in blue and the miRNA star in yellow. The percent of nucleotides in the alignment that match the consensus for each position is graphed below. Perfectly conserved nucleotides are marked with an asterisk (*).
Identification of a novel X-linked miRNA cluster in monotremes
We investigated tissue expression patterns of a subset of the novel monotreme miRNAs by comparing total normalized cloning frequencies across tissues (Fig. 3B). Strikingly, a large number of miRNAs in both the monotreme- and platypus-specific sets was almost exclusively expressed in testis (Fig. 3B; Supplemental Table 3). Indeed, closer inspection revealed that 92 of the monotreme-specific miRNAs (51%), which constituted the majority of the highly expressed species (Warren et al. 2007), fell into seven testis-expressed miRNA groups (miR-1347, miR-1376, miR-1414, miR-1419, miR-1420, miR-1421, and miR-1422) with related precursor hairpins (Fig. 3C). miRNAs were named based on the classification of their precursors, that is, miRNAs processed from a miR-1421-related hairpin were called miR-1421a through miR-1421am.
The testis-expressed miRNAs arose from nine large miRNA clusters on chromosome X1 and contigs 1754, 7160, 7359, 8388, 11,344, 22,847, 198,872, and 191,065. Physical mapping of six of these contigs placed five on the long arm of chromosome X1, raising the possibility that the testis miRNAs are derived from a single large cluster on chromosome X1 (Fig. 3D). The miRNA clusters are also related at the sequence level and appear to have arisen from a complex series of tandem duplications.
One miRNA cluster on chromosome X1 is ∼40 kb long and encodes at least 27 mature miRNAs (Fig. 3E; Supplemental Table 3). Although the miRNAs arising from this cluster are from highly related precursors, they are cloned at a range of frequencies (Fig. 3E; Supplemental Table 3), suggesting that there is differential regulation of miRNA expression, processing, or stability. Interestingly, although most miRNAs in this cluster are predominantly cloned from platypus testis, some are also detectable in echidna testis, and one small RNA mapping to this cluster is almost entirely restricted to echidna (Fig. 3E). This echidna small RNA is highly related to miR-1421 miRNAs but was not predicted as a miRNA because of the absence of a complete hairpin precursor in the platypus genome. This small RNA may be a miRNA that has been lost in platypus because of the insertion of a LINE element into the miRNA precursor since the divergence of platypus and echidna (Fig. 3E).
miRNAs are often classified into families on the basis of identity at the “seed” heptamer region at nucleotide positions 2–8 of the mature miRNA. The seed sequence participates in miRNA target recognition and binding, and 5′ end miRNA processing must be very precise to ensure the production of the correct seed. Interestingly, although the 92 testis-expressed monotreme miRNAs share only seven related precursor sequences, they generate a great diversity of seed sequences (55 unique seeds) (Fig. 3C; Supplemental Table 3). Indeed, the 51 miRNAs processed from miR-1421-related precursors give rise to 26 unique seed sequences (Fig. 3E).
To understand whether the unexpectedly high diversity of seed sequences in testis miRNAs arose from sequence divergence in the seed region or irregular processing, we mapped both the mature and star sequences for each of 53 miR-1421-related miRNAs on a multiple sequence alignment (Fig. 3F). Interestingly, the sites of processing are slightly offset for some miR-1421 miRNAs (Fig. 3F). In addition, although the mature miRNA is usually processed from the 5′ arm and the less frequently cloned star from the 3′ arm of the miR-1421 precursor, in 6 cases this orientation was switched, thus generating seed diversity (Fig. 3F).
The seed is usually the most highly conserved region of a miRNA hairpin. To examine sequence conservation in miR-1421 hairpins, we calculated percent identity to a consensus for each nucleotide. Overall, the miR-1421 hairpins were highly related and shared on average 82% identity across the entire hairpin (Fig. 3F). However, the most highly conserved positions were in the 5′ stem of the hairpin and 3′ end of the 5′ arm mature miRNA. Interestingly, the seed region of the 5′ arm mature miRNA is considerably less conserved, with an average of 76% identity with the consensus, than its foldback region at the 3′ end of the star sequence, which has an average of 88% identity (Fig. 3F). This is probably at least partly due to the predominance of A and G nucleotides in the seed region, both of which can base pair with uracil residues, and suggests that these miRNAs are exploring sequence space within the constraints of hairpin architecture (Fig. 3F).
