Abstract
Background
Chromatin dynamics and stability are both required to control normal development of multicellular organisms. Chromatin assembly factor CAF-1 is a histone chaperone that facilitates chromatin formation and the maintenance of specific chromatin states. In plants and animals CAF-1 is essential for normal development, but it is poorly understood which developmental pathways require CAF-1 function.
Results
Mutations in all three CAF-1 subunits affect Arabidopsis trichome morphology and lack of CAF-1 function results in formation of trichomes with supernumerary branches. This phenotype can be partially alleviated by external sucrose. In contrast, other aspects of the CAF-1 mutant phenotype, such as defective meristem function and organ formation, are aggravated by external sucrose. Double mutant analyses revealed epistatic interactions between CAF-1 mutants and stichel, but non-epistatic interactions between CAF-1 mutants and glabra3 and kaktus. In addition, mutations in CAF-1 could partly suppress the strong overbranching and polyploidization phenotype of kaktus mutants.
Conclusion
CAF-1 is required for cell differentiation and regulates trichome development together with STICHEL in an endoreduplication-independent pathway. This function of CAF-1 can be partially substituted by application of exogenous sucrose. Finally, CAF-1 is also needed for the high degree of endoreduplication in kaktus mutants and thus for the realization of kaktus' extreme overbranching phenotype.
Background
Chromatin stability and dynamics have to be well balanced to guarantee normal development. While flexibility of the chromatin structure permits developmental transitions necessary during the life cycle of an organism, epigenetic as well as genetic information has to be reliably propagated within a certain developmental phase. Various protein complexes have been described to be involved in chromatin regulation [1-3]. One biochemically well characterized complex involved in chromatin replication is Chromatin Assembly Factor CAF-1, which deposits histones H3 and H4 in a replication-dependent manner onto DNA (for review see [4,5]. This complex was initially identified as a negative supercoiling-inducing factor in human cell extracts [6,7] and is conserved among all major eukaryotic lineages. Homologs have been found in yeast (subunits CAC1, CAC2, CAC3; [8], in mammals (p150, p60, p48; [9], in insects (p180, p105/75, p55; [10-12] and in plants (FASCIATA (FAS) 1, FAS2, MSI1; [13,14].
Yeast CAF-1 mutants have impaired maintenance of silencing at mating type loci and near the telomeres, and exhibit increased sensitivity towards ultraviolet radiation [8,15-20]. In higher eukaryotes, CAF-1 is specific for replication-coupled deposition of the H3.1 variant, while other histone chaperones deposit the H3.3 variant (called H3.2 in plants) in a replication-independent way [21,22]. Because mostly H3.3 and much less H3.1 is found in active chromatin [23], it has been proposed that CAF-1-mediated assembly of chromatin facilitates transcriptional repression through H3.1 deposition [24]. A recent report that H3.1-containing nucleosomes are more stable than H3.3-containing nucleosomes supports this model [25]. Replication-coupled deposition of H3.1 by CAF-1 is essential in metazoans, because loss of CAF-1 function causes severe defects in chromatin metabolism and eventual cell death in mouse and human cells [26-30]. Loss of CAF-1 causes developmental arrest in Xenopus laevis [31], Drosophila [32] and zebrafish [33].
Arabidopsis thaliana is the only higher eukaryote for which viable CAF-1 mutants are available (for review see [34]). Mutants deficient in FAS1 and FAS2, the two larger subunits of Arabidopsis CAF-1, were originally isolated for their altered phyllotaxis and their flattened and bifurcated stems [35,36], which is a phenotype known as fasciation [37]. Fasciation is associated with altered expression of WUSCHEL, which is a key regulatory gene that defines the stem cell niche in the shoot apical meristem (SAM) [13]. Misspecification of the WUSCHEL domain alters size and shape of the meristem, which subsequently changes primordia spacing and therefore causes distortion of phyllotaxis. In contrast to null mutants of FAS1 and FAS2 that are viable null mutants of the smallest CAF-1 subunit MSI1 are lethal [38]. This lethality is not caused by loss of CAF-1 function, however, but by loss of the FERTILIZATION INDEPENDENT SEED DEVELOPMENT (FIS) complex, of which MSI1 is a subunit as well [39].
Initial research with fas mutants focused on CAF-1 function in meristematic tissue [13,35,36] Recent studies showed, however, that CAF-1 is also needed for complete compaction of heterochromatin and maintenance of transcriptional gene silencing [40,41], homologous recombination [42,43], regulation of endoreduplication [34], and cell differentiation [44].
Trichomes or leaf hairs protrude from the leaf surface to protect the plant against adverse environmental conditions and herbivorous insects [45,46]. Depending on the plant species and function, trichomes are uni- or multicellular, metabolically active or inactive structures. In Arabidopsis thaliana, trichomes are single, living cells with a complex structure, which makes them well suited to study cell determination and differentiation. Trichomes originate from the epidermal cell layer and are evenly spaced by lateral inhibition (for an overview see: [47]). After determination, the trichome progenitor cell stops division and switches to endoreduplication. The cell enlarges and protrudes from the epidermal cell layer. On rosette leaves, two branching events give trichomes their characteristic three-ended morphology. Genetic analyses have revealed a complex regulatory network that controls trichome spacing and differentiation. Two major groups of genes control branching. Some of the genes influence branching directly, while others control branch number in an endoreduplication-dependent manner (reviewed by: [48]).
