Abstract
Gene families associated with the ethylene signal transduction pathway in ripening kiwifruit (Actinidia deliciosa [A. Chev.] C.F. Liang et A.R. Ferguson var. deliciosa cv. Hayward) were isolated from a kiwifruit expressed sequence tag (EST) database, including five ethylene receptor genes, two CTR1-like genes, and an EIN3-like gene AdEIL1. All were differentially expressed among various kiwifruit vine tissues, and none was fruit specific. During fruit development, levels of transcripts of AdERS1a, AdETR3, and the two CTR1-like genes decreased, whereas those of AdERS1b and AdETR2 peaked at 97 d after full bloom. In ripening kiwifruit, there was a diverse response of the ethylene receptor family to internal and external ethylene. AdERS1a, AdETR2, and AdETR3 expression increased at the climacteric stage and transcripts were induced by external ethylene treatment, while AdERS1b showed no response to ethylene. AdETR1 was negatively regulated by internal and external ethylene in ripening fruit. The two CTR1-like genes also had different expression patterns, with AdCTR1 increasing at the climacteric stage and AdCTR2 undergoing little change. 1-Methylcyclopropene treatment prevented the ethylene response of all components, but transient down-regulation was only found with AdETR2 and AdCTR1. Similar gene and ethylene responses were found in both fruit flesh and core tissues. The ethylene-induced down-regulation of AdETR1 suggests that it may have a role in sensing ethylene and transmitting this response to other members of the receptor family, thus activating the signal transduction pathway.
Keywords: Actinidia, ethylene receptor, ethylene response, ethylene signal transduction, fruit ripening, gene expression, kiwifruit, 1-MCP
Introduction
Genetic studies of ethylene action in higher plants, especially in Arabidopsis and tomato, have established a linear ethylene signal transduction model, in which ethylene is perceived by a receptor family, and the signal is mediated downstream by members of different gene families including CTR1, EIN2, EIN3/EILs, and ERFs (Chang and Stadler, 2001; Chen et al., 2005). Ethylene receptors have high similarity to bacterial two-component signal systems, with ethylene binding, histidine kinase, and receiver domains, except that ERS-type ethylene receptors lack a receiver domain (Chang and Stadler, 2001). These receptors, along with CTR1, which encodes a Raf protein kinase, act as negative regulators. In contrast, the remaining components in the pathway have been identified as positive regulators, and loss of function or overexpression of them can lead to ethylene insensitivity or a constitutive ethylene response in air, respectively (Chao et al., 1997; Solano et al., 1998; Alonso et al., 1999). EIN3/EILs are considered to be positive regulators, stimulating other transcription factors, such as ERF1 (Solano et al., 1998).
Ethylene receptors are encoded by a multigene family, with at least five identified in Arabidopsis (C Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998), five in rice (Yau et al., 2004), and, amongst fruit species, six in tomato (Wilkinson et al., 1995; Lashbrook et al., 1998; Tieman and Klee, 1999), five in apples (Tatsuki et al., 2007; Wiersma et al., 2007), four in pears (El-Sharkawy et al., 2003), three in peach (Rasori et al., 2002; Trainotti et al., 2006), three in persimmon (Pang et al., 2007), and two in melon (Sato-Nara et al., 1999).
All of the ethylene receptors have a very similar structure (Chang and Stadler, 2001). However single ethylene receptor mutants show little or no effect on seedling phenotype (Hua and Meyerowitz, 1998), suggesting that the receptors appear to have a degree of functional redundancy. However, a single AtETR1 or AtERS1 mutant can also enhance the ethylene response (Cancel and Larsen, 2002; Qu et al., 2007), while a dual mutation of the two genes displays a constitutive ethylene response phenotype, suggesting that the ETR1 subfamily may have a profound effect on ethylene response (Qu et al., 2007). Furthermore, there is much evidence to show that ethylene receptors are differentially expressed in plant tissues. In Arabidopsis, AtETR1 and AtERS1 are ubiquitously expressed, but three other receptor genes are differentially expressed. For example, AtETR2 is expressed strongly in flower and leaf tissue (Hua et al., 1998; Sakai et al., 1998). Some genes also display specific expression patterns associated with development stages, for example NR, CmETR1, CmERS1, and MdERS1 mRNAs have been shown to increase during ripening of tomato, melon, and apple fruit (Wilkinson et al., 1995; Sato-Nara et al., 1999; Dal Cin et al., 2006). A further feature of the receptor pathway is differential sensitivity of receptors and the capacity of the pathway for amplification. Some genes such as AtERS1, AtETR2, AtERS2, NR, LeETR4, LeETR6, PpERS1, OsETR2, and OsERS1 can be strongly up-regulated by ethylene, whereas AtETR1, AtEIN4, LeETR1, LeETR2, and PpETR1 show a weaker response (Hua et al., 1998; Rasori et al., 2002; Yau et al., 2004; Kevany et al., 2007). There are also suggestions that genes such as OsERS2 can be slightly down-regulated by ethylene treatment (Yau et al., 2004).
So far, only one CTR1-like gene, AtCTR1, has been found in Arabidopsis (Kieber et al., 1993) and four isolated from tomato (Lin et al., 1998; Leclercq et al., 2002; Adams-Phillips et al., 2004). In tomato, four CTR1-like genes have been shown to be differentially expressed: TCTR2, like AtCTR1, is constitutively expressed and unresponsive to ethylene (Alexander and Grierson, 2002); LeCTR1 is up-regulated by ethylene and increases during ripening (Leclercq et al., 2002; Adams-Phillips et al., 2004); and LeCTR3 and LeCTR4 are expressed more highly in leaves than fruit and do not respond to ethylene (Adams-Phillips et al., 2004).
Despite the large amount of work published on the receptor and CTR1-like genes, there is still an imperfect understanding of how they respond and interact in ripening fruit. This is particularly the case in relation to differential responses to ethylene, and in relation to physiological changes occurring during fruit ripening. Kiwifruit is particularly useful in this regard. It is a typical climacteric fruit, with respiration and ethylene production rates increasing at late stages of softening. It is also very sensitive to ethylene, with fruit softening increasing upon exposure to extremely low concentrations of exogenous ethylene (0.1 μl l−1) (McDonald and Harman, 1982). There is also information suggesting that system II ethylene can provide a positive feedback regulating ethylene synthesis-related genes in kiwifruit (Whittaker et al., 1997; Xu et al., 1998). While there is good information on ethylene production in kiwifruit and response of the fruit to ethylene (Antunes and Sfakiotakis, 2002; Boquete et al., 2004; Koukounaras and Sfakiotakis, 2007), there has been no systematic attempt to relate this to the response of genes controlling the ethylene signal transduction pathway. This fruit species provides us with the opportunity to obtain new information on the interplay between various components of the pathway, particularly ethylene response, and the relationship to fruit behaviour.