Overall, the lack of conservation in processing site, mature strand choice, and seed sequence, as well as the striking divergence in miRNA repertoire between platypus and echidna suggest that testis miRNAs are fast evolving and may regulate a diverse set of targets.
piRNAs in platypus
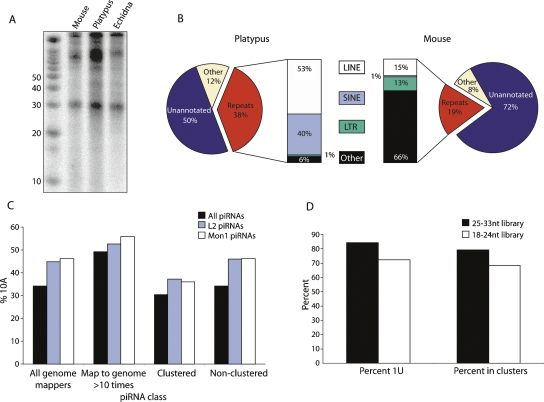
piRNAs are a class of germline-expressed small RNAs that interact with Piwi proteins (Aravin et al. 2007a). Many piRNAs function as the specificity determinants of an RNA-based innate immune system that protects the germline from transposable elements and other nucleic acid parasites (Aravin et al. 2007a). In eutherian mammals, piRNAs are ∼26–30 nt long, tend to start with a uracil, and are abundantly expressed from discrete genomic intervals in testis (Aravin et al. 2007a). Radioactive end labeling of total RNA from platypus, echidna, and mouse testes revealed an abundant class of ∼30-nt piRNAs in monotreme testis (Fig. 4A).
Figure 4.
piRNAs in platypus. (A) Total RNA labeling from mouse, platypus, and echidna testis reveals a distinct ∼30-nt piRNA species in monotremes. RNA was dephosphorylated, end labeled with 32P, and run on a denaturing 15% polyacrylamide gel. Sizes of markers (left) are in nucleotides. (B) Comparison of platypus and mouse piRNA libraries. A platypus piRNA library was generated by cloning small RNAs in a 25- to 33-nt size window from total platypus testis RNA. Sequences from this library and from an equivalent mouse piRNA library (Girard et al. 2006) were classed as repeat (red), unannotated (blue), or other (beige). The repeat piRNAs were broken down into classes of repeat (bars). (C) 10A profiles for platypus piRNAs. An A at piRNA position 10 is a signature of an active piRNA amplification system. “Clustered” piRNAs are defined as those that map to the genome once and fall into one of 50 piRNA clusters (Supplemental Table 4), while “non-clustered” piRNAs are those that map to the genome once but do not fall into a cluster. 10A percentages for all piRNAs are in black, those that are derived from LINE2 elements in blue, and those from Mon1 SINE elements in white. (D) Characteristics of “small” piRNAs. “Percent 1U” includes all small RNAs that match the genome; “percent in clusters” includes only small RNAs that map only once to the genome.
To investigate the relationships between the Piwi/piRNA pathways of monotremes and eutherians, we cloned and deeply sequenced RNAs sized between 25 and 33 nt from platypus testis. Seventy-two percent of uniquely mapping RNAs in this size range could be assigned to one of 50 large intervals in the platypus genome (Supplemental Table 4). These appeared similar to piRNA clusters observed in eutherian mammals, being an average of ∼30 kb long and exhibiting a strong strand bias for small RNA production. Based on these characteristics and their 5′ U bias, we conclude that these species are likely platypus piRNAs. As is the case in eutherian mammals, no piRNA clusters were observed on X chromosomes in platypus; however, as the platypus genome is incomplete and many of the platypus piRNA clusters are located on unmapped contigs, we cannot rule out the possibility that platypus has piRNA clusters on sex chromosomes (Supplemental Table 4).
Platypus piRNAs were annotated based on genome mapping (Supplemental Table 5). In mouse, piRNAs expressed in prepubertal testis are repeat-rich and have a function in transposon control, but piRNAs expressed in adult testis are repeat poor and have no known function (Aravin et al. 2007a, b). Interestingly, the platypus piRNA library contained a much larger proportion of repeat-associated species than an equivalent piRNA library from adult mouse testis (Girard et al. 2006) (Fig. 4B). Furthermore, the repeat composition of adult platypus and mouse repeat-associated piRNAs was quite distinct. Whereas in platypus the majority of repeat-associated piRNAs were from LINE and SINE elements, these repeats only made up 16% of the repeat-derived adult mouse piRNAs (Fig. 4B). As LINE and SINE elements, specifically L2 and Mon1, are thought to be active in platypus (Warren et al. 2007), we speculated that perhaps a difference in transposon expression may underlie the differential contribution of these repeat elements to the piRNA populations of adult testis in monotremes and eutherian mammals.