We have previously reported that trichome differentiation requires a functional CAF-1 complex, but it remained open in which genetic pathway CAF-1 acts during this process [44]. Here we provide evidence that CAF-1 and STICHEL (STI), which encodes a protein with similarity to ATP-binding eubacterial DNA-polymerase III-subunits [49], together control trichome differentiation in an endoreduplication-independent pathway.
Results
Sucrose suppresses the CAF-1 mutant trichome phenotype
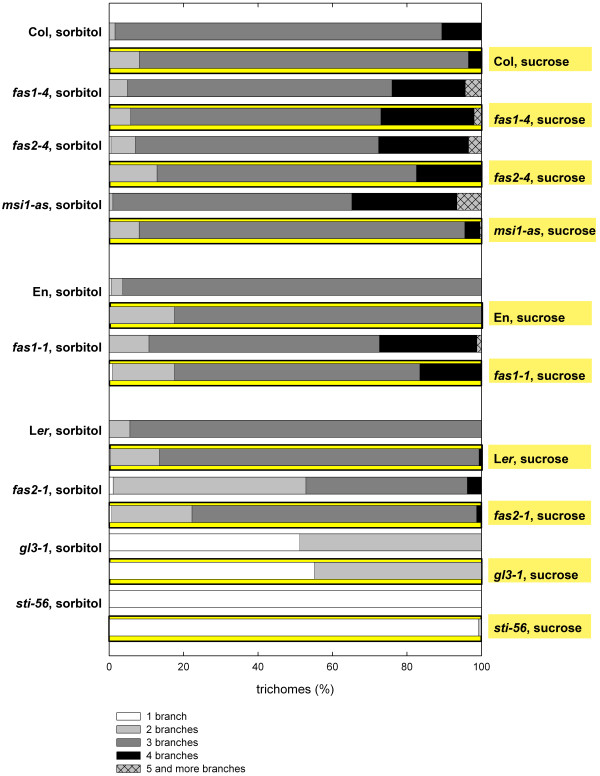
During the analysis of trichome development in CAF-1 mutants we observed that fas2-1 seedlings had fewer trichomes with supernumerary branches when grown on MS medium containing sucrose than on MS medium alone (data not shown). Carbohydrates control cell cycle activity and are known to influence plant development and organ formation (for review see: [50,51]), but a role in trichome development has not been reported. To test whether sucrose generally influences trichome development, wild type and CAF-1 mutant plants were grown on MS medium with 1% sucrose. Control plants were grown on MS medium containing 1% of the non-metabolizable sugar sorbitol. The number of trichome branches was recorded for the first and second rosette leaves (Fig. 1). In wild type plants of Columbia (Col), Enkheim (En) and Landsberg erecta (Ler) accessions, sucrose caused a small but consistent shift towards trichomes with fewer branches. This decrease in branch number was statistically significant (chi-squared test, p < 0.05) for Col, fas2-4, msi1-as, En and fas2-1. In CAF-1 mutants, sucrose suppressed, at least partially, the supernumerary branch phenotype. The effect was strongest in msi1-as, and weakest in fas1-4. Mutations of STI and GLABRA3 (GL3), which positively regulate trichome branching through the endoreduplication-independent and endoreduplication-dependent pathway, respectively, usually produce trichomes without branching (sti) or only a single branching event (gl3). Both mutants were unaffected by sucrose (Fig. 1). Thus, sucrose affects branching during trichome differentiation and can partially substitute for loss of CAF-1.
Figure 1.
Sucrose influences trichome morphology in CAF-1 mutants. The overbranching of rosette leaf trichomes in CAF-1 mutants is reduced on medium containing sucrose. Trichome branch number was assessed on the first and second primary leaves of wild-types Col (294, 342), En (161, 136) and Ler (119, 449) and the mutants fas1-4 (223, 171), fas2-4 (164, 121), fas1-1 (123, 98), fas2-1 (66, 124),gl3-1 (42, 55) and sti-56 (129, 171). Figures in parentheses represent the number of trichomes analyzed on sorbitol and sucrose, respectively. Note that gl3-1 produces only a limited number of trichomes on the primary rosette leaves. Plants were grown on MS medium supplied with either 1% sorbitol (unmarked bars) or 1% sucrose (bars highlighted in yellow).
Sucrose does not generally attenuate CAF-1 mutant phenotypes
It is possible that sucrose generally suppresses CAF-1 mutant phenotypes. Detailed analysis of CAF1 mutants showed, however, that only trichome branching but not other aspects of the CAF-1 mutant phenotype were attenuated by sucrose. In fact, distortion of phyllotaxis was strongly enhanced in fas2-1 mutants grown on MS medium with sucrose (Fig. 2). The angles between successive leaves were highly irregular, and some primordia did not complete differentiation into leaves but showed weak radialization (data not shown). In addition, internodes elongated and the usual compact appearance of a rosette was lost (Table 1). Furthermore, even after fas2-1 seedlings were transferred from sucrose medium to soil, about 10% of the plants showed defects in flower development (Fig. 2F, G). These plants produced flowers with missing or severely malformed petals and stamens, and unfused carpels. Additionally, ectopic ovules were sometimes produced at the margin of cauline leaves. Such phenotypes were not observed in control plants. This strong enhancement of the mutant phenotype was not observed in the fas1-1, fas1-4 and fas2-4 CAF-1 mutant alleles, suggesting that Ler is especially sensitive to loss of CAF-1 function when additional factors such as sucrose perturb early development.
