Abstract
DNA double strand breaks (DSBs) can be repaired by non-homologous end joining (NHEJ) or homology-directed repair (HR). HR requires nucleolytic degradation of 5′ DNA ends to generate tracts of single-stranded DNA (ssDNA), which are also important for the activation of DNA damage checkpoints. Here we describe a quantitative analysis of DSB processing in the budding yeast Saccharomyces cerevisiae. We show that resection of an HO endonuclease-induced DSB is less extensive than previously estimated and provide evidence for significant instability of the 3′ ssDNA tails. We show that both DSB resection and checkpoint activation are dose-dependent, especially during the G1 phase of the cell cycle. During G1, processing near the break is inhibited by competition with NHEJ, but extensive resection is regulated by an NHEJ-independent mechanism. DSB processing and checkpoint activation are more efficient in G2/M than in G1 phase, but are most efficient at breaks encountered by DNA replication forks during S phase. Our findings identify unexpected complexity of DSB processing and its regulation, and provide a framework for further mechanistic insights.
Keywords: cell cycle, checkpoints, DNA damage, DNA replication
Introduction
Double strand breaks (DSBs) are among the most dangerous of chromosomal lesions, and can lead to cell death and genomic rearrangements (Hoeijmakers, 2001). Two major pathways, non-homologous end joining (NHEJ) and homology-directed repair (HR), compete for the repair of DSBs (Paques and Haber, 1999; Daley et al, 2005). During the first step of HR, breaks undergo nucleolytic degradation of their 5′-ending strands, a process known as resection. This generates 3′-ended single-stranded tails, which are required for the downstream events in HR (Paques and Haber, 1999). Resection also prevents NHEJ, thus acting as a switch between repair pathways (Frank-Vaillant and Marcand, 2002; Daley and Wilson, 2005). The choice between NHEJ and HR is regulated by cyclin-dependent kinases (CDKs), and thus influenced by the cell cycle stage (Ira et al, 2004). Cells are proficient for NHEJ in G1 when CDK activity is low, but not in G2/M, when CDK activity is high and HR is predominant (Aylon et al, 2004; Ira et al, 2004). The molecular mechanism underlying these CDK-dependent effects is important but still obscure (Aylon et al, 2004; Ira et al, 2004; Lisby et al, 2004).
DSBs induced at a single HO endonuclease cleavage site (HOcs) can activate the DNA damage checkpoint, resulting in cell cycle arrest and a number of other processes important for cell survival (Rouse and Jackson, 2002; Longhese et al, 2006). Checkpoint activation by a single HOcs is generally quite slow and correlates with the generation of single-stranded DNA (ssDNA) by resection. Although the mechanisms of signalling initiation are still poorly understood, it is widely believed that the ssDNA binding protein RPA has a crucial role in this process. (Garvik et al, 1995; Lee et al, 1998; Zou and Elledge, 2003; Lisby et al, 2004; O'Connell and Cimprich, 2005).
Several approaches have been used to analyse the generation of ssDNA from DSBs (White and Haber, 1990; Fishman-Lobell et al, 1992). The first of these approaches measures resection indirectly in a genetic assay. In this assay (Fishman-Lobell et al, 1992), a DSB is introduced with the HO endonuclease between two direct repeats. Repair can occur by deletion of the intervening sequences in a process known as single strand annealing (SSA) (Fishman-Lobell et al, 1992; Aboussekhra et al, 1996; Vaze et al, 2002). It is believed that generation of ssDNA at the repeats is a critical intermediate in the repair reaction; however, it is unknown whether this ssDNA is generated entirely by resection or whether other mechanisms, for example DNA unwinding by a DNA helicase, contribute. In addition, whether or not there is some processing of the 3′ overhang before SSA is unknown. The second set of approaches involves the direct detection of ssDNA in purified DNA. For example, one assay (White and Haber, 1990) is based on the inability of many restriction enzymes to cleave ssDNA. As resection proceeds beyond restriction sites, additional, slower migrating bands appear in DNA blot hybridisation analysis of DNA extracted following DSB formation and resection. In a second assay, sequences around the HOcs are used to probe for ssDNA using non-denaturing slot blots (Aylon et al, 2003, 2004). These assays are useful because they directly visualise resection. However, because they involve a variety of factors that are difficult to measure (e.g. the electrophoretic migration of mixed species of DNA, differential transfer of different-sized DNA fragments and differential probe hybridisation), they can provide a general picture of resection but are less useful for quantifying resection rates. To help clarify these issues, quantitative assays for ssDNA generation are required. In one such assay, quantitative amplification of ssDNA, tagged oligonucleotide primers are first annealed to ssDNA at low temperature and extended (Booth et al, 2001; Zubko et al, 2006). The tagged product is then isolated and subjected to quantitative PCR. We have developed a simpler but highly accurate method to quantify ssDNA generated at a DSB introduced by the site-specific HO endonuclease and have applied this assay to examine factors influencing resection and checkpoint activation.