Enrichment for A residues at position 10 of piRNAs is a signature of their participation in ongoing transposon defense (Aravin et al. 2007a; Brennecke et al. 2007). In other systems, piRNA populations with a 10A bias are secondary piRNAs and largely arise from transcripts from active transposons (Brennecke et al. 2007). If LINE and SINE elements are active in platypus, a higher proportion of 10A piRNAs might be expected from these elements than from the bulk piRNA population. In accord with this prediction, the percentage of 10A was significantly greater in L2 and Mon1 piRNAs (Fig. 4C; P < 10−125). We also reasoned that the more times a repeat-derived piRNA matched the genome, the greater the likelihood that it is derived from an active transposon, and thus greater the likelihood of 10A. To test whether we could see this effect in the platypus piRNA population, we calculated the 10A bias for piRNAs that mapped to the genome at least ten times. Strikingly, 10A was at 49% in this population and increased to 53% and 56% in L2 and Mon1 piRNAs, respectively (Fig. 4C; P < 10−4). Moreover, piRNAs that mapped once to the genome but lay outside of the 50 piRNA clusters (Supplemental Table 4), and thus presumably represented individual active transposons recognizable through their divergent sequences, had a higher proportion of 10A, particularly if they were derived from L2 or Mon1 elements (Fig. 4C). Overall, this analysis provides evidence that piRNA pathways in platypus are active in transposon defense against L2 and Mon1 elements, and this may underlie the high proportion of LINE and SINE piRNAs in platypus.
One characteristic feature of piRNAs is that they are generally larger than miRNAs and siRNAs. However, a surprisingly large proportion of the platypus testis 18- to 24-nt library was composed of small RNAs with piRNA-specific properties. Indeed, 68% of small RNAs in this library that mapped to the genome once were derived from one of the 50 piRNA clusters (Fig. 4D). Furthermore, 79% of small RNAs in this library that mapped to the genome at least once had a U at position 1 (Fig. 4D). Further studies will be required to determine whether this small piRNA class has unique biogenesis or Piwi binding characteristics as compared with the abundant, larger piRNA species.
Discussion
Overall, these studies indicate that the RNAi machinery and its functions in genome defense and gene regulation are conserved in monotremes. Cloning and deep sequencing of miRNAs and piRNAs in monotremes have revealed small RNAs and regulatory systems similar to those found in other mammals, as well as monotreme-specific components.
In general, we found conservation of both sequence and expression patterns of miRNAs in monotremes, chicken, and eutherian mammals. Some platypus miRNAs had unique conservation patterns. For example, orthologs of platypus miR-458 were found only in fish, but not in chicken or mammals. We also identified four miRNAs that are shared between platypus and chicken but that are not shared with other mammals (Fig. 2; Supplemental Table 3). As these miRNAs appear to have been lost from some lineages, they are presumably involved in biological functions that have either become redundant or obsolete in some branches of mammalian evolution.
We have identified 180 novel miRNA candidates unique to platypus and echidna, including at least one large miRNA cluster on platypus chromosome X1. These clustered miRNAs were almost exclusively expressed in testis among the six tissues we analyzed. There are several examples of such expanded fast-evolving testis-expressed miRNA clusters on X chromosomes in other mammals. For instance, the MIRN506 to MIRN514 cluster on the X chromosome of humans and other primates contains 10 rapidly evolving miRNAs whose testis-specific expression and developmental regulation suggest a role in spermatogenesis (Bentwich et al. 2005; Zhang et al. 2007). Likewise, the Mirn743 to Mirn465 cluster is a testis-expressed cluster of 19 miRNAs on the rodent X chromosome (Yu et al. 2005; Ro et al. 2007). A large miRNA cluster has also been described on the X chromosome of the marsupial opossum Monodelphis domestica (Devor and Samollow 2007). Male reproduction is generally fast-evolving in mammals, and genes on sex chromosomes, present as single copy in males, are particularly susceptible to positive selection for male reproductive success (Wyckoff et al. 2000; Delbridge and Graves 2007). Identification of miRNA target genes and analysis of miRNAs in additional vertebrate groups will illuminate the significance of such fast-evolving miRNA clusters in spermatogenesis.
The testis-expressed miRNAs in monotreme testis generate at least 92 mature miRNAs with 55 unique seed sequences. Although most of these seeds are novel, seven of them are shared with miRNAs in mouse, human, or chicken (Supplemental Table 3). Interestingly, six of the shared seeds are found in uniquely mammalian clustered miRNAs, which tend to be expressed in embryonic, placental, and reproductive tissues. These include a subset of miRNAs in the mouse Mirn290 to Mirn295 cluster, Mirn467a, Mirn764, and members of the human MIRN512 to MIRN373 cluster (Houbaviy et al. 2003, 2005; Bentwich et al. 2005). It is unclear whether these seed similarities imply functional or evolutionary relatedness.