Figure 2.
Sucrose influences the phenotype of fas2-1 mutants. Wild-type Ler and fas2-1 mutant seedlings were grown on medium supplied with 1% sorbitol or with 1% sucrose. A: Ler grown on sorbitol. B: Ler grown on sucrose. C: fas2-1 grown on sorbitol. D: fas2-1 grown on sucrose. Note the severely distorted phyllotaxis. E: fas2-1 grown on sucrose exhibiting internode elongation. F and G: Flowers of fas2-1 plants grown for 2.5 weeks on sucrose and later on soil. H: Flowers of a Ler plant grown for 2.5 weeks on sucrose and later on soil. Scale bars: 0.5 mm.
Table 1.
Sucrose strongly alters the phenotype of fas2-1 but not of the other CAF-1 mutants or wild-type plants.
| Genotype, treatment | Wild-type phenotype | fas mutant phenotype, rosette habit | fas mutant phenotype, elongated internodes | |||
| Plants | % | Plants | % | Plants | % | |
| En, sorbitol | 14 | 93.3 | 1 | 6.7 | 0 | 0 |
| En, sucrose | 38 | 97.4 | 1 | 2.6 | 0 | 0 |
| fas1-1, sorbitol | 0 | 0 | 17 | 100 | 0 | 0 |
| fas1-1, sucrose | 0 | 0 | 36 | 100 | 0 | 0 |
| Ler, sorbitol | 27 | 100 | 0 | 0 | 0 | 0 |
| Ler, sucrose | 39 | 97.5 | 0 | 0 | 1 | 2.6 |
| fas2-1, sorbitol | 0 | 0 | 29 | 100 | 0 | 0 |
| fas2-1, sucrose | 0 | 0 | 26 | 18.3 | 116 | 81.7 |
| Col, sorbitol | 38 | 100 | 0 | 0 | 0 | 0 |
| Col, sucrose | 36 | 100 | 0 | 0 | 0 | 0 |
| fas1-4, sorbitol | 0 | 0 | 35 | 100 | 0 | 0 |
| fas1-4, sucrose | 0 | 0 | 37 | 100 | 0 | 0 |
| fas2-4, sorbitol | 0 | 0 | 25 | 100 | 0 | 0 |
| fas2-4, sucrose | 0 | 0 | 39 | 100 | 0 | 0 |
| msi1-as, sorbitol | 0 | 0 | 44 | 100 | 0 | 0 |
| msi1-as, sucrose | 0 | 0 | 42 | 100 | 0 | 0 |
Shown are the number of seedlings scored in a given category and the percentage.
Mutations in CAF-1 partially suppress the kaktus supernumerary branching phenotype
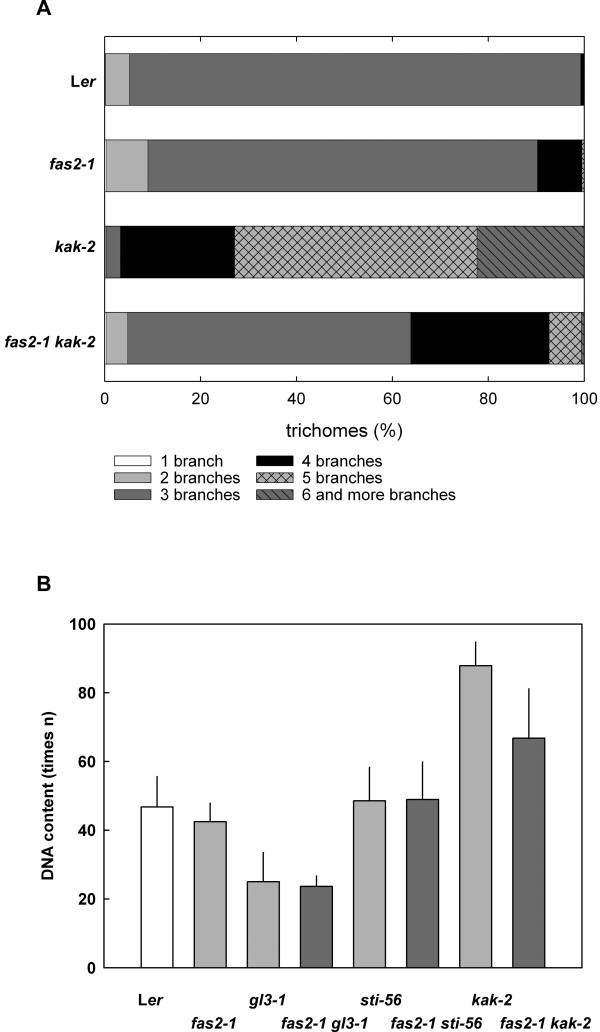
We previously suggested that CAF-1 controls trichome branching via an endoreduplication-independent pathway [44]. To further test this hypothesis, we first analyzed fas2-1 kak-2 double mutants. KAKTUS (KAK) encodes a putative HECT-domain E3 ligase [52], and kak mutant trichomes have increased ploidy levels and highly supernumerary branches [53]. Characterization of the trichome morphology on rosette leaves of fas2-1 kak-2 double mutant plants revealed that the two alleles were not epistatic (Fig. 3A). This result is consistent with the hypothesis that CAF-1 controls trichome branching independent of the KAK-containing pathway. However, the branching phenotype of fas2-1 kak-2 trichomes was intermediate to the two single mutants rather than additive, suggesting that KAK and CAF-1 can influence each other.