Results
A quantitative assay for DSB resection
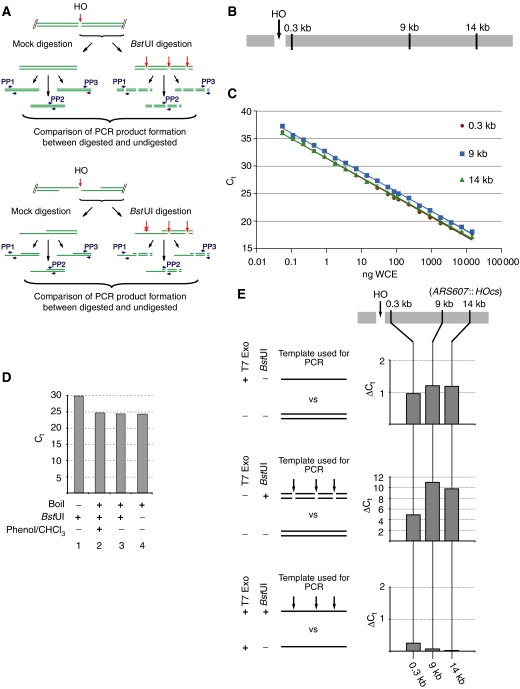
We developed a quantitative real-time PCR (qPCR) assay to analyse DNA turnover at a site-specific DSB generated by HO endonuclease (Figure 1A). A strain was constructed in which an HOcs was inserted into chromosome VI. Total amounts of DNA at three specific sites distal to the break (at 0.3, 9 and 14 kb; Figure 1B) were determined by qPCR and normalised to an amplicon on chromosome XIII. The fraction of DNA which is single stranded can then be determined by qPCR of DNA samples digested with a restriction endonuclease (BstUI) that cleaves within each amplicon but is unable to cleave ssDNA (Figure 1A). Threshold amplification cycles were determined as described in Figure 1 and Materials and methods. A difference of ‘1' in threshold cycle values (ΔCt=1) between two reactions corresponds to a two-fold difference in template levels. As shown in Figure 1C, a plot of Ct against log[DNA] was linear for each set of primers over a very wide range of template concentrations. Digestion of double-stranded DNA (dsDNA) with BstUI prior to amplification resulted in at least five-fold increase of Ct, corresponding to a reduction in amplifiable DNA amounts of at least 32-fold (Figure 1D, bars 1 and 4; Figure 1E, middle panel). In contrast, no such difference was observed when single-stranded (boiled) DNA was digested with BstUI and used for qPCR, confirming the resistance of ssDNA to BstUI digestion (Figure 1D, bars 3 and 4). No active restriction enzyme was carried into the PCRs, because extraction of samples with phenol/chloroform did not result in changes in Ct during the following qPCR (Figure 1D, bars 2 and 3).
Figure 1.
Overview of the assay for the quantification of ssDNA and control experiments. (A) Overview of the assay. PP1–3: primer pairs 1–3. See main text for description. (B) Schematic representation of the three sites analysed for ssDNA formation at ARS607::HOcs. (C) Extensive linear range of qPCR amplification. Serial dilutions of whole-cell DNA extract of strain YCZ64 were used in qPCR of the amplicons at the indicated positions. WCE, whole cell extract. (D) No BstUI activity is retained in the qPCRs and ssDNA is resistant to BstUI digestion. DNA extracted from strain YCZ64 was digested with BstUI and used for qPCR (column 1). Another sample of the same extract was boiled and two-thirds of this was digested with BstUI whereas one-third was mock digested before use in qPCR (column 4). The digested sample was split in two and one half was extracted with phenol/chloroform before use in qPCR (columns 2 and 3). qPCR was performed for all four samples using oligonucleotides OCZ125/OCZ126/OCZ140, amplifying the region 0.3 kb from the HOcs. (E) Analysis of in vitro resection using T7 exonuclease. DNA was extracted from strain YCZ64 after 1 h of HO induction. Top panel: Both strands are amplified with similar efficiencies. DNA was either digested or mock-digested with T7 exonuclease and used as template in qPCR. The graphs show the difference in Ct values between the two reactions. Middle panel: BstUI digestion of dsDNA interferes with subsequent PCR amplification. DNA was mock-digested with T7 exonuclease and subsequently digested or mock-digested with BstUI. Graphs represent comparisons of Ct values. Bottom panel: ssDNA is resistant to BstUI digestion. DNA was digested with T7 exonuclease and subsequently either digested or mock-digested with BstUI. DNA was then used in qPCRs. The graphs show comparisons of the Ct values obtained in the two reactions.
To mimic 5′–3′ resection, HO-cleaved genomic DNA was treated with T7 exonuclease (a 5′–3′ exonuclease) in vitro. As shown in Figure 1E (top), treatment with T7 exonuclease resulted in a ΔCt of ‘1' at all three positions relative to the break consistent with the removal of just one of the two strands of DNA and indicating that at all three positions, both DNA strands are amplified equally well. Figure 1E (middle) shows that BstUI digestion of HO cut DNA results in a 5- to 10-fold increase in ΔCt; however, BstUI digestion of T7 exo-treated DNA (bottom) had no effect on ΔCt. Taken together, these results indicate that this assay can efficiently discriminate between ssDNA and dsDNA and can accurately quantify both.
Characterisation of DSB processing in G1 and G2/M phases
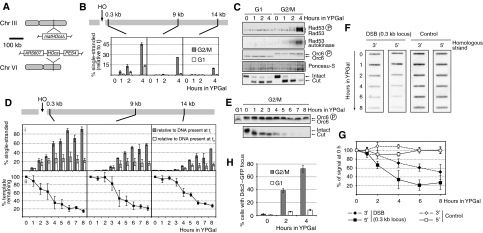
We measured ssDNA formation in strains containing an HOcs on chromosome VI but lacking the HO sites normally present at the MAT locus. This strain thus contains just one accessible HOcs. Cells were arrested in either G1 with α factor or in G2/M with nocodazole, and HO was expressed from a galactose-inducible promoter. To assess the quality of cell cycle arrests, immunoblotting was performed with antibodies against Orc6 (Figure 2C), a target of Clb-CDK that migrates with reduced gel mobility when phosphorylated (Liang and Stillman, 1997). Southern blot analysis was used to confirm break formation (Figure 2C). Using qPCR, we determined the amount of ssDNA relative to total DNA for each position present at each time point (Figure 2B). It had previously been estimated that DSB resection occurs at a rate of ∼4 kb/h (Fishman-Lobell et al, 1992; Vaze et al, 2002). Consistent with this, we found evidence for extensive resection using Southern blot analysis after restriction enzyme digestion (Supplementary Figure 1); however, quantitative analysis showed that fewer than 30% of G2/M-arrested cells (grey bars) had ssDNA at the 9 kb locus and fewer than 15% had ssDNA at the 14 kb locus after 4 h. Indeed, even at the break-proximal 0.3 kb locus in G2/M, only ∼50% of the DNA was found to be single stranded after 4 h (Figure 2B). Confirming published results (Aylon et al, 2004; Ira et al, 2004), we found considerably less DSB resection in G1-arrested cells than in G2/M-arrested cells even at the 0.3 kb locus, suggesting that initiation of break resection is less efficient in G1-arrested cells (Figure 2B).