Monotremes are unique among mammals for their multiple sex chromosomes, which form a chain during meiosis (Grutzner et al. 2004; Rens et al. 2004). It is unclear whether the ten paired X and Y chromosomes of monotremes undergo meiotic sex chromosome inactivation and sex body formation as is observed in eutherian mammals and marsupials (Handel 2004; Namekawa et al. 2007). Abundant testis expression of at least one X1-linked miRNA cluster in monotremes suggests that either this cluster is expressed in premeiotic or somatic cells of the testis or that sex chromosomes are not completely silenced in monotremes.
The testis-clustered miRNAs identified in platypus and echidna are processed irregularly. In some cases, the mature strand and star strands are switched, and the RNase III processing sites are slightly offset between individual hairpins (Fig. 3F). These differences may reflect subtle differences in hairpin structure or sequence. Alternatively, monotremes may possess unique components of the miRNA processing machinery that are able to process miRNA hairpins in an unusual manner.
The platypus genome encodes three proteins of the PIWI family involved in piRNA biogenesis and function, PIWIL1, PIWIL2, and PIWIL4 (Fig. 1B). The conservation pattern and longer branch length of PIWIL2 suggest that it might encode the ancestral PIWI protein in vertebrates and that subsequent PIWI family members may have arisen by gene duplication events. This theory is consistent with observations that PIWIL2 binds a smaller size range of piRNAs, more akin to the piRNAs found in invertebrates, and that it is the only PIWI protein in mammals that is expressed in both the embryonic and adult gonad (Kuramochi-Miyagawa et al. 2001, 2004; Aravin et al. 2006).
Although platypus piRNAs are organized in large strand-biased clusters similar to those found in other mammals, the piRNA system appears to be actively engaged in combating transposon activity in mature platypus testis. This is in contrast to piRNAs in adult testis of other mammals, which are depleted of repeat sequences and have no known function (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). The piRNAs of platypus adult testis have features of both fish and eutherian piRNA systems (Houwing et al. 2007) and suggest that mammalian piRNA clusters that are expressed late in germ cell development may have progressively lost repeat content as they acquired novel functions (Aravin et al. 2007b; Carmell et al. 2007).
The small RNA complement of platypus has highlighted not only unique and conserved roles for miRNAs and piRNAs in monotremes but also provides insight into the evolution of small RNA pathways in mammals.
Methods
Tree building
The phylogenetic trees of EIF2C and PIWI families in multiple species were constructed using the Phylip package (version 3.67) (Felsenstein 1997). The full-length coding region of each gene was used to calculate the distance, and a neighbor-joining method was used to obtain the topology of the tree.
Small RNA cloning, sequencing, and annotation
RNA was isolated by a standard TRizol method from snap-frozen tissue of adult male platypus (O. anatinus), echidna (T. aculeatus), and chicken (G. gallus) heart, liver, lung, kidney, brain, and testis. Animals (platypus and echidna) were captured at the Upper Barnard River, New South Wales, Australia, during breeding season (AEC permit no. S-49-2006 to F.G.). miRNA (18–24 nt) and piRNA (25–33 nt) fractions of total RNA were used to clone libraries as described (Brennecke et al. 2007). Small RNA libraries were sequenced on the Illumina 1G sequencer. In the case of miRNA libraries, 3′ linkers were clipped using a dynamic alignment algorithm, which required perfect matches at positions 25–30 but allowed mismatches toward the end of the read. The piRNA library was uniformly clipped to 26 nt in length. Platypus and echidna small RNAs with perfect matches to the platypus genome were annotated using Ensembl and RepeatMasker platypus genome annotations. piRNA clusters were defined manually and may be incomplete.