In the present research, eight components associated with the ethylene signal transduction pathway were isolated from an Actinidia expressed sequence tag (EST) database. Real-time and end-point PCR were used to estimate tissue- and development-specific gene expression in kiwifruit. Treatment of fruit with ethylene and the receptor inhibitor 1-methylcyclopropene (1-MCP; Blankenship and Dole, 2003) was used to follow responses of these genes in relation to fruit ripening.
Materials and methods
Plant materials and treatments
Kiwifruit (Actinidia deliciosa [A. Chev.] C.F. Liang et A.R. Ferguson var. deliciosa cv. Hayward) were harvested from a commercial orchard, Patumahoe, South Auckland, New Zealand, when the fruit were at a mean soluble solids concentration (SSC) of 6.7%.
Fruit in the size range 90–130 g, with no visible defects, were divided into three batches of 400 fruit each, and for post-harvest treatments, each batch was sealed into a 306 l chamber and held at 20 °C. The first batch was exposed to C2H4 (100 μl l−1, 24 h); the second was treated with 1-MCP (0.5 μl l−1, 24 h; SmartFresh™); and the last batch had no treatment. Previous work from our laboratory (Kim et al., 2001), and elsewhere (Boquete et al., 2004; Koukounaras and Sfakiotokis, 2007), has shown that 0.5 μl l−1 1-MCP is an effective concentration to inhibit ethylene perception and production in kiwifruit. To eliminate the accumulation of CO2 over the 24 h, ∼0.5 kg of hydrated lime (McDonald's Lime Ltd.) was placed in each chamber, packed into a paper bag. Fans were used in the chambers to maintain air circulation. After treatment, fruit were held at 20 °C until reaching a firmness of <2 N (0.2 kgf) when they were considered to be eating-ripe. This did not apply to the 1-MCP-treated fruit which did not soften appreciably over the experimental period. At each sampling time, 10 fruit were taken from each treatment to provide tissue for RNA analysis. Each fruit was divided into core and flesh (no seeds or skin), and the two tissue types separately bulked for the 10 fruit. The fruit tissue samples were then frozen in liquid nitrogen, and stored at –80 °C for further use.
To provide samples for a fruit development series, young ‘Hayward’ kiwifruit at different developmental stages were picked from the HortResearch orchard, Te Puke, New Zealand. Full blossom was on 15 November 2006, and there were six subsequent sampling times: 20, 57, 97, 124, 159, and 181 days after full bloom (DAFB). At the final harvest at 181 DAFB, when the fruit had reached commercial maturity, fruit had a mean firmness of 78.7 N.
For analysis of different kiwifruit plant tissues, samples of root, stem, leaf, and petal were collected at anthesis from ‘Hayward’ vines planted at Wuyi, Zhejiang, PR China. The samples were frozen in liquid nitrogen and transported to the laboratory at Zhejiang University, Hangzhou. Tissue samples were ground in liquid nitrogen and stored at –80 °C for further use.
Fruit evaluation
Eight jars (0.5 l), each containing two fruit, were used for evaluating ethylene production rates of fruit from each treatment. The sealed jars were held at 20 °C for 1 h, and then 1 ml of head-space gas was removed from each jar using a syringe, and ethylene was measured by gas chromatography using a gas chromatograph (Hewlett-Packard 5890 series II), fitted with an FID (Philips PU4500, Unicam) and an alumina F1 column (1.5 m×6 mm). The injector, detector, and oven temperatures were 160, 200, and 130 °C, respectively.
Fruit firmness was measured using a hand-held penetrometer (Effegi) with a 7.9 mm diameter head. Firmness was measured on each of 10 fruit at two 90° positions at the equator of the fruit, after a 1 mm thick slice of peel was removed. SSC was measured by slicing both ends of each of the same 10 fruit, and three drops of juice from each slice were then applied to an Atago digital hand-held refractometer for measurement.
RNA extraction and cDNA synthesis
Total RNA was isolated from the frozen tissues following the protocol described by S Chang et al. (1993). Approximately 1.8 g of tissue was extracted in 15 ml of extraction buffer. TURBO DNase (Ambion) was used for removing contaminating DNA. The cDNA was synthesized from 2.5 μg of DNA-free RNA with a SuperScript III First Strand Synthesis kit (Invitrogen) following the manufacturer's protocol. Ten-fold diluted cDNA was used in real-time PCR and end-point PCR analysis. For each time point, three batches of RNA were isolated for separate cDNA synthesis.
Gene isolation and sequence analysis
Five ethylene receptor genes and two CTR1-like genes were found in the HortResearch EST database of Actinidia spp. using the BLAST algorithm. Isolation of an EIN3-like gene was achieved using degenerate primers described as follows: forward primer, 5′-T[G/T]GAGA[G/A]GAGGATGTGGAG[A/G]GAC and reverse primer, 5′-ATAAT[A/G]GCAAGCCA[A/T/G]GT[A/T]GCAC. The 3′-untranslated region (UTR) of candidate sequences was obtained by 3′-RACE (rapid amplification of cDNA ends). Alignments were carried out on Vector NTI (v. 9.0.0, Invitrogen, using Clustal W) and phylogenetic trees were generated (MEGA v. 3.1) (Kumar et al., 2004).
Oligonucleotide primers
The oligonucleotide primers for real-time analysis were designed with primer3 (v. 0.4.0, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The size of all PCR products ranged from 150 bp to 227 bp. Gene specificity and quality tests for primers were performed as described by Zhang et al. (2006). Primers were tested as follows: (i) a BLAST search in the Actinidia EST database to confirm that no other similar sequence of primers existed in kiwifruit; (ii) each primer set was amplified using the plasmids which contain sequence from 3′-RACE of each gene, to confirm primer-specific amplification only of the target gene; (iii) the melting peak and dissociation curve from use of the LightCycler 1.5 instrument (Roche) was used to confirm that there was no dimer generation in the primers; (iv) PCR product size was analysed on 1% agarose gels stained with ethidium bromide; and (v) PCR products were re-sequenced to confirm that the sequence of the amplification region was the same as from the 3′-RACE results. The sequences of all primers used for real-time and end-point PCR are described in Table 1.