Figure 2.
Analysis of DSB processing and checkpoint activation. Cells of strains YCZ101 (1cs, ARS607::HOcs matHOcsΔ, panels B and C), YCZ70 (1cs, ARS607::HOcs matHOcsΔ DDC2-GFP, panel H) and YHHD180 (2cs, ARS607::HOcs, panels D–G) were used. (A) Overview of the strain used. (B) Analysis of ssDNA formation. (C) Immunoblot, Rad53 autokinase assay and ARS607::HOcs DSB assay by Southern blot analysis. Note that in the Southern blot panel, the disappearance of the band corresponding to the cut locus is due to DSB processing. (D, E) Analysis of DSB processing in a longer G2/M experiment: (D) quantification of DSB processing; (E) immunoblot and DSB assay of the experiment shown in panel D. (F, G) Denaturing slot blot analysis confirming the instability of both strands at a DSB: (F) results from one representative experiment; (G) quantifications of the results. (H) Analysis of Ddc2–GFP focus formation.
To quantify resection in more detail, we carried out a longer time course in G2/M-arrested cells (Figure 2D and E). We used two methods to calculate the relative amounts of ssDNA. First, we calculated ssDNA as a percentage of the total amount of DNA at each locus present before HO induction (‘relative to t0', white bars in Figure 2D). Second, we calculated it as a percentage of the total amount of DNA at each locus at that time point (‘relative to ti' grey bars in Figure 2D). Comparison of these two methods revealed a novel feature of resection (Figure 2D). At 0.3 kb, the large majority (>80%) of DNA molecules remaining at 6–8 h were single-stranded (grey bars ‘relative to ti'). However, when compared with total DNA before HO induction (white bars ‘relative to t0'), the amount of ssDNA peaked at approximately 30% between 4 and 6 h and began to drop off after that. Similar results were seen at the more distal loci although to a lesser extent and with delayed kinetics. This suggested that significant loss of DNA was occurring at later time points. This was confirmed by examining changes in total template levels with time by qPCR (Figure 2Dii). This showed that at all three positions, template levels were reduced to well below 50% at later time points (the maximum reduction expected if only 5′–3′ resection occurred), approaching 20% at even the most distal locus. Therefore, in addition to degradation of the 5′ strand, substantial loss of the 3′ strand must also occur. To confirm this, we used strand-specific probes in a slot blot hybridisation assay. Figure 2F and G shows that both the 5′ and 3′ strands disappear with time specifically after HO cleavage. The 5′ strand disappears more quickly than the 3′ strand. Taken together, these experiments show that both strands are resected after HO cleavage, although the 5′ strand is processed faster than the 3′ strand.
DSB resection and checkpoint activation
We monitored activation of the DNA damage checkpoint in the experiment described in Figure 2B. We were interested in the correlation between DSB resection and checkpoint activation. Estimates from previous experiments (Pellicioli et al, 2001) indicated that resection tracts corresponding to tens of kilobases correlated with checkpoint activation. We used the activation of Rad53, an essential DNA damage checkpoint kinase, as a marker for checkpoint activation. We analysed both the appearance of a phosphorylation-dependent gel mobility shift of Rad53 and the kinase activity using an in-blot assay (Pellicioli et al, 1999). Consistent with previous results (Pellicioli et al, 2001), we detected Rad53 activation in G2/M-arrested cells but not in G1-arrested cells after cleavage of one HOcs (Figure 2C). We also examined localisation of the checkpoint protein Ddc2 tagged with GFP to DNA repair foci to gain single-cell-based information on checkpoint activation. Ddc2 is required for virtually all checkpoint responses (Paciotti et al, 2000; Rouse and Jackson, 2000) and had previously been shown to localise to DNA damage-specific foci (Melo et al, 2001). Figure 2H shows that, after HO induction, Ddc2–GFP foci were largely absent from G1-arrested cells, but appeared relatively early in G2/M-arrested cells. By 2 h, approximately 40% of G2/M cells had detectable Ddc2 foci. At this time point, there was virtually no ssDNA at either of the distal loci and only approximately 15% ssDNA at the very proximal 0.3 kb locus (Figure 2B). Therefore, detectable Ddc2 foci form in at least some cells before resection proceeds past 0.3 kb. By 4 h, ∼80% of cells showed at least one focus, indicating that the DNA damage checkpoint was active in the majority of cells, even though long resection tracts were largely absent.
Dose dependence of checkpoint activation and DSB resection
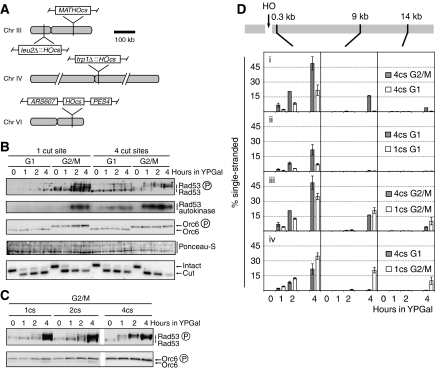
We next examined the effect of DSB dose on DSB response. We were interested in this because one HOcs will generate one DSB in G1 phase, but two DSBs after chromosome replication in G2/M phase. Moreover, previous work has shown that a single HO-induced DSB cannot activate the DNA damage checkpoint in G1 phase, but doses of gamma irradiation sufficient to generate multiple DSBs can activate the DNA damage checkpoint in G1 phase (Lisby et al, 2004). To examine this, we used a strain that contained the endogenous HOcs at MAT in addition to the HOcs on chromosome VI, thereby increasing the DSB dose to 2 in G1 phase. The silent HML and HMR loci were deleted in this strain. No Rad53 activation was detected upon break formation in this strain in G1 (Supplementary Figure 2A), indicating that dosage alone cannot explain the cell cycle differences. The addition of a third HOcs was also insufficient to allow Rad53 activation in G1 (Supplementary Figure 2B). However, when four HOcs were introduced (Figure 3A), Rad53 phosphorylation and autokinase activity were observed in G1-arrested cells (Figure 3B), indicating that Rad53 activation in response to HO-induced DSBs is possible in G1, albeit requiring at least four DSBs.