miRNA prediction
Clipped small RNAs of 18 nt or longer from platypus/echidna or chicken 18- to 24-nt libraries were mapped to the platypus or chicken genomes, respectively, requiring perfect sequence identity. Echidna small RNAs were mapped to the platypus genome because of the lack of availability of an echidna genome. The potential for each position of the genome to be a 5′ end of a mature miRNA sequence was evaluated by counting the total number of sequence counts starting at that position. Two windows of 150 base pairs were folded around the potential mature miRNA, one starting 30 bases upstream (for a mature in the left arm) and one starting 100 bases upstream (for a mature in the right arm). Folding was done by sampling 500 structures using RNAsubopt from the Vienna RNA Package (Wuchty et al. 1999) and taking the one that had the greatest number of paired arm bases. The longest pair of substrings whose bases only matched to each other and whose starts and ends were paired were defined as the two arms, and each hairpin was trimmed to the ends of the arms. Only hairpins that had at least 20 paired bases in the arms were considered. It was required that at least 18 of 20 bases following the potential mature 5′ end overlapped with the intended arm and that none of the bases overlapped with the opposite arm. We considered only hairpins for which at least 40% of the reads within the hairpin and 50% of reads within 10 bases of the putative 5′ end of the mature belonged to the putative 5′ end of the mature miRNA. To avoid repeat associated sequences, no read within the hairpin could have more than five total matching positions in the genome, and 99% of the reads must not have more than three matching positions. It was required that there be no more than six positions that had more than 1% of the hairpin reads and no more than four positions that had more than 10% of the hairpin reads. Candidate miRNAs that were sequenced only once were discarded. The 2350 hairpins from platypus and echidna that fit these criteria were inspected manually to remove piRNAs, repeats, degradation products, and other types of noncoding RNAs, leaving 332 candidate miRNAs. A total of 999 hairpins were predicted for chicken, which were not manually curated. Chicken orthologs of platypus miRNAs were assigned by BLASTN requiring at least 16-nt identity in the mature miRNA. In 13 cases in which chicken orthologs were cloned but not predicted (e.g., because of missing genome sequence), the cloned chicken sequences are shown in Supplemental Table 3. The most frequently cloned small RNA in the candidate miRNA hairpin was designated as the mature sequence, and the most frequently cloned sequence from the other arm was designated as the miRNA* sequence. Platypus and echidna orthologs of known miRNAs were identified by BLASTN with the Rfam miRNA database, requiring at least 16 nucleotide matches. miRNAs were named according to their ortholog; novel miRNAs were given unique names. Four miRNAs cloned only from echidna (oan-miR-1331, oan-miR-1336, oan-miR-1352, and oan-miR-1384) were given platypus names because of the lack of availability of the echidna genome.
Heatmap construction
Orthologs for candidate mature miRNAs in platypus brain, heart, liver, and testis (for comparison with human) or platypus brain, heart, liver, testis, kidney, and lung (for comparison with mouse and chicken) were found by BLASTN using the human or mouse Rfam database and 999 predicted chicken miRNAs, requiring at least 16 matched bases. miRNA read counts in platypus and those in corresponding tissues in chicken and in a human and mouse miRNA expression atlas (Landgraf et al. 2007) were first normalized to the tissue with the least number of total miRNA reads. miRNAs that did not have at least 10 normalized reads in both platypus and human/mouse/chicken were discarded. Each miRNA in each tissue was assigned an expression score, a ratio of the number of reads in that tissue versus all tissues in that organism. The miRNAs were then clustered by their tissue expression ratios in platypus using the repeated bisection option in the Cluto clustering package (Zhao and Karypis 2005). The distance between the confidence intervals of the expression ratios for each platypus/human, platypus/mouse, or platypus/chicken pair of miRNAs was computed after Wilson correction with Z = 2 (Brown et al. 2001) and was displayed as a difference score on the heatmap. The following human and mouse libraries from the miRNA expression atlas (Landgraf et al. 2007) were used for comparison with platypus: brain, hsa_Frontal-cortex-adult (human), average mmu_Brain-WT, and mmu_Cortex (mouse); heart, hsa_Heart (human) and mmu_Heart (mouse); liver, hsa_Liver (human) and mmu_Liver (mouse); testis, hsa_Testis (human) and mmu_Testis (mouse); lung, mmu_Lung (mouse); kidney, mmu_Kidney (mouse). For platypus/echidna expression profiles, the same procedure was used, except read counts for candidate mature miRNAs in platypus for brain, kidney, lung, heart, liver, and testis were compared with those for identical miRNAs found in echidna. Missing orthologs may in some cases reflect a failure to predict the miRNA rather than absence.
Multiple sequence alignment
Fifty-three miR-1421 related precursor sequences were aligned using ClustalW. The most highly sequenced small RNA from the most abundantly cloned arm of the hairpin was designated as the mature miRNA, and the small RNA with the greatest number of reads from the other arm was designated as the miRNA star. The consensus sequence indicates the most frequently occurring nucleotide for each position.
Platypus cell culture
Mitotic metaphase chromosomes were prepared from established platypus and echidna fibroblast cell lines. Primary cultures were set up from toe web from animals captured at the Upper Barnard River, New South Wales, Australia, during breeding season (AEC permit no. S-49-2006 to F.G.) following standard procedures. The cells where maintained in AmnioMAX C100 medium (GIBCO).