Figure 3.
Trichome phenotype in fas2-1 kak-2 double mutants. A: Trichome branching in Ler, fas2-1, kak-2 and fas2-1 kak-2. The double mutants have an intermediate number of branches per trichome compared to the single mutants. B: Nuclear DNA content of trichomes from Ler, fas2-1, kak-2 and fas2-1 kak-2 leaves. The DNA content of trichome nuclei of fas2-1 kak-2 mutants is in between the DNA content of the single mutants.
Loss of CAF1 function restricts DNA endoreduplication in kak-2 mutants
While fas2-1 mutants and wild-type plants have the same DNA content of trichome nuclei [44], mutations in KAK allow additional rounds of endoreduplication in leaf hair nuclei [53]. However, CAF-1 function is needed for chromatin integrity and has been suggested to be required during cell cycle progression [40]. It was therefore possible that loss of CAF-1 function in the fas2-1 kak-2 mutant restricts the kak endoreduplication potential and thus limits trichome branching in the fas2-1 kak-2 mutant. Analysis of the DNA content revealed that trichomes of fas2-1 kak-2 mutants had on average one third less nuclear DNA than trichomes of kak-2 single mutants (Fig. 3B). This level was between the numbers of endocycles observed in fas2-1 and kak-2. One possible explanation is that CAF-1 is needed for efficient progression through the endocycle in trichomes. Such a limitation would be consistent with the proposed slower progression through S-phase in CAF-1 mutants [40,54].
CAF-1 and STICHEL act together in the endoreduplication-independent pathway of trichome differentiation
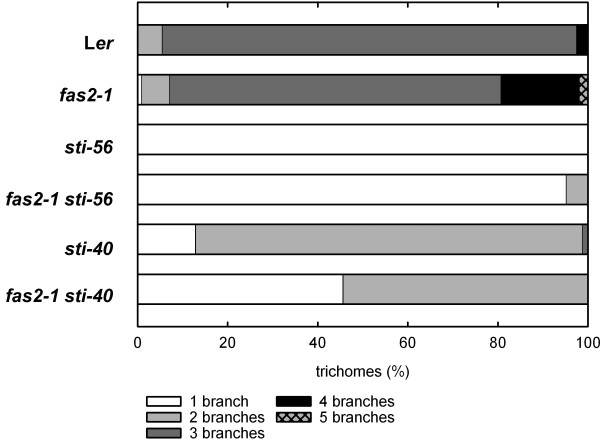
Analysis of fas2-1 kak-2 (this work) and fas2-1 gl3-1 [44] double mutants suggested that FAS2 acts in a pathway parallel to KAK and GL3 and controls trichome branching in an endoreduplication-independent manner. STICHEL (STI), a protein with similarity to eubacterial DNA-polymerase III-subunits [49], also controls trichome branching in an endoreduplication-independent pathway. To test whether CAF-1 functions in the STI-pathway for trichome differentiation, fas2-1 was crossed with a strong and a weak sti allele. While sti-56 almost completely abolishes trichome branching, sti-40 develops many trichomes with one branching event [49,55]. Analysis of trichome morphology of the fas2 sti double mutants revealed strong, although not complete epistasis of the sti-56 null allele over fas2 (Fig. 4). Interestingly, fas2 fortifies the weak phenotype of the hypomorphic sti-40 allele. Together, these results suggest that FAS2 and STI function together in the same pathway for trichome differentiation.
Figure 4.
Mutations in STI are epistatic over fas2. Trichome branching in Ler, fas2-1, sti-40, sti-56, fas2-1 sti-40 and fas2-1 sti-56. Double mutants between two sti alleles and fas2-1 exhibit the same branching phenotype as the two sti alleles alone.
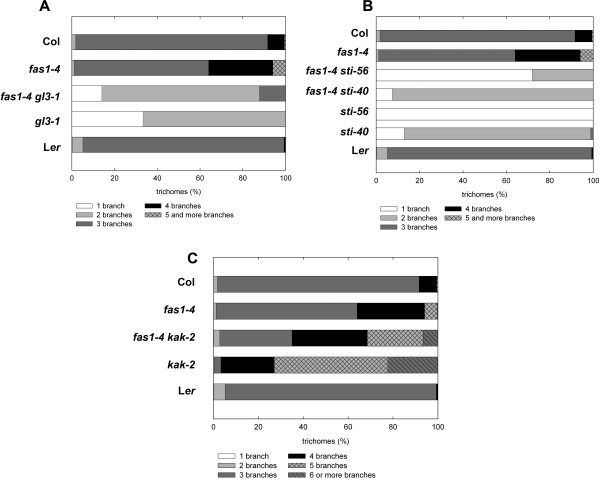
We have reported earlier that FAS2 controls trichome branching in the context of the CAF-1 complex [44]. To test the hypothesis that CAF-1 and STI function in the same pathway, we generated double mutants of fas1 and gl3, kak and sti. The trichome branching phenotypes of the various double mutants with fas1 and fas2 were similar: fas1-4 gl3-1 exhibited intermediate phenotypes (Fig. 5A) compared with the single mutants, while fas1-4 sti-40 and fas1-4 sti-56 again showed strong epistasis of sti over fas1 (Fig. 5B). Furthermore, fas1-4 kak-2 double mutants had a similar partial suppression of the kak phenotype as did the fas2-1 kak-2 double mutants (Fig. 5B). These results are consistent with our view that CAF-1 and STI function in the same pathway of trichome differentiation.