Figure 3.
Checkpoint activation and DSB resection are dose-dependent processes. Cells of strains YCZ173 (1cs, ARS607::HOcs matHOcsΔ), YCZ64 (2cs, ARS607::HOcs MATHOcs), YCZ172 (4cs, ARS607::HOcs trp1Δ::HOcs leu2Δ::HOcs MATHOcs) were used. 1cs, one HO cut site; 2cs, two HO cut sites; 4cs, four HO cut sites. (A) Overview of the 4HOcs strain. (B, C) Immunoblot, Rad53 autokinase and DSB assays. (D) Analysis of DSB processing.
Even in G2/M-arrested cells, Rad53 activation showed dose dependence. When four HOcs were present, Rad53 activation was faster and resulted in the entire pool of Rad53 shifting to the slow-migrating form, suggesting that almost all Rad53 was activated (Figure 3C). However, different thresholds exist for checkpoint activation in G1 and G2/M as two DSBs (one HOcs) were sufficient for Rad53 activation in G2/M but not in G1 (two HOcs; Supplementary Figure 2A).
We next investigated whether DSB dose affected DSB resection (Figure 3D). Figure 3D shows that ssDNA formation from the chromosome VI HOcs in both G1- and G2/M-arrested cells was greater in the 4HOcs strain than in the 1HOcs strain (Figure 3Dii and iii). However, this effect was restricted to the region analysed closest to the break and in neither case could we detect significant resection at the 9 kb locus in G1 phase. Furthermore, although resection was enhanced in G1 when more breaks were introduced, it was still not as efficient as resection in G2/M. Therefore, although resection is stimulated at a given DSB when other breaks are present within the genome, this effect is confined to regions close to the ends. These experiments provide further evidence that Rad53 can be activated without generation of extensive ssDNA tracts.
DSB resection is affected by both NHEJ-dependent and NHEJ-independent processes in G1
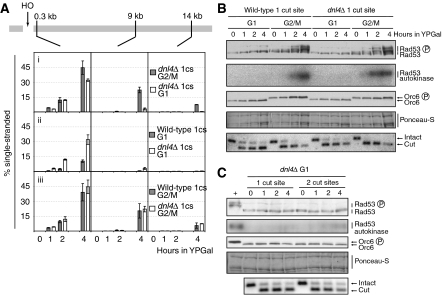
We examined the possibility that resection in G1 phase is prevented by competition with NHEJ, which is more efficient in G1 than G2/M phase. We analysed DSB processing in a strain lacking DNL4, the ligase required for NHEJ (Schar et al, 1997; Teo and Jackson, 1997; Wilson et al, 1997). Figure 4A shows that resection was indeed increased in the dnl4Δ strain when compared with the wild-type control. There was little or no effect in G2/M-arrested cells, but the effect in G1-arrested cells was more pronounced. This effect in G1, however, was restricted to the region closest to the DSB, where resection was similar to G2/M (Figure 4Ai and ii). This suggests that, in the absence of NHEJ, more resection events are initiated in G1. Because this increase in resection was not propagated to regions further away from the break, either the rate or the processivity of resection in G1-arrested cells is reduced relative to G2/M even in the absence of NHEJ. These results suggest that resection of a DSB in G1 phase is reduced because it is inhibited by NHEJ and also because the rate and/or processivity of the 5′–3′ degradation machinery is reduced.
Figure 4.
DSB resection is regulated by both NHEJ-dependent and NHEJ-independent mechanisms in G1. Cells of strains YCZ101 (1cs, ARS607::HOcs matHOcsΔ), YCZ136 (1cs, ARS607::HOcs matHOcsΔ dnl4Δ) and YCZ202 (2cs, ARS607::HOcs MATHOcs dnl4Δ) were used. 1cs, one HO cut site. (A) Analysis of DSB processing. (B, C) Immunoblot, Rad53 autokinase and DSB assays.
Loss of NHEJ results in an increased checkpoint response to DSBs in an asynchronous population (Lee et al, 1998). We therefore also investigated the effect of deletion of DNL4 on checkpoint activation in our synchronised cultures. Interestingly, although deletion of DNL4 increased resection close to the break, it did not make G1-arrested cells proficient for checkpoint activation (Figure 4B). We also analysed G1 cells containing two HOcs to increase the DSB dose to the same level as that of the G2/M cells in the previous experiment. As before, the absence of Dnl4 did not make cells proficient for checkpoint activation in G1 (Figure 4C). These findings therefore show that the inefficient response to DSBs in G1 phase does not arise from inhibitory effects of NHEJ on resection and checkpoint activation, even when compensated for DSB dose.