Table 1.
Primers for real-time PCR and end-point PCR analysis
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Regiona |
| AdERS1a | TTACCCAACTGGGTCCAAG | GAGCTCGGAGCCACAACAT | UTR |
| AdERS1b | CGAGGATGTTGCTGAAAGC | GCACAGAGCCACAACATATTTC | UTR |
| AdETR1 | AGCATGGTGTGTTGGAAGC | CGTCGCTGCTTCCTATTGT | UTR |
| AdETR2 | AGGCATCTCTGAAAGTGGAG | GGAGATCCAAAAGCAAAACCTGA | CR |
| AdETR3 | TGAAGTAGTAAACCAAGAAGA | AATCATGATGAGGTTTGCCT | UTR |
| AdCTR1 | CGGATAATTCAGGACCCAA | TGTGTAATTCCGGCCAGTTCACTCT | UTR |
| AdCTR2 | TCTGTTCATCGTGCCTTCAG | GCACCAAGCTGCATTTGATT | UTR |
| AdEIL1 | TCAGATTTCATGGAGCCACA | ACATTCCAAAAGCAGGGTTG | UTR |
CR, coding region.
Real-time PCR
The PCR mixture (10 μl total volume) comprised 2 μl of 5×LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche), 0.5 μl of each primer (10 μM), 1 μl of cDNA, and 6 μl of PCR-grade water. PCRs were performed on a LightCycler 1.5 instrument (Roche), initiated by 5 min at 95 °C, then followed by 40–45 cycles of 95 °C for 5 s, 60 °C for 5 s, and 72 °C for 10 s, and completed with a melting curve analysis program. No-template controls and melting curve analyses were included in every reaction. Kiwifruit actin was used as the housekeeping gene as a positive control, with forward primer 5′-TGCATGAGCGATCAAGTTTCAAG-3′ and reverse primer 5′-TGTCCCATGTCTGGTTGATGACT-3′ (Zhang et al., 2006). Expression levels were expressed as a ratio relative to the day of harvesting for the ripening fruit series and to 181 DAFB for the fruit development series, ratios at both these time points being set at 1. Relative levels of gene expression were calculated from standard curves for each gene, established by four dilutions of cDNA: 2-, 10-, 100-, and 1000-fold.
For the analysis of tissue-specific gene expression, 20 μl total reaction volume comprised 10 μl of 2×SYBR Green PCR Master Mix (Bio-Rad), 1 μl of each primer (10 μM), 1 μl of cDNA, and 7 μl of PCR-grade water. The PCR was performed on an iCycler iQ real-time PCR instrument (Bio-Rad). The PCR program was initiated by 5 min at 94 °C, then followed by 45 cycles of 94 °C for 10 s and 60 °C for 30 s. The no-template and positive controls were the same as described above for the fruit experiment. Relative expression levels were calculated using the 1 MAA (month after anthesis) young fruit set at 1.
End-point PCR
End-point PCR was used for confirming the real-time PCR results. Both primers and cDNA used for real-time analysis were also used for end-point PCR. The PCR mixture (25 μl) comprised 2.5 μl of 10×PCR buffer, 0.5 μl of 10 mM dNTP, 1 μl of each primer (10 μM), 1 μl of 50 mM MgCl2, 0.2 μl of Invitrogen Platinum® Taq (Invitrogen), 1 μl of cDNA, and 17.8 μl of PCR-grade water. PCR was performed on a Mastercycler PCR machine (Eppendorf) with the following protocol: 94 °C for 5 min, followed by 28 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 10 s, and a final extension at 72 °C for 6 min. AdEIL1 transcripts were much more abundant and so only 25 cycles were used for this gene. A 9 μl aliquot of PCR products plus 1 μl of 10×loading dye were analysed on 1% agarose gels stained with ethidium bromide, and the results were recorded by photography with fixed parameters (Bio-Rad).
Results
Gene isolation and analysis
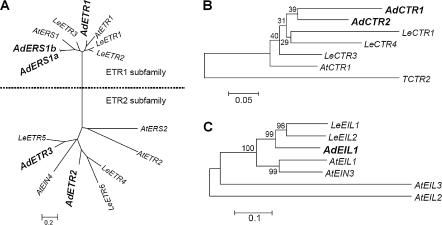
The components of three different levels of the ethylene signal transduction pathway were characterized from ripening kiwifruit. Five ETR1-like and two CTR1-like sequences were isolated from the HortResearch Actinidia EST database. Five receptor gene family members were designated as AdERS1a (EU170626), AdERS1b (EU170627), AdETR1 (EU170628), AdETR2 (EU170629), and AdETR3 (EU170630) based on a phylogenetic tree (Fig. 1A). Phylogenetic analysis of deduced amino acid sequences suggested that AdERS1a, AdERS1b, and AdETR1 could be clustered into the ETR1 subfamily, while AdETR2 and AdETR3 belonged to the ETR2 subfamily (Fig. 1A). As with tomato, there was no ERS2-type ethylene receptor detected in kiwifruit. AdERS1a and AdERS1b were 95% identical based on amino acid sequences in alignable regions. Another small family consisted of two putative CTR1-like genes, designated as AdCTR1 (EU170631) and AdCTR2 (EU170632). AdCTR1 and AdCTR2 were similar to each other at 75% amino acid identity. Although the short sequence length of AdCTR1 precludes a comparison with other gene sequences, AdCTR2 was 83% identical with LeCTR1 and 88% with LeCTR4 (Fig. 1B). Using a degenerate oligonucleotide primer, an EIN3-like gene, AdEIL1 (EU170633), was cloned from ripening kiwifruit (Fig. 1C).
Fig. 1.
Phylogenetic analysis of AdETRs (A), AdCTRs (B), and AdEIL1 (C). The amino acid sequences were analysed with Vector NTI (v. 9.0.0, Invitrogen) and the phylogenetic tree constructed with MEGA (v. 3.1) using a bootstrap test of phylogeny with minimum evolution test and default parameters. The amino acid sequences of Arabidopsis and tomato were obtained from the NCBI (National Center for Biotechnology Information) database, and accession numbers are as follows: AtETR1 (AAA70047), AtERS1 (NP_181626), AtETR2 (NP_188956), AtERS2 (AAC62209), AtEIN4 (AAD02485), AtCTR1 (AAA32780), AtEIN3 (NP_188713), AtEIL1 (NP_180273), AtEIL2 (NP_197611) and AtEIL3 (NP_177514) in Arabidopsis; LeETR1 (AAC02213), LeETR2 (AAC02214), LeETR3 (AAC49124), LeETR4 (AAU34076), LeETR5 (AAD31397), LeETR6 (AAL86614), LeCTR1 (AAL87456), TCTR2 (CAA06334), LeCTR3 (AAR89820), LeCTR4 (AAR89822), LeEIL1 (AAK58857) and LeEIL2 (AAK58858) in tomato.