Physical mapping of BAC clones
A total of 100 ng of DNA from BAC clones was directly labeled with spectrum orange or spectrum green (Vysis, Abbot Molecular) using random primers and Klenow polymerase. Physical mapping by fluorescence in situ hybridization to male platypus metaphase chromosomes was performed under standard conditions. Briefly, the slides were treated with 100 μg/mL RNase A/2× SSC at 37°C for 30 min and with 0.01% pepsin in 10 mM HCl at 37°C for 10 min. After refixing for 10 min in 1× PBS, 50 mM MgCl2, 1% formaldehyde, the preparations were dehydrated in an ethanol series. Slides were denatured for 2.5 min at 75°C in 70% formamide, 2× SSC at pH 7.0 and again dehydrated. For hybridization of one-half slide, 10 μL of probe DNA was coprecipitated with 10–20 μg of boiled genomic platypus DNA as competitor and 50 μg salmon sperm DNA as carrier and dissolved in 50% formamide, 10% dextran sulfate, 2× SSC. The hybridization mixture was denatured for 10 min at 80°C. Preannealing of repetitive DNA sequences was carried out for 30 min at 37°C. The slides were hybridized overnight in a moist chamber at 37°C and thereafter washed three times for 5 min in 50% formamide, 2× SSC at 42°C and once for 5 min in 0.1× SSC. Chromosomes and cell nuclei were counterstained with 1 μg/mL DAPI in 2× SSC for 1 min and mounted (Vectashield). Images were taken with a Zeiss AxioImager Z.1 epifluorescence microscope equipped with a CCD camera and Zeiss Axiovision software.
Acknowledgments
The v5.0.1 (ornAna1) platypus genome sequence and assembly was produced by the Genome Sequencing Center at Washington University, St. Louis. We thank Michelle Rooks, Michael Regulski, Laura Cardone, and Danea Rebolini for assistance with miRNA cloning and sequencing; Rachel O’Neill and Dawn Carone (University of Connecticut), Michael Darre (UCONN Poultry Farm), and Russell Jones (Newcastle University) for help with the tissue collection; John Wallis (Washington University) and Natasha McInnes (University of Adelaide) for assistance with the BAC end database and BAC identification; and Sam Griffiths-Jones (miRBase) for help with miRNA predictions. Facilities were provided by Macquarie Generation and Glenrock Station, New South Wales. Approval to collect animals was granted by the New South Wales National Parks and Wildlife Services, New South Wales Fisheries, and the Animal Ethics Committee, University of Adelaide. E.P.M. is a Sir Keith Murdoch Fellow of the American Australian Association. F.G. is an Australian Research Council Research Fellow. A.S. was supported in part by the Schering AG/Ernst Schering Foundation and in part by the Human Frontier Science Program Organization (HFSPO). P.K. was supported in part by a National Science Foundation Graduate Research Fellowship. This work was funded in part by grants from the NIH (G.J.H.) and by a kind gift from Kathryn W. Davis (G.J.H.).
Footnotes
[Supplemental material is available online at www.genome.org. The microRNA sequence data from this study have been submitted to miRBase under accession nos. MI0006658–MI0007078 (http://microrna.sanger.ac.uk/sequences/).
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073056.107.
References
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Kuramochi-Miyagawa S., Nakano T., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Hannon G.J., Brennecke J., Hannon G.J., Brennecke J., Brennecke J. The Piwi/piRNA pathway: Adaptive defense for the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J., Girard A., Fejes-Toth K., Hannon G.J., Fejes-Toth K., Hannon G.J., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Ason B., Darnell D.K., Wittbrodt B., Berezikov E., Kloosterman W.P., Wittbrodt J., Antin P.B., Plasterk R.H., Darnell D.K., Wittbrodt B., Berezikov E., Kloosterman W.P., Wittbrodt J., Antin P.B., Plasterk R.H., Wittbrodt B., Berezikov E., Kloosterman W.P., Wittbrodt J., Antin P.B., Plasterk R.H., Berezikov E., Kloosterman W.P., Wittbrodt J., Antin P.B., Plasterk R.H., Kloosterman W.P., Wittbrodt J., Antin P.B., Plasterk R.H., Wittbrodt J., Antin P.B., Plasterk R.H., Antin P.B., Plasterk R.H., Plasterk R.H. Differences in vertebrate microRNA expression. Proc. Natl. Acad. Sci. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O.R., Cardillo M., Jones K.E., MacPhee R.D., Beck R.M., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Cardillo M., Jones K.E., MacPhee R.D., Beck R.M., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Jones K.E., MacPhee R.D., Beck R.M., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., MacPhee R.D., Beck R.M., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Beck R.M., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Price S.A., Vos R.A., Gittleman J.L., Purvis A., Vos R.A., Gittleman J.L., Purvis A., Gittleman J.L., Purvis A., Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Kellis M., Sachidanandam R., Hannon G.J., Sachidanandam R., Hannon G.J., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brown L.D., Cai T.T., DasGupta A., Cai T.T., DasGupta A., DasGupta A. Interval estimation for a binomial proportion. Stat. Sci. 2001;16:101–133. [Google Scholar]