DNA replication greatly enhances DNA damage checkpoint activation
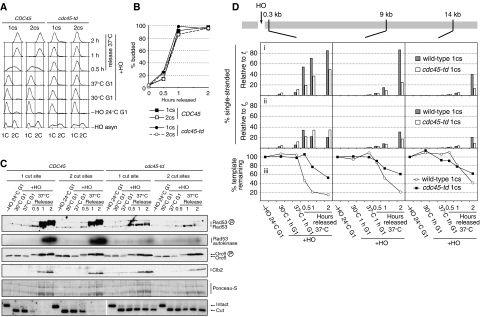
As DSBs are not very effective in activating the DNA damage checkpoint in G1 phase, cells with DSBs will occasionally enter S phase, and, consequently, replication forks will encounter these DSBs. Furthermore, gaps or nicks in only one of the two strands resulting from intermediates of repair of base and nucleotide damage will be transformed into DSBs when met by a replication fork. This is arguably the most physiologically relevant mechanism of DSB formation, as such repair events occur abundantly in the absence of exogenous DNA damage (Lindahl and Barnes, 2000; Lisby et al, 2001). Therefore, it is possible that DSBs trigger specialised signalling and/or processing events during S phase. To investigate this, we analysed cells that were released into S phase following DSB formation in G1. To distinguish the effects of DNA replication forks from other S-phase effects, we also examined DSBs in cells that were deficient in the initiation of DNA replication. Previous work has shown that the temporal order of other cell cycle events that are not coupled to replication, such as CDK activation and subsequent entry into a reductive mitosis occur with normal timing in the absence of DNA replication (Kelly et al, 1993; Piatti et al, 1995; Diffley et al, 2000; Tercero et al, 2000; Stern and Murray, 2001). For this purpose, a strain that carried the temperature-sensitive cdc45-td degron allele of CDC45, a gene required for both the initiation and elongation of DNA replication, was utilised (Tercero et al, 2000). Cells of the cdc45-td strain and CDC45 control strains, each containing either one or two HOcs, were arrested in G1 at the permissive temperature in the absence of HO expression (Figure 5A). HO was induced and Cdc45-td was inactivated by shifting the temperature to 37°C before cultures were released from the G1 arrest into nocodazole-containing medium to prevent mitosis. In cells containing even a single HOcs that were proficient for DNA replication (i.e. CDC45+), Rad53 was activated much faster and to higher levels than in cells in which replication had been prevented with the Cdc45 degron (Figure 5C). Within 2 h of release from the G1 block, essentially all of the Rad53 had shifted to the slower migrating form in the CDC45+ strain (Figure 5C), a situation never observed in G2/M cells containing a comparable number of DSBs (Figure 3C) or in the cdc45-td strains (Figure 5C). This effect was not a consequence of the higher incubation temperature of 37°C, as it was also observed during replication at 30°C (Figure 6A and Supplementary Figure 3E). Furthermore, increasing the temperature to 37°C in nocodazole-arrested cells did not result in increased Rad53 activation (Supplementary Figure 3B). All cultures were released from the G1 arrest with similar kinetics, as determined by the appearance of budded cells (Figure 5B), and flow cytometry confirmed replication in the CDC45 strains and its absence in the cdc45-td strains (Figure 5A). As judged by Orc6 phosphorylation, Clb-CDK activity appeared synchronously in all the strains (Figure 5C). Lastly, Clb2, the major mitotic cyclin in budding yeast (Nasmyth, 1996), appeared at similar times in all the strains (Figure 5C). We thus conclude that replication in the presence of a DSB induces a substantially stronger checkpoint response than that observed in either G1 or G2/M phase.
Figure 5.
DNA replication is required for efficient checkpoint activation and stimulates DSB processing. Strains YCZ158 (1cs, ARS607::HOcs matHOcsΔ), YCZ151 (2cs, ARS607::HOcs MATHOcs), YCZ166 (1cs, ARS607::HOcs matHOcsΔ cdc45-td) and YCZ171 (2cs, ARS607::HOcs MATHOcs cdc45-td) were used. 1cs, one HO cut site. Strains were released from G1 arrest into media containing nocodazole to prevent entry into a second cell cycle. (A) Flow cytometry. (B) Budding index. (C) Immunoblot, Rad53 autokinase and DSB assays. (D) Analysis of DSB processing.
Figure 6.
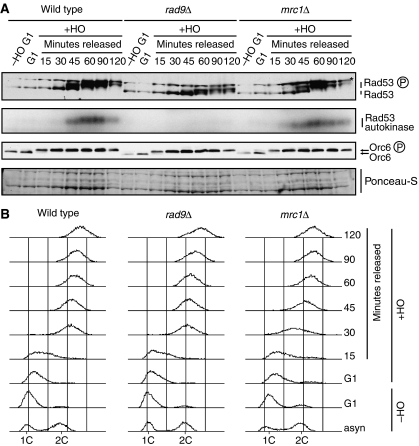
The increased checkpoint activation by DSBs during DNA replication is dependent on Rad9 but not on Mrc1. Cultures of strains YCZ158 (ARS607::HOcs matHOcsΔ), YCZ200 (ARS607::HOcs matHOcsΔ mrc1Δ) and YCZ201 (ARS607::HOcs matHOcsΔ mrc1Δ) were grown in YPRaff and arrested in G1. HO was induced by shifting the culture to YPGal for 1 h, and cells were subsequently released from the G1 arrest. (A) Immunoblot and Rad53 autokinase analysis. (B) Flow cytometry analysis of DNA content.
Two alternative adaptor proteins are required for mediating activation of Rad53 (Longhese et al, 2003, 2006). In response to general DNA damage, Rad9 is the primary mediator, whereas activation in response to replication stress is mediated by Mrc1 (Alcasabas et al, 2001). As Mrc1 is a component of the replication apparatus (Katou et al, 2003), it is possible that a fork encountering a DSB initiates an Mrc1-dependent signalling event. We therefore tested mrc1Δ and rad9Δ strains for their ability to support Rad53 activation during replication in the presence of a DSB (Figure 6). In this experiment, deletion of MRC1 had little or no effect on Rad53 autokinase activity or Rad53 phospho-shift (Figure 6A and B). By contrast, Rad53 autokinase activity was completely lost in the RAD9 deletion strain (Figure 6A and B). The residual Rad53 phospho-shift is a DNA damage-independent consequence of G2/M arrest and does not correspond to kinase activation (Tercero et al, 2003). Thus, the Mrc1-dependent pathway for sensing DNA replication stress makes little or no contribution to the enhanced DSB response during S phase, which appears to be entirely Rad9-dependent, similar to the situation in G2/M.