Tissue specificity
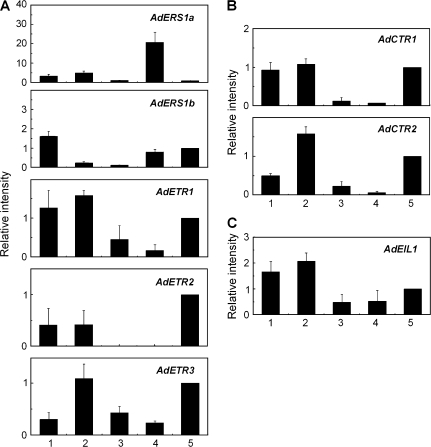
Tissue-specific expression analysis revealed that the genes were differentially expressed in various kiwifruit tissues (Fig. 2). Within the ethylene receptor family, AdETR1, AdETR2, and AdETR3 had similar expression patterns, with transcripts strongly expressed in root, stem, and fruit tissues, and more weakly in petals and leaves, although AdETR3 transcripts were also low in roots (Fig. 2A). The closely related AdERS1a and AdERS1b genes showed different expression patterns, with AdERS1a higher in petal tissue and much more abundant, and AdERS1b expressed at the highest level in roots and at a low level in stem and leaf tissue.
Fig. 2.
Expression of different components of the ethylene signalling pathway in various kiwifruit tissues: 1, root; 2, stem; 3, leaf; 4, petal; 5, fruitlet 1 month after anthesis (1 MAA). Real-time PCR was used to analyse AdETR (A), AdCTR (B), and AdEIL1 (C) expression patterns. Each column height indicates relative mRNA abundance, and 1 MAA (5) was set to 1. Error bars on each column indicate SEs from three replications.
The two CTR1-like genes exhibited similar expression patterns (Fig. 2B), and AdEIL1 transcripts were more abundant in root and stem tissue (Fig. 2C). None of the genes was specifically expressed in the fruit.
Expression patterns during fruit development
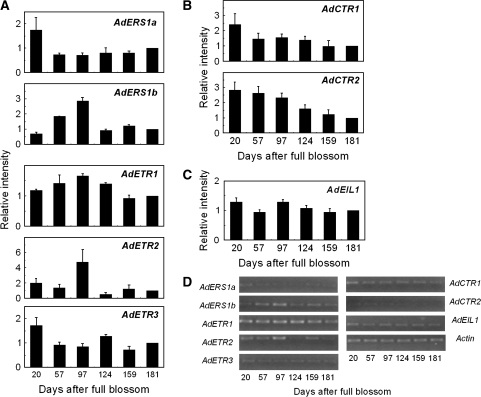
During fruit development from 20 DAFB through to maturity (181 DAFB), transcripts levels of AdERS1a and AdETR3 were similar, with a relatively high abundance in fruit at the early stages of development (20 DAFB) and then lower but constant expression until 181 DAFB (Fig. 3A). AdERS1b and AdETR2 had strong peaks in expression at 97 DAFB. AdETR1 was the most constantly expressed throughout fruit development.
Fig. 3.
Expression of different components of the ethylene signalling pathway during kiwifruit development. Real-time PCR was used to analyse AdETR (A), AdCTR (B), and AdEIL1 (C) expression patterns. Each column height indicates relative mRNA abundance, and 181 DAFB was set to 1. Error bars on each column indicate SEs from three replications. (D) End-point PCR analysis of gene expression.
The two members of the CTR1-like gene family showed similar decreasing expression patterns (Fig. 3B), and AdEIL1 was constitutively expressed throughout development (Fig. 3C). End-point PCR results confirmed the real-time PCR results (Fig. 3D).
Fruit ripening
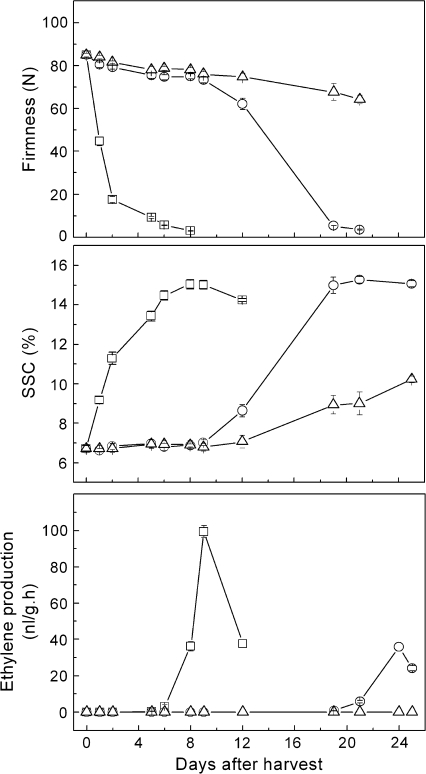
Fruit held at 20 °C softened gradually after harvest, reaching an ‘eating-ripe’ stage of firmness of ∼10 N by 19 d (Fig. 4). This was concomitant with an increase in soluble solids that peaked at about 19 d, followed by the climacteric peak in ethylene production. The rates of fruit softening and soluble solids accumulation were substantially faster in ethylene-treated fruit, reaching the same ripe stage by ∼8 d along with a climacteric peak at 9 d. In contrast, treatment with 1-MCP slowed down fruit softening, with only a slight decrease from 85 N to 61.3 N after 25 d at 20 °C; SSC also increased at about this time. 1-MCP inhibited ethylene production, and no ethylene was detectable from 1-MCP-treated fruit over the whole experimental period.
Fig. 4.
Effects of exogenous C2H4 and 1-MCP on post-harvest kiwifruit at 20 °C. Two different batches of fruit were treated with ethylene (100 μl l−1, open squares) or 1-MCP (0.5 μl l−1, open triangles) for 24 h. The third batch of fruit was sealed in a similar container of the same volume for 24 h as a control (open circles). All containers had an air circulation system and lime to absorb CO2. After treatment, fruit were held at 20 °C. Harvest day is described as 0 d. Error bars indicate SEs from 10 replications.