Figure 5.
Genetic interactions of fas1-4 with gl3-1, si-40, sti-56 and kak-2. A: Trichome branching on rosette leaves of fas1-4, gl3-1 and fas1-4 gl3-1. The double mutant is intermediate to the single mutants. B: Trichome branching on rosette leaves of fas1-4, sti-40, sti-56, fas1-4 sti-40 and fas1-4 sti-56. The two sti alleles are epistatic over fas1-4. C: Trichome branching on rosette leaves of fas1-4, kak-2 and fas1-4 kak-2. The strong overbranching phenotype of kak-2 is partially suppressed by fas1-4.
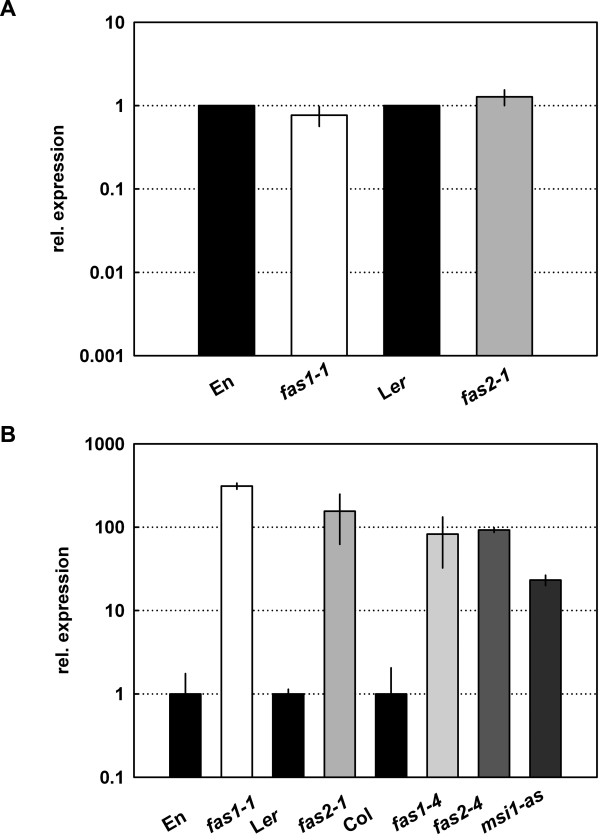
Because sti showed epistasis over CAF-1 mutant alleles, it is likely that STI acts downstream of CAF-1. One possibility is that CAF-1 is needed for correct STI expression during trichome differentiation. To test this hypothesis we measured STI mRNA levels in CAF-1 mutants by quantitative RT-PCR. However, STI transcript levels were not significantly increased in fas1 and fas2 trichomes (Fig. 6). Similar results were obtained for STI expression in fas1 and fas2 seedlings and apices (data not shown). These results suggest that CAF-1 affects STI function instead of modulating STI expression.
Figure 6.
H3.2 but not STI expression is changed in fas1-1 and fas2-1 trichomes. A: STI transcript levels were measured by quantitative RT-PCR in En, Ler, fas1-1 and fas2-1. Expression is shown relative to the corresponding wild type. B: H3.2 transcript levels were measured by quantitative RT-PCR in En, Ler, Col, fas1 and fas2 mutants and msi1-as. Expression is shown relative to the corresponding wild type.
H3.2 is up-regulated in fasciata mutant trichomes
We previously showed that transcription of the gene for replacement histone variant H3.2 was upregulated in fas1-1, fas2-1 and msi1-as seedlings [40]. H3.2 is incorporated by a CAF-1 independent pathway into nucleosome of chromatin found mostly in transcriptionally active, less compact chromosome regions (reviewed by [4,56]). We asked whether altered trichome differentiation in CAF-1 mutants was correlated with increased expression of H3.2 in trichomes. RNA was extracted from trichomes of wild-type, CAF-1 mutants and msi1-as plants, and mRNA levels of the H3.2 gene At1g13370 were determined by quantitative RT-PCR. This analysis showed that H3.2 transcript levels were indeed increased by about 100-fold in trichomes of CAF-1 mutants and msi1-as plants (Fig. 6B). These results show that loss of CAF-1 function causes increased expression of H3.2 not only in whole seedlings but also in trichomes. Thus, it is likely that chromatin of CAF-1 mutant trichomes contains increased amounts of the H3.2 variant histone.
Discussion
Trichome cell specification and maturation provide a good model system to study cell differentiation in Arabidopsis. Analysis of trichome differentiation has revealed a complex gene network that directs and controls the cell determination, specification and differentiation process [48,57,58]. Here, we report the effects of mutations in the chromatin remodeling complex CAF-1 on trichome development and the genetic interaction of CAF-1 mutant alleles with the trichome regulators GL3, STI and KAK.