DSB processing during S phase
We next analysed whether replication into a DSB would affect its processing. Consistent with the inefficient resection of DSBs in G1, very little ssDNA was detected in both the cdc45-td and the CDC45+ cells before release from the G1 block (Figure 5Di). As soon as cells were released from the arrest, extensive ssDNA formation and DNA degradation were detected in the wild-type cells, but not the cdc45-td cells, showing that replication into a DSB induces rapid turnover of DNA ends (Figure 5Di and iii). Resection was also much faster in replicating cells than in cells arrested in G2/M. For example, in cells released into S phase in the presence of a DSB, most of the DNA that was present at the site 0.3 kb from the DSB was single-stranded after only 2 h (Figure 5Di). Similar levels of ssDNA at this locus were reached only after 6 h in G2/M-arrested cells (Figure 2D). Moreover, much more of the DNA was single-stranded at position 9 kb from the break (Figures 2B, D and 5Di). When we analysed the percentage of ssDNA relative to the starting material, again we observed that at times when most of the DNA present was single-stranded, these molecules corresponded to a smaller fraction of the starting DNA in the replicating strain (Figure 5Di and ii). As described above, this indicates degradation of both strands. In support of this, whole template levels were reduced to well below the 50% value expected if degradation of only one strand occurred (Figure 5Diii). This effect was far less prominent in the cdc45-td strain. Furthermore, Southern blot-based DSB assays also clearly show enhanced disappearance of the HO ‘cut' band following DNA replication (Figure 5C, bottom panel) consistent with rapid processing during S phase. As before, the elevated incubation temperature of 37°C was not responsible for any of these effects. We observed no difference in DSB processing when we compared 30 and 37°C cultures in either G2/M (Supplementary Figure 3C) or in a G1 arrest/release experiment (Supplementary Figure 3F). Virtually identical findings were obtained with the 2HOcs strain (see Supplementary Figure 4). Taken together, these findings show that after replication in the presence of a DSB, resection is much more extensive than in any other situation.
Discussion
We have developed a quantitative assay to measure the generation of single-stranded DNA from a site-specific DSB, which has allowed us to investigate factors affecting DSB processing and their relation to checkpoint activation. Our results indicate that the overall rate of break resection is somewhat slower and less synchronous than previously estimated. We have provided evidence indicating that, in addition to degradation of the 5′ strand, resection also involves a slower or delayed degradation of the 3′ strand. We have also identified and characterised four factors affecting the turnover of DSBs: NHEJ, cell cycle stage, DSB dose and the presence of DNA replication forks.
Previous work indicated that resection from an HOcs is highly efficient and proceeds at approximately 4 kb/h. These estimates were based primarily on a genetic assay in which an HOcs was placed between two direct repeats, and the time required for repair by deletion of the intervening sequences, interpreted as single-strand annealing, was measured (Fishman-Lobell et al, 1992; Vaze et al, 2002). Our quantitative, physical assay paints a subtly different picture of resection. Our results show that some breaks must be processed at approximately this rate. For example, the appearance of ssDNA at the 14 kb position after 4 h requires a resection rate of at least 3.5 kb/h (Figure 2B and D), very similar to the rate calculated from the genetic experiment. However, this accounts for only 10% of DSBs. Therefore, the majority of breaks seem to be processed considerably more slowly. This could be because resection actually proceeds at different rates in different subsets of breaks. Alternatively, and we feel more likely, processive resection of all breaks may proceed at a similar rate once initiated, but may initiate stochastically, at a slower rate.
At present, we do not know why our estimate is lower than the estimate from the genetic assay. It is possible that these differences reflect genuine strain differences. Alternatively, an attractive possibility is that some ssDNA at DSBs is not generated by exonucleases, but instead generated by DNA helicases, perhaps followed by occasional endonuclease cleavage. Because complementary DNA strands would re-anneal during DNA purification, such ssDNA would be ‘invisible' in any physical assay but ‘visible' in the genetic assay.
Our results also show that resection is not limited to the 5′ strand: the 3′ strand is also degraded, although this resection lags behind the 5′ strand resection. At present, we do not know if this is because 3′ strand resection is generally slower or because it initiates less frequently. The extensive tracts of ssDNA generated from unreparable DSBs are generally not required for repair and are probably of limited physiological significance. For example, during yeast mating type switching, the region of homology between MAT and its recombination donors is only ∼150 bp (Haber, 1998). Resection of the 3′ strand might aid recombination or SSA by ensuring there is a 3′ end close to the strand invasion event. In Escherichia coli, RecBCD processes DSBs by a mechanism involving DNA unwinding before DNA cleavage. Interestingly, RecBCD can cleave both the 3′ and the 5′ strands, which is regulated by encountering chi sequences in the DNA (Wigley, 2007). Perhaps a similar mechanism exists in budding yeast.
Several nucleases have been implicated in DSB processing. Most prominent among these are Mre11 and Sae2/Com1 (Paques and Haber, 1999). Although both Mre11 and Sae2 have nuclease activity, neither is the 5′–3′ exonuclease predicted to be required for 5′ strand resection (D'Amours and Jackson, 2002; Lengsfeld et al, 2007). Sae2 has been shown to have endonuclease activity especially effective on hairpin structures, whereas Mre11, a component of the MRN complex, has 3′–5′ exonuclease activity as well as endonuclease activity. However, it is unclear how important these activities are for DSB processing because SAE2 nuclease mutants have not yet been tested in vivo and MRE11 nuclease mutants do not show resection defects (Llorente and Symington, 2004). It will be of interest to determine whether MRE11 nuclease-deficient mutants are defective for the 3′ strand processing we have described here.