Expression of ETR family genes in ripening kiwifruit flesh
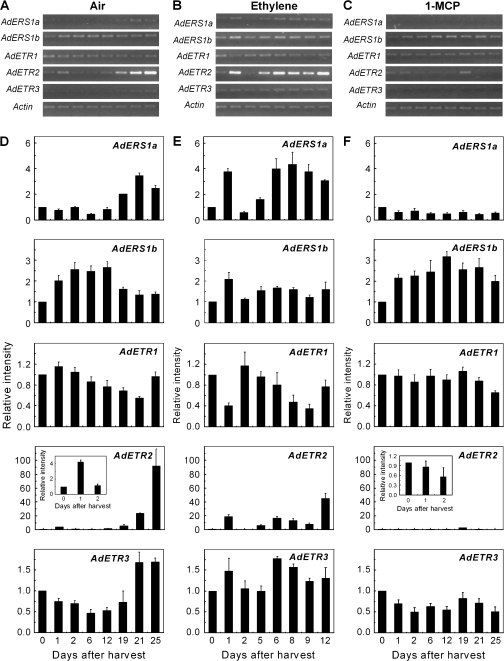
The transcript abundance of AdERS1a, AdETR2, and AdETR3 increased concomitantly with the increase in ethylene production at ∼19 d (Fig. 5A, D). Levels of AdETR2 were substantially greater than those of the other two genes. Expression of AdETR1 followed an opposite pattern, decreasing during ripening, except at the post-climacteric stage where fruit would be becoming senescent (Fig. 5D). Transcripts of AdERS1b developed higher levels up to 12 d and then they decreased to stable levels for the rest of the ripening period.
Fig. 5.
Expression of the AdETR gene family in kiwifruit flesh during fruit ripening at 20 °C. Gene transcript accumulation was evaluated by real-time PCR (D, E, F) and end-point PCR (A, B, C). Fruit were held at 20 °C and treated with air (A, D), 100 μl l−1 ethylene (B, E), or 0.5 μl l−1 1-MCP (C, F). Harvest time (0 d) was set at 1. Error bars indicate SEs from three replications.
The three genes, AdERS1a, AdETR2, and AdETR3, which showed increases associated with ethylene production in untreated fruit, were also the most responsive to ethylene treatment, all showing a transient increase 1 d after the treatment (Fig. 5B, E). The largest stimulation (∼20-fold) was with AdETR2, which also had the largest increase in the control treatment. AdETR1 was transiently down-regulated by ethylene 1 d after treatment, but then the expression patterns from day 2 onwards were similar to those from fruit in air (Fig. 5B, E).
The association of gene expression with ethylene production in the control fruit was confirmed by the 1-MCP treatment, which substantially inhibited the increases in AdERS1a, AdETR2, and AdETR3 transcript levels towards the end of the ripening period (Fig. 5C, F). The data suggest that the transient ethylene effects were consistent with the general association of the various genes with ethylene production. 1-MCP also inhibited the 1 d increase in AdETR2 transcript levels found in the control fruit. Relatively higher expression levels in 1-MCP-treated fruit were only found with AdERS1b. End-point PCR data confirmed the real-time results (Fig. 5A–C).
Expression of CTR1-like genes and AdEIL1 in ripening kiwifruit flesh
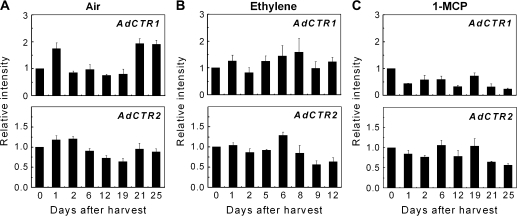
The transcript abundance of AdCTR1 increased with the ethylene climacteric (Fig. 6) but was unresponsive to the external ethylene treatment. AdCTR2 was relatively unresponsive during fruit ripening, showing no response to exogenous ethylene and 1-MCP treatments.
Fig. 6.
Expression of the AdCTR gene family in kiwifruit flesh during fruit ripening at 20 °C. Gene transcript accumulation was evaluated by real-time PCR (A, B, C). Fruit were held at 20 °C and treated with air (A), 100 μl l−1 ethylene (B), or 0.5 μl l−1 1-MCP (C). Harvest time (0 d) was set at 1. Error bars indicate SEs from three replications.
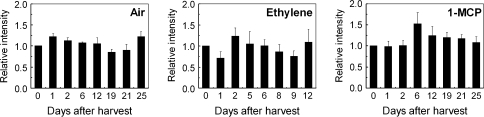
The expression pattern of AdEIL1 (Fig. 7) was relatively unchanged during ripening. Ethylene slightly decreased transcript levels after 1 d.
Fig. 7.
Expression of AdEIL1 in ripening kiwifruit flesh at 20 °C. The histograms represent the average data from three replications using real-time PCR analysis, with 0 d set at 1. Error bars indicate SEs from three replicates.
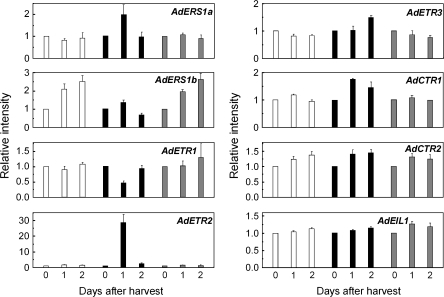
Gene expression in ripening kiwifruit core tissue
In order to confirm the expression pattern observed in the flesh, analysis of core tissue of the same fruit was included in the study (Fig. 8). Expression patterns of AdERS1a, AdERS1b, AdETR1, AdETR2, AdCTR2, and AdEIL1 at the first three time points, in the absence or presence of ethylene, were the same in core and flesh. However, there were some differences in expression with AdETR3 and AdCTR1. There was a delay in the response to ethylene with AdETR3 compared with that in the flesh, and AdCTR1 levels in the core were enhanced by the ethylene treatment while there was no response in the flesh. 1-MCP had no effect on any gene expression in the core.
Fig. 8.
Expression of different components of the ethylene signalling pathway in ripening kiwifruit core tissue. White histograms represent controls, black histograms a 100 μl l−1 ethylene treatment, and grey histograms treatment of fruit with 0.5 μl l−1 1-MCP. 0 d for each treatment was set as 1, and error bars indicate SEs from three replications.