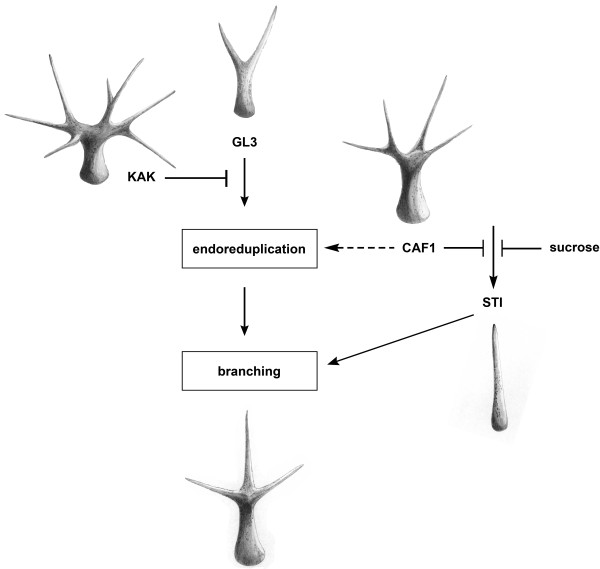
Because CAF-1 mutants have increased trichome branching but normal endoreduplication [44], CAF-1 limits branching during trichome maturation independent of endoreduplication. Genetic evidence suggests that CAF-1 acts parallel to the GL3-KAK pathway (Fig. 7), which promotes trichome branching through the control of endoreduplication ([44,59], this work). Nevertheless, CAF-1 is needed for the GL3-KAK pathway to function normally, because the kak phenotype is partially suppressed in kak-2 fas2-1 double mutants. The kak-2 fas2-1 double mutants do not only have less trichome branching but also a lower DNA content than kak-2 single mutants. These results suggest that CAF-1 is needed for the increased endoreduplication cycles in kak-2 trichomes. One possible explanation for this observation is that the slower progression through the S phase in the mitotic cell cycle, which we proposed for CAF-1 mutants earlier [40], impedes the increased endoreduplication activity in kak-2 mutant trichomes. In seedlings and leaves, CAF-1 restricts endoreduplication [34,42-44], and it is possible that lack of CAF-1 triggers additional endocycles in certain cell types with low endoreduplication, but that CAF-1 is also needed to sustain multiple rounds of endocycles in cells types with high endoreduplication such as kak-2 trichomes.
Figure 7.
Model of CAF-1 function in trichome branching. The initiation of branching is regulated by two independent pathways. In the first pathway, Gl3 is a postive regulator and KAK is a negative regulator. In this pathway, endoreduplication triggers trichome branching. In the second pathway, STI is a positive regulator and CAF-1 is a negative regulator. Exogenous sucrose can partly substitute the negative function of CAF-1. CAF-1 is also required for extensive endoreduplication such as in the kak-2 mutant. Images represent trichome phenotypes in the respective mutants.
Exogenous sucrose alleviates the CAF-1 mutant trichome branching phenotype and weakly suppresses trichome branching in wild-type plants. Since the branching phenotype of CAF-1 mutants grown on soil, which constitutes a less defined but rich medium, was much more similar to the trichome phenotype of CAF-1 mutants grown on sorbitol than on sucrose (data not shown), we suggest that the suppression of trichome branching results from sucrose signaling rather than a starvation effect. Sucrose is known as a potent signaling molecule that controls gene expression, cell cycle and development [50,51]. However, to our knowledge no effect of sucrose on trichome development has been reported before. Sucrose promotes cell cycle progression [60] and can induce endoreduplication [61], but these effects most likely do not explain the observed reduced trichome branching. More rapid progression through the cell cycle and faster growth on sucrose-containing medium could amplify defects associated with chromatin assembly during S-phase in CAF-1 mutants. We found that sucrose greatly enhances the organ development phenotype of fas2-1 in Ler, and mildly enhances this phenotype of other CAF-1 mutant alleles. We propose that specifically during trichome development, sucrose signals can partially substitute for the CAF-1 requirement by a currently unknown mechanism.
Conclusion
Together, we observed (i) that CAF-1 mutants in a wild-type background have increased trichome branching but no increased endoreduplication, (ii) that CAF-1 mutants and gl3 mutants (defective in the endoreduplication-dependent pathway) show an additive interaction, (iii) that CAF-1 mutants and sti-56 null mutants (defective in the endoreduplication-independent pathway) show an epistatic interaction, (iv) that CAF-1 mutants enhance the phenotype of the hypomorphic sti-40 allele (partially defective in the endoreduplication-independent pathway) and (v) that CAF-1 mutants and kak mutants (defective in the endoreduplication-dependent pathway) do not show an epistatic interaction. We conclude that the most parsimonious model to explain all results is that CAF-1 acts together with STI in an endoreduplication-independent pathway that is parallel to the endoreduplication-dependent pathway of GL3 and KAK (Fig. 7). In addition, while CAF-1 is not needed for the normal endoreduplication in WT trichomes, CAF-1 is needed for the extranumerous rounds of endoreduplication that occur in kak mutants. The genetic evidence places CAF-1 in the same pathway with STI, an activator of trichome branching that does not affect DNA content [49]. STI shares sequence similarity with the ATP-binding subunit of eubacterial DNA-polymerase III, but the functional relevance of this similarity has not yet been established, and it is not known if STI is a nuclear protein. CAF-1 does not affect trichome branching by modulating STI expression, but acts as a negative regulator in the STI pathway (Fig. 7). It is not known how CAF-1 can negatively regulate the STI pathway. One possibility is that CAF-1 mediated chromatin assembly and compaction [40] are directly needed for normal trichome maturation. Alternatively, it is possible that CAF-1 represses expression of other, limiting components of the STI pathway. CAF-1 mutants have increased expression of H3.2, which is incorporated into chromatin independently of CAF-1. If chromatin of other genes in the STI pathway was enriched in H3.2, the less stable nucleosomes that are formed as a result could facilitate increased transcription, eventually causing increased activity of the STI pathway. In summary, we conclude that CAF-1 is required to support the exceptionally high endoreduplication of kak-2 trichomes but not for normal endoreduplication of wild-type trichomes. In wild-type trichomes, CAF-1 restricts the activity of the STI pathway.