Our findings confirm and extend the observation that DSB processing is less efficient in G1 than in G2/M (Aylon et al, 2004; Ira et al, 2004). Previous work has shown clearly that CDKs are crucial for this difference. Our work has identified several other factors contributing to this. We found that inefficient resection during G1 phase is partially due to the inhibitory effect of NHEJ (Figure 4A). This probably reflects competition for the DSB between NHEJ and resecting factors. In the absence of NHEJ, break-proximal sequences are resected at rates similar to those seen in G2/M. This suggests that NHEJ may be the primary rate-limiting factor for the initiation of DSB processing during G1 phase. However, it is not the only reason for reduced DSB processing during G1. Resection of break-distal sequences occurs at greatly reduced rates in this cell cycle stage, even in the absence of NHEJ (Figure 4A). These results suggest that at least two features of DSB processing are regulated during the cell cycle: elevated NHEJ during G1 phase prevents the initiation of resection; and a second, NHEJ-independent mechanism prevents extensive resection during G1 phase. Further work is required to determine how these processes are regulated by CDKs.
DSB processing as well as checkpoint activation can be increased in G1 by the formation of additional HO-induced breaks (Figure 3B and D). As is the case with NHEJ-deficient cells, the increased processing during G1 phase caused by multiple DSBs is limited to break-proximal sequences. This may suggest that some component of NHEJ becomes limiting in the presence of multiple breaks. This dose-dependent processing may help to explain some discrepancies in the recent literature. In studies using a single HOcs, it was reported that G1 cells cannot process the DSB efficiently, and are unable to activate the DNA damage checkpoint (Pellicioli et al, 1999; Ira et al, 2004). In contrast, studies using ionising radiation (IR) to induce DSBs have shown that ssDNA formation and checkpoint activation can occur in G1 (Lisby et al, 2004; Barlow et al, 2008). The IR doses used in the latter study resulted in the formation of two DSBs per cell, on average. It is interesting that our results show that four HO-induced DSBs can stimulate break-proximal resection and can activate Rad53 during G1 phase. Consistent with this, recent work by Barlow et al (2008) has indicated that IR-induced damage and HO cleavage are processed differently during G1 phase. They have attributed this to differences between a clean break (HOcs) and breaks with ragged ends (IR). Our results suggest that simple break dosage may also contribute to these differences.
When a DSB is present during DNA replication, it is very efficiently processed and induces a robust checkpoint response (Figure 5C and D). Efficient processing and checkpoint activation are not properties of the S-phase milieu per se but rather require active DNA replication, suggesting that collision of replication forks with the DSB is required. This replication-dependent checkpoint activation, however, does not require Mrc1 and instead is completely dependent on the classic Rad9-dependent DSB signalling pathway (Figure 6). The precise role of replication forks remains to be determined, but may involve delivery of factors required for DSB processing, mediators of chromatin remodelling (Shibahara and Stillman, 1999), histone modification (Masumoto et al, 2005) or sister chromatid cohesion (Uhlmann and Nasmyth, 1998).
Using our quantitative assay in combination with more extensive mutational analysis should provide us with new insights into the interconnectedness between DNA processing and checkpoint activation. Furthermore, it will be of great interest to elucidate both the pathway mediating degradation of the 3′ strand and the mechanistic basis for the stimulation of DSB processing and checkpoint activation by DNA replication.
Materials and methods
Strains and media
All yeast strains used were isogenic to the w303 background and grown at 30°C unless otherwise indicated. Gene deletions and HOcs integrations were performed by PCR-mediated gene replacement. All strains except YHHD180 were bar1Δ to prevent adaptation to α factor. The HOcs at MAT was deleted in such a way as to leave MATa1 gene that borders it intact. HO was expressed from pJH1097 (a gift from J Haber) integrated into ADE3. The HOcs at ARS607 was introduced as follows: a PCR product of the 117 bp HOcs (Haber, 2002) was introduced into pUG6 (Guldener et al, 1996) and amplified with another set of primers for integration. We observed that recombination with the silent mating-type locus HML led to mating-type switching and insensitivity to α factor in strains containing the endogenous HOcs at MAT (data not shown). We also noted that HO occasionally cleaved HML and HMR, although at a very low frequency (data not shown). Where relevant, strains therefore also contained deletions of HML and HMR. Supplementary Table 1 shows a list of strains used with their relevant genotype.
Growth conditions and cell cycle blocks were as described previously (Diffley et al, 1994). For HO inductions, cells were pre-grown to 107 cells/ml in YPRaff (1% yeast extract, 2% bacto peptone, 2% raffinose), arrested with either α factor (1 μg/ml) or nocodazole (5 μg/ml) in fresh YPRaff and then shifted to fresh medium containing 2% galactose instead of raffinose in addition to α factor or nocodazole. HO cutting was quantified by Southern blot analysis.
Immunoblotting and Rad53 autokinase assay
Extracts were prepared as described previously (Tercero et al, 2003). Rad53 protein was detected with antibody JDI48 (Tercero et al, 2003). SB49 antibody was used for detection of Orc6 (Weinreich et al, 1999). Clb2 was detected using antibody sc-9071 (Santa Cruz). Rad53 in situ autokinase assay was performed as described previously (Pellicioli et al, 1999).
Flow cytometry and fluorescence microscopy
Flow cytometry was performed as described previously (Diffley et al, 1994). Deltavision microscopy with a × 60/1.4 NA Planapochromat lens on an Olympus IX71 inverted microscope was used to examine Ddc2–GFP. Images were captured and manipulated with SoftWorx software (Applied Precision).
DNA extraction and preparation for qPCR
DNA was extracted from 108 cells. Cells were resuspended in 500 μl extraction buffer (1% SDS, 100 mM NaCl, 50 mM Tris–HCl pH 8.0, 10 mM EDTA) and 5 μl β-mercaptoethanol (Sigma) and 500 U lyticase (Sigma) were added. Cells were lysed by 6 min incubation at 37°C with shaking. DNA was then prepared by standard phenol/chloroform extraction. RNase A (0.05 μg/μl; Sigma) was used to break down RNA (45 min at 37°C). A 7.5 μl volume of each sample was digested with 10 U of BstUI for 1 h. Digested DNA was serially diluted in TE, first 1:4 and then twice 1:2.