Discussion
Analysis of the Actinidia EST database showed the presence of at least five receptor genes, and these were grouped into two subfamilies consistent with the subdivision of receptor genes for Arabidopsis and tomato. As in tomato, there was no ERS2-type ethylene receptor, but AdERS1b had 79% homology with AtERS1 and 80% with LeETR3 at the amino acid levels. AdCTR1 and AdCTR2 were closely matched with LeCTR1. An EIN3-like gene, AdEIL1, that was isolated from ripening kiwifruit, like other LeEIL genes, could be grouped with AtEIN3/AtEIL1. Although these components could all be separated into different groups or subfamilies by deduced amino acid sequences, it is not possible to predict gene function.
None of these kiwifruit genes was fruit specific, confirming data from other species. The most notable result was the consistently higher expression, with the exception of AdERS1a, in roots, stems, and fruit. The different pattern for AdERS1a suggests a specific role in flowers, and accentuates the differences between the closely homologous ERS genes. These differences are noted below for fruit development and ripening as well. The low transcript abundance in leaf and petal tissue of AdETR1, AdETR2, and AdETR3 shows similarities with data from tomato LeETR4 and LeETR5 genes (Tieman and Klee, 1999). The patterns of expression in different tissues found here are generally similar to those found in other species such as Arabidopsis and tomato for both the receptor family and CTR-like genes (Hua et al., 1998; Lashbrook et al., 1998; Tieman and Klee, 1999; Adams-Phillips et al., 2004). These patterns match the multiple functions of ethylene in higher plants (Bleecker and Kende, 2000).
The expression patterns of the receptor genes were different during fruit development, and can be associated with two specific stages of fruit development where ethylene might be involved. AdERS1a and AdETR3 were expressed more strongly in the first 3–4 weeks of development. This is a pattern similar to that found in plum (PdETR1 and PdERS1; Fernández-Otero et al., 2007), strawberry (FaETR1 and FaERS1; Trainotti et al., 2005), and persimmon (DkERS1; Pang et al., 2007) fruit. The major period of cell division in kiwifruit occurs in this period, suggesting an involvement of ethylene at this growth stage. Kiwifruit fruit growth is rapid after this period, mean weights being 61.27 g at 97 DAFB and 87.16 g at 181 DAFB in the present material. Generally, about two-thirds of the fruit volume and weight is gained over the first 10 weeks of growth (Beever and Hopkirk, 1990). Expression of AdERS1b and AdETR2 peaked at 97 DAFB, suggesting that these genes may be associated with an ethylene involvement in this rapid development, particularly involving cell expansion. A closely related gene in muskmelon, CmERS1, was also maximally expressed at a similar stage of fruit development (Sato-Nara et al., 1999). The higher expression levels of AdCTR1 and AdCTR2 in the early stages of fruit development coincided with the maximal expression of the receptor genes.
During fruit ripening, three of the receptor genes, AdERS1a, AdETR2, and AdETR3, showed a strong association with the ethylene climacteric. This was confirmed by the reduction by 1-MCP of this climacteric-associated stimulation and the positive transcript response to external ethylene. In the latter case, there was a transient stimulation as well as an increase with the climacteric. These data fit with those from other species such as tomato, where NR and LeETR4 also increase with ripening (Wilkinson et al., 1995; Kevany et al., 2007). In NR antisense plants, fruit ripened normally, but down-regulation of LeETR4 resulted in accelerated ripening, suggesting that NR is only associated with ripening whereas LeETR4 is an important regulator of ripening (Tieman et al., 2000). There are no data on specific function, but AdETR2 and AdETR3 are closer to LeETR4, and AdERS1a is closer to NR, and further fundamental analysis would determine whether the former have a stronger regulatory role.
It is notable that AdETR2, as distinct from AdERS1a and AdETR3, was markedly stimulated at the later post-climacteric stage, shown in both control and ethylene-treated fruit. It was also expressed at much higher levels, and together this suggests it has a stronger response to ethylene and perhaps is associated with senescence processes rather than those of earlier ripening.
One of the most interesting results from the present analysis was the down-regulation of AdETR1 during ripening and particularly in response to ethylene. This result was confirmed in transcripts from the core tissue. Similar results have been shown in two other studies. Ciardi et al. (2000) showed that LeETR2 levels were reduced in response to ethylene in tomato leaves, in contrast to an increase in NR and LeETR4 levels, and Kevany et al. (2007) showed similar results in tomato fruit, although these were not large. These results raise the possibility that AdETR1 has a different response to ethylene from the other receptor genes in kiwifruit. In tomato, the general increases found in receptor gene expression seem to be counter-intuitive to the concept of the need for increased sensitivity, since increasing receptor expression might increase the level of receptor-based suppression of the pathway through CTR-like genes, according to the negative regulator model. However, increased receptor levels would provide greater sensitivity for ethylene binding as well. Because unbound ethylene receptors are negative regulators of the ethylene response, the down-regulation of AdETR1 suggests that this may help activate the signal transduction pathway, making it more responsive. This is then coupled with greater binding capacity from up-regulation of other members of the receptor family, to stimulate ripening in the presence of adequate levels of ethylene. In this scenario, AdETR1 may act as the principal suppressor of the pathway, whereas AdERS1a, AdETR2, and AdETR3 may play a more prominent role in activating the signal transduction pathway once ethylene binding takes place. This fits with the concept of receptor renewal being critical in ethylene response since, with a half dissociation time of ∼12 h for AtETR1 and AtETR2 (O'Malley et al., 2005), new receptor synthesis is needed to amplify the signal. Recent results with tomato suggest that reduction in levels of LeETR4 and LeETR6 in antisense lines can result in early fruit ripening (Kevany et al., 2007). Exposure to ethylene caused a reduction in receptor protein concomitant with increased gene expression, showing that ethylene binding stimulated receptor protein degradation. It is possible that this may also be the case with AdETR1 in response to ethylene; down-regulation could presumably result in less receptor protein and subsequent de-repression of the pathway.