Methods
Plant material and growth conditions
Seeds of Columbia (Col), Landsberg erecta (Ler) and Enkheim (En) Arabidopsis thaliana wild-type accessions and of fas1-1 (accession En) [13,35], fas2-1 (accession Ler) [13,36], fas1-4 (accession Col) [44], fas2-4 (accession Col) [44] and gl3-1 (accession Ler) [62,63] mutants were obtained from the Nottingham Arabidopsis Stock Centre. Note that in addition to the used fas1-4 allele in Col and described first in [44], another fas1 allele was described under the same name (fas1-4) by Kirik and collaborators in accession C24 [43]. The msi1-as line has been described before [44]. The mutants kak-2 (accession Ler) [52], sti-40 (accession Ler) [55] and sti-56 (accession Ler) [49] were kindly provided by M. Hülskamp. Seeds were sown on sterile basal salts Murashige and Skoog (MS) medium (Duchefa, Brussels, Belgium), which was supplemented with 1% sucrose or 1% sorbitol when required. Plants were analyzed on plates or transferred to soil ("Einheitserde", H. Gilgen optima-Werke, Arlesheim, Switzerland) 10 days after germination. Alternatively, seeds were sown directly on soil. Plants were kept in Conviron growth chambers with mixed cold fluorescent and incandescent light (110 to 140 μmol/m2s, 21 ± 2°C) under long day (LD, 16 h light) photoperiods or were alternatively raised in green houses.
Analysis of trichome branching
To determine the branching pattern, all trichomes on the adaxial side of the first two leaves of an average of six plants were analyzed.
Ploidy analysis
Ploidy of trichome nuclei was determined as described [44,64]. Briefly, plant tissue was fixed in FAA (50% ethanol, 5% glacial acetic, 10% formaldehyde) and stained for 90 minutes with 130 μg/ml DAPI in McIlvaines buffer (60 mM citric acid, 80 mM sodium phosphate, pH 4.1). Samples were washed twice (15 minutes and 60 minutes) with McIlvaines buffer, and mounted in McIlvaines buffer with 50% glycerol. DAPI fluorescence was recorded with a MagnaFire CCD camera (Optronics, Goleta, CA), or with an Apogee Alta U32 CCD camera (Apogee Instruments, Roseville, CA). Images were quantified using ImageJ. Total fluorescence of at least 30 representative nuclei per experiment was determined and calibrated using guard cell nuclei (n ≥ 30), which are considered to be strictly diploid [64].
RNA isolation, RT-PCR and Real Time PCR
RNA was extracted from seedlings as previously described [65]. For RT-PCR analysis, 0.4–1 μg total RNA was treated with DNase I. The DNA-free RNA (0.2 – 1.0 μg) was reverse-transcribed using a RevertAid First Strand cDNA Synthesis Kit according to manufacturer's instructions (Fermentas, Nunningen, Switzerland). Trichomes for RNA extraction were harvested into a few microlitres RNA later (Ambion, Austin, TX) and then processed like the other samples. Aliquots of the generated cDNA were used as template for PCR with gene specific primers. For qPCR analysis, the Universal ProbeLibrary system (Roche Diagnostics, Rotkreuz, Switzerland) was used on a 7500 Fast Real-Time PCR instrument (Applied Biosystems, Lincoln, CA). Details of the assays used are in Table 2. Analysis of the results was performed according to the method described by Simon [66].
Table 2.
qPCR assays.
| Gene | Forward primer | Reverse primer | Universal Probe Library probe |
| STI, At2g02480 (target gene) | agctgagtttgctgggaaaa | ttttcatctgaaacaacaccaac | #9 (Arabidopsis) |
| H3.2, At1g13370 (target gene) | aaccgtcgctcttcgtga | ttggaatggaagtttacggttc | #99 (Arabidopsis) |
| PP2A, At1g13320, (reference gene1)) | ggagagtgacttggttgagca | cattcaccagctgaaagtcg | #82 (Arabidopsis) |
1) [67]
Shown are the analyzed genes, sequences of the primers and the identifier of the corresponding Universal ProbeLibrary probes.
Authors' contributions
VE performed plant analysis, carried out the molecular and genetic studies and drafted the manuscript. LH participated in the study design and helped to write the manuscript. WG helped to write the manuscript.
Acknowledgments
Acknowledgements
We thank Claudia Köhler and Cristina Alexandre for critical reading of the manuscript, and Bartosz Urbaniak for technical help with qPCR. Work in the authors' laboratory is supported by SNF project 3100AO-116060 and ETH project TH-16/05-2.
Contributor Information
Vivien Exner, Email: vexner@ethz.ch.
Wilhelm Gruissem, Email: Wilhelm.Gruissem@ethz.ch.
Lars Hennig, Email: lhennig@ethz.ch.