- Carmell M.A., Girard A., de van Kant H.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J., Girard A., de van Kant H.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J., de van Kant H.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J., Bestor T.H., de Rooij D.G., Hannon G.J., de Rooij D.G., Hannon G.J., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chen P.Y., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Sheridan R., John B., Marks D.S., Gaidatzis D., John B., Marks D.S., Gaidatzis D., Marks D.S., Gaidatzis D., Gaidatzis D., et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes & Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell D.K., Kaur S., Stanislaw S., Konieczka J.H., Yatskievych T.A., Antin P.B., Kaur S., Stanislaw S., Konieczka J.H., Yatskievych T.A., Antin P.B., Stanislaw S., Konieczka J.H., Yatskievych T.A., Antin P.B., Konieczka J.H., Yatskievych T.A., Antin P.B., Yatskievych T.A., Antin P.B., Antin P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Davis E., Caiment F., Tordoir X., Cavaille J., Ferguson-Smith A., Cockett N., Georges M., Charlier C., Caiment F., Tordoir X., Cavaille J., Ferguson-Smith A., Cockett N., Georges M., Charlier C., Tordoir X., Cavaille J., Ferguson-Smith A., Cockett N., Georges M., Charlier C., Cavaille J., Ferguson-Smith A., Cockett N., Georges M., Charlier C., Ferguson-Smith A., Cockett N., Georges M., Charlier C., Cockett N., Georges M., Charlier C., Georges M., Charlier C., Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Delbridge M.L., Graves J.A., Graves J.A. Origin and evolution of spermatogenesis genes on the human sex chromosomes. Soc. Reprod. Fertil. Suppl. 2007;65:1–17. [PubMed] [Google Scholar]
- Devor E.J., Samollow P.B., Samollow P.B. In vitro and in silico annotation of conserved and nonconserved microRNAs in the genome of the marsupial Monodelphis domestica. J. Hered. 2007;99:66–72. doi: 10.1093/jhered/esm085. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 1997;46:101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A., Sachidanandam R., Hannon G.J., Carmell M.A., Hannon G.J., Carmell M.A., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H., Beyret E., Wang Z., Lin H., Wang Z., Lin H., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzner F., Rens W., Tsend-Ayush E., El-Mogharbel N., O'Brien P.C., Jones R.C., Ferguson-Smith M.A., Marshall Graves J.A., Rens W., Tsend-Ayush E., El-Mogharbel N., O'Brien P.C., Jones R.C., Ferguson-Smith M.A., Marshall Graves J.A., Tsend-Ayush E., El-Mogharbel N., O'Brien P.C., Jones R.C., Ferguson-Smith M.A., Marshall Graves J.A., El-Mogharbel N., O'Brien P.C., Jones R.C., Ferguson-Smith M.A., Marshall Graves J.A., O'Brien P.C., Jones R.C., Ferguson-Smith M.A., Marshall Graves J.A., Jones R.C., Ferguson-Smith M.A., Marshall Graves J.A., Ferguson-Smith M.A., Marshall Graves J.A., Marshall Graves J.A. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004;432:913–917. doi: 10.1038/nature03021. [DOI] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Handel M.A. The XY body: A specialized meiotic chromatin domain. Exp. Cell Res. 2004;296:57–63. doi: 10.1016/j.yexcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A., Murray M.F., Sharp P.A., Sharp P.A. Embryonic stem cell-specific microRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Dennis L., Jaenisch R., Sharp P.A., Dennis L., Jaenisch R., Sharp P.A., Jaenisch R., Sharp P.A., Sharp P.A. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Filippov D.V., Blaser H., Raz E., Moens C.B., Blaser H., Raz E., Moens C.B., Raz E., Moens C.B., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Steiner F.A., Berezikov E., de Bruijn E., de van Belt J., Verheul M., Cuppen E., Plasterk R.H., Steiner F.A., Berezikov E., de Bruijn E., de van Belt J., Verheul M., Cuppen E., Plasterk R.H., Berezikov E., de Bruijn E., de van Belt J., Verheul M., Cuppen E., Plasterk R.H., de Bruijn E., de van Belt J., Verheul M., Cuppen E., Plasterk R.H., de van Belt J., Verheul M., Cuppen E., Plasterk R.H., Verheul M., Cuppen E., Plasterk R.H., Cuppen E., Plasterk R.H., Plasterk R.H. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006a;34:2558–2569. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., de Bruijn E., Kauppinen S., Plasterk R.H., Kauppinen S., Plasterk R.H., Plasterk R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006b;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T., Kimura T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T., Fujita Y., Sato M., Matsuda Y., Nakano T., Sato M., Matsuda Y., Nakano T., Matsuda Y., Nakano T., Nakano T. Two mouse piwi-related genes: miwi and mili. Mech. Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Ikawa M., Iwai N., Okabe M., Deng W., Iwai N., Okabe M., Deng W., Okabe M., Deng W., Deng W., et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rice A., Kamphorst A.O., Landthaler M., Kamphorst A.O., Landthaler M., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Nakano T., Bartel D.P., Kingston R.E., Bartel D.P., Kingston R.E., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Hannon G.J., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T., Patkaniowska A., Dorsett Y., Teng G., Tuschl T., Dorsett Y., Teng G., Tuschl T., Teng G., Tuschl T., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Namekawa S.H., VandeBerg J.L., McCarrey J.R., Lee J.T., VandeBerg J.L., McCarrey J.R., Lee J.T., McCarrey J.R., Lee J.T., Lee J.T. Sex chromosome silencing in the marsupial male germ line. Proc. Natl. Acad. Sci. 2007;104:9730–9735. doi: 10.1073/pnas.0700323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D., Mecklenbrauker I., Das P.P., Santana A., Koenig U., Enright A.J., Miska E.A., Tarakhovsky A., Mecklenbrauker I., Das P.P., Santana A., Koenig U., Enright A.J., Miska E.A., Tarakhovsky A., Das P.P., Santana A., Koenig U., Enright A.J., Miska E.A., Tarakhovsky A., Santana A., Koenig U., Enright A.J., Miska E.A., Tarakhovsky A., Koenig U., Enright A.J., Miska E.A., Tarakhovsky A., Enright A.J., Miska E.A., Tarakhovsky A., Miska E.A., Tarakhovsky A., Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes & Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R.H. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Rens W., Grutzner F., O'Brien P.C., Fairclough H., Graves J.A., Ferguson-Smith M.A., Grutzner F., O'Brien P.C., Fairclough H., Graves J.A., Ferguson-Smith M.A., O'Brien P.C., Fairclough H., Graves J.A., Ferguson-Smith M.A., Fairclough H., Graves J.A., Ferguson-Smith M.A., Graves J.A., Ferguson-Smith M.A., Ferguson-Smith M.A. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc. Natl. Acad. Sci. 2004;101:16257–16261. doi: 10.1073/pnas.0405702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S., Park C., Sanders K.M., McCarrey J.R., Yan W., Park C., Sanders K.M., McCarrey J.R., Yan W., Sanders K.M., McCarrey J.R., Yan W., McCarrey J.R., Yan W., Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T., Rich T.H., Vickers-Rich P., Springer M., Woodburne M.O., Rich T.H., Vickers-Rich P., Springer M., Woodburne M.O., Vickers-Rich P., Springer M., Woodburne M.O., Springer M., Woodburne M.O., Woodburne M.O. The oldest platypus and its bearing on divergence timing of the platypus and echidna clades. Proc. Natl. Acad. Sci. 2008;105:1238–1242. doi: 10.1073/pnas.0706385105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Berezikov E., Yamashita Y., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Berezikov E., Yamashita Y., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Yamashita Y., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Lagos-Quintana M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Kloosterman W.P., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Enomoto M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Hatanaka H., Fujiwara S., Watanabe H., Soda M., Fujiwara S., Watanabe H., Soda M., Watanabe H., Soda M., Soda M., et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren W.C., Hillier L.W., Marshall Graves J.A., Birney E., Ponting C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Hillier L.W., Marshall Graves J.A., Birney E., Ponting C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Marshall Graves J.A., Birney E., Ponting C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Birney E., Ponting C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Ponting C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Belov K., Miller W., Clarke L., Chinwalla A.T., Miller W., Clarke L., Chinwalla A.T., Clarke L., Chinwalla A.T., Chinwalla A.T., et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2007;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Okuno T., Sasaki H., Minami N., Imai H., Sasaki H., Minami N., Imai H., Minami N., Imai H., Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Horvitz H.R., Kauppinen S., Plasterk R.H., Kauppinen S., Plasterk R.H., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wuchty S., Fontana W., Hofacker I.L., Schuster P., Fontana W., Hofacker I.L., Schuster P., Hofacker I.L., Schuster P., Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Wyckoff G.J., Wang W., Wu C.I., Wang W., Wu C.I., Wu C.I. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang X., Du Z., Li N., Wang X., Du Z., Li N., Du Z., Li N., Li N. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 2006;580:3610–3616. doi: 10.1016/j.febslet.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P., Shih I.H., Bartel D.P., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yu Z., Raabe T., Hecht N.B., Raabe T., Hecht N.B., Hecht N.B. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol. Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- Zhang R., Peng Y., Wang W., Su B., Peng Y., Wang W., Su B., Wang W., Su B., Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Karypis G., Karypis G. Data clustering in life sciences. Mol. Biotechnol. 2005;31:55–80. doi: 10.1385/MB:31:1:055. [DOI] [PubMed] [Google Scholar]