Another significant finding from the present analysis was the difference in expression patterns between the two ERS genes. AdERS1b was very similar in amino acid sequence to AdERS1a, but their expression patterns were quite different in developing and ripening kiwifruit and in response to ethylene. Whereas AdERS1a showed patterns similar to those of AdETR2 and AdETR3, AdERS1b exhibited no ethylene response, but its expression pattern was strongly associated with fruit softening. This trend was consistent in control and ethylene-treated fruit, and also in 1-MCP-treated fruit where relatively higher expression levels were maintained concomitant with delayed softening. This particular expression pattern has not been shown in other published fruit data. During late ripening or senescence stages of tomato (Lashbrook et al., 1998), melon (Sato-Nara et al., 1999), and peach (Dal Cin et al., 2006), where softening is presumably increasing, only increases in expression of receptor genes have been found. Except in pear fruit (El-Sharkawy et al., 2003), these two particular genes have not been distinguished. In the latter case, expression differences in the closely related PcERS1a and PcERS1b were not followed. Despite the close homology with AdERS1a, the relative insensitivity to ethylene of AdERS1b and its pattern associated with softening invites the speculation that it might have a separate involvement in promoting some aspect of later fruit ripening or softening.
Published data on the CTR-like genes show a range of expression changes with ripening and in response to ethylene. For instance, PcCTR1 expression increased during pear fruit ripening and in response to ethylene treatment (El-Sharkawy et al., 2003). MdCTR1 and PpCTR1 showed little change in expression during apple and peach fruit ripening, with different responses to 1-MCP (Dal Cin et al., 2006). PpCTR1 showed no response to 1-MCP, and MdCTR1 was suppressed at a late stage in ripening. Generally, the present data on the two CTR-like genes are more similar to those of Arabidopsis in showing low levels of transcriptional response to ethylene than the four tomato genes which have been shown to be transcriptionally more active (Adams-Philips et al., 2004). To date, no more than two CTR-like genes can be found in our kiwifruit database.
AdEIL1 also exhibited no response to ethylene and 1-MCP, and was constitutively expressed during fruit development and ripening, except in root and stem tissues. Similar results have been found with VrEIL and NtEIL genes in mung bean (Lee and Kim, 2003) and tobacco (Mariani and Weterings, 2003), respectively. However, ACC treatment results in accumulation of EIN3 protein in Arabidopsis (Yanagisawa et al., 2003), and EIN3/EIL genes can bind the primary ethylene response element (PERE) motif, which is part of the promoter of some ERFs, such as AtERF1 (Solano et al., 1998), and ripening-related genes such as E4 (Montgomery et al., 1993). AdEIL1 would be expected to have some similar role in kiwifruit.
In many other fruit, the ethylene signal transduction pathway genes have been shown to be differentially expressed in different fruit parts. For example, CmERS1 and CmETR1 showed slightly different patterns in the outer, mid, and inner parts of melon fruit flesh (Sato-Nara et al., 1999), and there were also differences in ETR gene expression between epicarp and mesocarp tissue of plum fruit (Fernández-Otero et al., 2007). In kiwifruit, gene expression patterns in the core reflected those of the flesh, with some delay in up- or down-regulation which might be due to delays in effects of ethylene and 1-MCP in penetrating the fruit tissue. The value in comparing core data is that the major patterns in expression have been confirmed in the two tissue types.
In conclusion, expression studies of the five ethylene receptor genes and two CTR1-like genes encoding the first two levels of ethylene signal transduction in ripening kiwifruit, show different expression patterns, and confirm that there is a complex interplay among the receptor genes in terms of ethylene response. In particular, one gene (AdETR1) is down-regulated by ethylene, with implications for responsiveness of the ethylene signal transduction pathway. The marked expression differences between two closely related genes, AdERS1a and AdERS1b, suggest a difference in roles in terms of ripening. These differences in gene expression patterns, in comparison with some of the published data in other fruit species, help show why kiwifruit are particularly sensitive to ethylene, and why this species may be a good model for further study of control of the ethylene signal transduction pathway.
Acknowledgments
We wish to thank Dr J Burdon for advice, Mr R Craig for providing fruit, and Dr M Montefiori for collecting samples during fruit development. This research was supported by the National Nature Science Foundation in China (no. 30571248), The University Doctoral Foundation of China (no. 20040335022), and the 111 project (no. B06014). It is part of a collaborative programme between Zhejiang University, China, and HortResearch, NZ.
Glossary
Abbreviations
- CR
coding region
- DAFB
days after full bloom
- EST
expressed sequence tag
- MAA
month after anthesis
- PERE
primary ethylene response element
- SSC
soluble solids concentration
- UTR
untranslated region
- 1-MCP
1-methylcyclopropene
References
- Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J. Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Molecular Biology. 2004;54:387–404. doi: 10.1023/B:PLAN.0000036371.30528.26. [DOI] [PubMed] [Google Scholar]
- Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. Journal of Experimental Botany. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Antunes MDC, Sfakiotakis EM. Ethylene biosynthesis and ripening behaviour of ‘Hayward’ kiwifruit subjected to some controlled atmospheres. Postharvest Biology and Technology. 2002;26:167–179. [Google Scholar]
- Beever DJ, Hopkirk G. Fruit development and fruit physiology. In: Warrington IJ, Weston GC, editors. Kiwifruit: science and management. Auckland: New Zealand Society for Horticultural Science; 1990. pp. 97–126. [Google Scholar]
- Blankenship SM, Dole JM. 1-Methylcyclopropene: a review. Postharvest Biology and Technology. 2003;28:1–25. [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annual Review of Cell and Developmental Biology. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Boquete EJ, Trinchero GD, Fraschina AA, Vilella F, Sozzi GO. Ripening of ‘Hayward’ kiwifruit treated with 1-methylcyclopropene after cold storage. Postharvest Biology and Technology. 2004;32:57–65. [Google Scholar]
- Cancel JD, Larsen PB. Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in arabidopsis. Plant Physiology. 2002;129:1557–1567. doi: 10.1104/pp.003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Stadler R. Ethylene hormone receptor action in Arabidopsis. BioEssays. 2001;23:619–627. doi: 10.1002/bies.1087. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen Y-F, Etheridge N, Schaller GE. Ethylene signal transduction. Annals of Botany. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardi JA, Tieman DM, Lund ST, Jones JB, Stall RE, Klee HJ. Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiology. 2000;123:81–92. doi: 10.1104/pp.123.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin V, Rizzini FM, Botton A, Tonutti P. The ethylene biosynthetic and signal transduction pathways are differently affected by 1-MCP in apple and peach fruit. Postharvest Biology and Technology. 2006;42:125–133. [Google Scholar]