References
- Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- Santoro R, De Lucia F. Many players, one goal: how chromatin states are inherited during cell division. Biochem Cell Biol. 2005;83:332–343. doi: 10.1139/o05-021. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Hennig L, Gruissem W. Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiol. 2002;130:1090–1101. doi: 10.1104/pp.006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-X. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking Chromatin Assembly Factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Bulger M, Ito T, Kamakaka RT, Kadonaga JT. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc Natl Acad Sci USA. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT, Bulger M, Kaufman PD, Stillman B, Kadonaga JT. Postreplicative chromatin assembly by Drosophila and human Chromatin Assembly Factor 1. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Bulger M, Kamakaka RT, Kobayashi R, Kadonaga JT. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- Ach RA, Taranto P, Gruissem W. A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell. 1997;9:1595–1606. doi: 10.1105/tpc.9.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- Monson EK, de Bruin D, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, Berman J. Chromatin Assembly Factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Kaufman PD. Role of Saccharomyces cerevisiae Chromatin Assembly Factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/S1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio T, Casella JF, Heidmann T. A truncated form of the human CAF-1 p150 subunit impairs the maintenance of transcriptional gene silencing in mammalian cells. Mol Cell Biol. 2001;21:1953–1961. doi: 10.1128/MCB.21.6.1953-1961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M, Stillman B. Chromatin Assembly Factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci USA. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatiyan A, Krude T. Silencing of Chromatin Assembly Factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol Cell Biol. 2004;24:2853–2862. doi: 10.1128/MCB.24.7.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–351. doi: 10.1016/S1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Quivy JP, Grandi P, Almouzni G. Dimerization of the largest subunit of Chromatin Assembly Factor 1: importance in vitro and during Xenopus early development. EMBO J. 2001;20:2015–2027. doi: 10.1093/emboj/20.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, He F, Xie G, Guo X, Xu Y, Chen Y, Liang X, Stagljar I, Egli D, Ma J, Jiao R. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev Biol. 2007;311:213–222. doi: 10.1016/j.ydbio.2007.08.039. [DOI] [PubMed] [Google Scholar]
- Fischer S, Prykhozhij S, Rau MJ, Neumann CJ. Mutation of zebrafish caf-1b results in S phase arrest, defective differentiation, and p53-mediated apoptosis during organogenesis. Cell Cycle. 2007;6:2962–2969. doi: 10.4161/cc.6.23.4950. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholz E. Radiation induced mutants showing changed inflorescence characteristics. Arab Inf Serv. 1966;3:19–20. [Google Scholar]
- Leyser H, Furner I. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 1992;116:397–403. [Google Scholar]
- Worsdell W. Fasciation: its meaning and origin. New Phytol. 1905;4:55–74. doi: 10.1111/j.1469-8137.1905.tb05881.x. [DOI] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy O, Hennig L, Breuninger H, Laux T, Köhler C. Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development. 2007;134:3639–3648. doi: 10.1242/dev.009027. [DOI] [PubMed] [Google Scholar]
- Schönrock N, Exner V, Probst A, Gruissem W, Hennig L. Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J Biol Chem. 2006;281:9560–9568. doi: 10.1074/jbc.M513426200. [DOI] [PubMed] [Google Scholar]
- Ono T, Kaya H, Takeda S, Abe M, Ogawa Y, Kato M, Kakutani T, Mittelsten Scheid O, Araki T, Shibahara K. Chromatin Assembly Factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells. 2006;11:153–162. doi: 10.1111/j.1365-2443.2006.00928.x. [DOI] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006;25:5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Pecinka A, Wendeler E, Reiss B. The Chromatin Assembly Factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell. 2006;18:2431–2442. doi: 10.1105/tpc.106.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Taranto P, Schönrock N, Gruissem W, Hennig L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development. 2006;133:4163–4172. doi: 10.1242/dev.02599. [DOI] [PubMed] [Google Scholar]
- Johnson H. Plant pubescence: an ecological perspective. Bot Rev. 1975;41:233–258. doi: 10.1007/BF02860838. [DOI] [Google Scholar]
- Mauricio R, Rausher M. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution. 1997;51:1435–1444. doi: 10.2307/2411196. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M, Uhrig J. Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem Soc Trans. 2007;35:146–148. doi: 10.1042/BST0350146. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Dev Biol. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua NH, Jürgens G, Hülskamp M. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol. 2003;131:643–655. doi: 10.1104/pp.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003;35:729–742. doi: 10.1046/j.1365-313X.2003.01844.x. [DOI] [PubMed] [Google Scholar]
- El Refy A, Perazza D, Zekraoui L, Valay JG, Bechtold N, Brown S, Hülskamp M, Herzog M, Bonneville JM. The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol Genet Genomics. 2003;270:403–414. doi: 10.1007/s00438-003-0932-1. [DOI] [PubMed] [Google Scholar]
- Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. Essential role of Chromatin Assembly Factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell. 2007;18:129–141. doi: 10.1091/mbc.E06-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers U, Berger J, Hülskamp M. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124:3779–3786. doi: 10.1242/dev.124.19.3779. [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne AM, Bonneville JM. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics. 1999;152:461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD. Molecular genetic analysis of trichome development in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:137–163. doi: 10.1146/annurev.arplant.48.1.137. [DOI] [PubMed] [Google Scholar]
- Hülskamp M. Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JA. Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol. 2000;20:4513–4521. doi: 10.1128/MCB.20.13.4513-4521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnacker L, Ferwerda M. Effects of sucrose on polyploidization in early callus cultures of Solanum tuberosum. Plant Cell. 1990;21:153–17. [Google Scholar]
- Koornneef M, Dellaert LW, VeJ van der. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003;130:2555–2565. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]