Quantitative real-time PCR
qPCR was performed with the ABI7000 Sequence Detection System and corresponding software (Applied Biosystems). We utilised the TaqManR fluorogenic probe system (Heid et al, 1996). TaqManR probes were synthesised at Applied Biosystems and, together with primer sequences and reaction concentrations, are listed in Supplementary Table 2. qPCR ROX master mixes were obtained from Abgene. The following programme was used for all reactions: 95°C 15 min → 45 × (95°C 15 s → 60°C 1 min). A 4 μl volume of each of the three DNA dilutions (see above) was used in a total reaction volume of 40 μl. We used these template dilutions, rather than doing triplicate reactions with identical DNA concentrations, to check for linearity of the PCRs. An average threshold cycle (De Sanctis et al, 2001) value was then determined for each sample. To calculate ssDNA as the percentage of the DNA present at each time point (‘relative to ti'), we used the formula
where ΔCt is the difference in average cycles between digested template and undigested template of a given time point and f is the fraction cut by HO.
A different formula was used to calculate the amounts of ssDNA as percentage of the DNA present at the start of the experiment (‘relative to t0'):
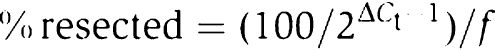 |
In this case, ΔCt describes the difference in average cycles between digested template at ti and undigested template at t0.
All Ct values were corrected for different DNA concentrations, as determined by qPCR of an amplicon on a different chromosome, not containing any BstUI sites (Supplementary Table 2).
For mimicking 5′–3′ resection, T7 exonuclease obtained from NEB was used according to the manufacturer's instructions.
Strand-specific slot blot analysis of DSB turnover
DNA was extracted as described for the qPCR assay. Equal amounts of the samples from each time point were digested with HindIII and denatured by boiling for 5 min followed by snap cooling on ice. The DNA was diluted in a large excess of 0.5 M NaOH and 10 mM EDTA and incubated at 42°C for 15 min. Equal amounts of each sample were transferred to Hybond XL membranes (GE Healthcare) using the Bio-Dot SF apparatus (Bio-Rad). The membranes were dried and DNA was UV crosslinked. Hybridisation was carried out at 61°C and as suggested in the Hybond XL handbook. The probes that were used were essentially longer versions of the probes used in qPCR for the 0.3 kb locus and the control locus and were labelled with 32P using polynucleotide kinase (New England Biolabs). They are summarised in Supplementary Table S3. Membranes were exposed to storage phosphor screens and read using a Typhoon 9400 scanner (GE Healthcare). Bands were quantified using ImageQuant TL (GE Healthcare).
Supplementary Material
Supplementary data 1
Acknowledgments
We are grateful to Jim Haber and members of the Diffley lab for helpful discussions. We are also grateful to Anne Early for critical reading of the manuscript and to Hiro Funabiki for discussions, help and cooperativity during the revision process. This work was supported by Cancer Research UK and EU Contract RTN (HPRN-CT-2002-00238).
References
- Aboussekhra A, Vialard JE, Morrison DE, de la Torre-Ruiz MA, Cernakova L, Fabre F, Lowndes NF (1996) A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J 15: 3912–3922 [PMC free article] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M (2003) Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol 23: 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Griffith E, Brady G, Lydall D (2001) Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res 29: 4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39: 431–451 [DOI] [PubMed] [Google Scholar]
- Daley JM, Wilson TE (2005) Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol 25: 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sanctis V, Bertozzi C, Costanzo G, Di Mauro E, Negri R (2001) Cell cycle arrest determines the intensity of the global transcriptional response of Saccharomyces cerevisiae to ionizing radiation. Radiat Res 156: 379–387 [DOI] [PubMed] [Google Scholar]
- Diffley JF, Bousset K, Labib K, Noton EA, Santocanale C, Tercero JA (2000) Coping with and recovering from hydroxyurea-induced replication fork arrest in budding yeast. Cold Spring Harb Symp Quant Biol 65: 333–342 [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303–316 [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J, Rudin N, Haber JE (1992) Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol 12: 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell 10: 1189–1199 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE (1998) Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet 32: 561–599 [DOI] [PubMed] [Google Scholar]
- Haber JE (2002) Uses and abuses of HO endonuclease. Methods Enzymol 350: 141–164 [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6: 986–994 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P (1993) The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74: 371–382 [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Stillman B (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev 11: 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE (2000) Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 65: 127–133 [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Clerici M, Lucchini G (2003) The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat Res 532: 41–58 [DOI] [PubMed] [Google Scholar]
- Longhese MP, Mantiero D, Clerici M (2006) The cellular response to chromosome breakage. Mol Microbiol 60: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K (1996) At the heart of the budding yeast cell cycle. Trends Genet 12: 405–412 [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Cimprich KA (2005) G2 damage checkpoints: what is the turn-on? J Cell Sci 118: 1–6 [DOI] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP (2000) The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev 14: 2046–2059 [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE (2001) Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell 7: 293–300 [DOI] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18: 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J 14: 3788–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2000) LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J 19: 5801–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2002) Interfaces between the detection, signaling, and repair of DNA damage. Science 297: 547–551 [DOI] [PubMed] [Google Scholar]
- Schar P, Herrmann G, Daly G, Lindahl T (1997) A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev 11: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Stillman B (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585 [DOI] [PubMed] [Google Scholar]
- Stern BM, Murray AW (2001) Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol 11: 1462–1467 [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP (1997) Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J 16: 4788–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JF (2000) DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J 19: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11: 1323–1336 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K (1998) Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385 [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B (1999) The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci USA 96: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CI, Haber JE (1990) Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J 9: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley DB (2007) RecBCD: the supercar of DNA repair. Cell 131: 651–653 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zubko MK, Maringele L, Foster SS, Lydall D (2006) Detecting repair intermediates in vivo: effects of DNA damage response genes on single-stranded DNA accumulation at uncapped telomeres in budding yeast. Methods Enzymol 409: 285–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data 1