- El-Sharkawy I, Jones B, Li ZG, Lelièvre JM, Pech JC, Latché A. Isolation and characterization of four ethylene perception elements and their expression during ripening in pears (Pyrus communis L.) with/without cold requirement. Journal of Experimental Botany. 2003;54:1615–1625. doi: 10.1093/jxb/erg158. [DOI] [PubMed] [Google Scholar]
- Fernández-Otero CI, Torre FDL, Iglesias R, Rodríguez-Gacio MC, Matilla AJ. Stage- and tissue-expression of genes involved in the biosynthesis and signalling of ethylene in reproductive organs of damson plum (Prunus domestica L. subsp. insititia) Plant Physiology and Biochemistry. 2007;45:199–208. doi: 10.1016/j.plaphy.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. The Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Dal Cin V, Klee HJ. Ethylene receptor degradation controls the timing of ripening in tomato fruit. The Plant Journal. 2007;51:458–467. doi: 10.1111/j.1365-313X.2007.03170.x. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim HO, Hewett EW, Lallu N. Softening and ethylene production of kiwifruit reduced with 1-methylcyclopropene. Acta Horticulturae. 2001;553:167–170. [Google Scholar]
- Koukounaras A, Sfakiotakis E. Effect of 1-MCP prestorage treatment on ethylene and CO2 production and quality of ‘Hayward’ kiwifruit during shelf-life after short, medium and long term cold storage. Postharvest Biology and Technology. 2007;46:174–180. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ. Differential regulation of the tomato ETR gene family throughout plant development. The Plant Journal. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Leclercq J, Adams-Phillips LC, Zegzouti H, Jones B, Latché A, Giovannoni JJ, Pech J-C, Bouzayen M. LeCTR1, a tomato CTR1-like gene, demonstrates ethylene signalling ability in Arabidopsis and novel expression pattern in tomato. Plant Physiology. 2002;130:1132–1142. doi: 10.1104/pp.009415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Kim WT. Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiology. 2003;132:1475–1488. doi: 10.1104/pp.103.022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hackett RM, Payton S, Grierson D. A tomato sequence, TCTR2 (accession no. AJ005077), encoding an Arabidopsis CTR1 homologue. Plant Physiology. 1998;117:1126. [Google Scholar]
- Mariani IRC, Weterings K. Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. Journal of Experimental Botany. 2003;54:2239–2244. doi: 10.1093/jxb/erg240. [DOI] [PubMed] [Google Scholar]
- McDonald B, Harman JE. Controlled-atmosphere storage of kiwifruit. I. Effect on fruit firmness and storage life. Scientia Horticulturae. 1982;17:113–123. [Google Scholar]
- Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proceedings of the National Academy of Sciences, USA. 1993;90:5939–5943. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Rodriguez FI, Esch JJ, Binder BM, O'Donnell P, Klee HJ, Bleecker AB. Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. The Plant Journal. 2005;41:651–659. doi: 10.1111/j.1365-313X.2004.02331.x. [DOI] [PubMed] [Google Scholar]
- Pang JH, Ma B, Sun H-J, Ortiz GI, Imanishi S, Sugaya S, Gemma H, Ezura H. Identification and characterization of ethylene receptor homologs expressed during fruit development and ripening in persimmon (Diospyros kaki Thumb.) Postharvest Biology and Technology. 2007;44:195–203. [Google Scholar]
- Qu X, Hall BP, Gao ZY, Schaller GE. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutation in the ethylene receptors ETR1 and ERS1. BMC Plant Biology. 2007;7:3. doi: 10.1186/1471-2229-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasori A, Ruperti B, Bonghi C, Tonutti P, Ramina A. Characterization of two putative ethylene receptor genes expressed during peach fruit development and abscission. Journal of Experimental Botany. 2002;53:2333–2339. doi: 10.1093/jxb/erf097. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Nara K, Yuhashi K-I, Higashi K, Hosoya K, Kubota M, Ezura H. Stage- and tissue-specific expression of ethylene receptor homolog genes during fruit development in muskmelon. Plant Physiology. 1999;119:321–329. doi: 10.1104/pp.120.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signalling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes and Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Endo A, Ohkawa H. Influence of time from harvest to 1-MCP treatment of apple fruit quality and expression of genes for ethylene biosynthesis enzymes and ethylene receptors. Postharvest Biology and Technology. 2007;43:28–35. [Google Scholar]
- Tieman DM, Klee HJ. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiology. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proceedings of the National Academy of Sciences, USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L, Bonghi C, Ziliotto F, Zanin D, Rasori A, Casadoro G, Ramina A, Tonutti P. The use of microarray μPEACH1.0 to investigate transcriptome changes during transition from pre-climacteric to climacteric phase in peach fruit. Plant Science. 2006;170:606–613. [Google Scholar]
- Trainotti L, Pavanello A, Casadoro G. Different ethylene receptors show an increased expression during the ripening of strawberries: does such an increment imply a role for ethylene in ripening of these non-climacteric fruits? Journal of Experimental Botany. 2005;56:2037–2046. doi: 10.1093/jxb/eri202. [DOI] [PubMed] [Google Scholar]
- Whittaker DJ, Smith GS, Gardner RC. Expression of ethylene biosynthetic genes in Actinidia chinensis fruit. Plant Molecular Biology. 1997;34:45–55. doi: 10.1023/a:1005789220668. [DOI] [PubMed] [Google Scholar]
- Wiersma PA, Zhang H, Lu C, Quail A, Toivonen PMA. Survey of the expression of genes for ethylene synthesis and perception during maturation and ripening of ‘Sunrise’ and ‘Golden Delicious’ apple fruit. Postharvest Biology and Technology. 2007;44:204–211. [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Xu ZC, Ikoma Y, Yano M, Ogawa K, Hyodo H. Varietal differences in the potential to produce ethylene and gene expression of ACC synthase and ACC oxidase between ‘Kui mi’ and ‘Hong xin’ of Chinese kiwifruit. Journal of the Japanese Society for Horticultural Science. 1998;67:204–209. [Google Scholar]
- Yanagisawa S, Yoo S-D, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature. 2003;425:521–525. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- Yau CP, Wang L, Yu M, Zee SY, Yip WK. Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. Journal of Experimental Botany. 2004;55:547–556. doi: 10.1093/jxb/erh055. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen K, Bowen J, Allan A, Espley R, Karunairetnam S, Ferguson I. Differential expression within the LOX gene family in ripening kiwifruit. Journal of Experimental Botany. 2006;57:3825–3836. doi: 10.1093/jxb/erl151. [DOI] [PubMed] [Google Scholar]