Abstract
Rice (Oryza sativa) varieties that are arsenate-tolerant (Bala) and -sensitive (Azucena) were used to conduct a transcriptome analysis of the response of rice seedlings to sodium arsenate (AsV) in hydroponic solution. RNA extracted from the roots of three replicate experiments of plants grown for 1 week in phosphate-free nutrient with or without 13.3 μM AsV was used to challenge the Affymetrix (52K) GeneChip Rice Genome array. A total of 576 probe sets were significantly up-regulated at least 2-fold in both varieties, whereas 622 were down-regulated. Ontological classification is presented. As expected, a large number of transcription factors, stress proteins, and transporters demonstrated differential expression. Striking is the lack of response of classic oxidative stress-responsive genes or phytochelatin synthases/synthatases. However, the large number of responses from genes involved in glutathione synthesis, metabolism, and transport suggests that glutathione conjugation and arsenate methylation may be important biochemical responses to arsenate challenge. In this report, no attempt is made to dissect differences in the response of the tolerant and sensitive variety, but analysis in a companion article will link gene expression to the known tolerance loci available in the Bala×Azucena mapping population.
Keywords: Arsenate tolerance, candidate genes, glutathione-S-transferase, microarray, Oryza sativa
Introduction
In the environment, arsenic (As) is present in both organic and inorganic forms; the inorganic species arsenate [As(V)] and arsenite [As(III)] are more abundant in soils compared with the organic As species monomethylarsonic acid (MMAA) and dimethylarsinic acid (DMAA) (Takamatsu et al., 1982), with the concentrations of As(III) and methylated species rising in anaerobic soils (Abedin et al., 2002). The mechanism by which plants take up the inorganic As species differs. As(V) has been shown to be taken up by the high affinity phosphate uptake system (Ullrich-Eberius et al., 1989), while As(III) uptake is thought to be through aquaporins in the roots (Meharg and Jardine, 2003). Both forms of inorganic As are toxic to plants. As(III) can react with sulphydryl groups of enzymes and proteins, which leads to loss of function and can cause cell death (Requejo and Tena, 2005). As(V) can compete with phosphate, replacing it in key molecules, including ATP (Scott et al., 1993; Meharg and Hartley-Whitaker, 2002; Quaghebeur and Rengel, 2003). Also, when As(V) enters the plant, a proportion of it is reduced to As(III), a process thought to lead to oxidative stress (Hartley-Whitaker et al., 2001). Exposure of plants to inorganic As leads to the synthesis of phytochelatins that complex As(III), these complexes being transported across the tonoplast by ABC-type cassette transporters (see, for example, Bleeker et al., 2006).
Mylona et al. (1998) have shown that maize enzymes involved in reactive oxygen scavenging [catalase, superoxide dismutase, and glutathione-S-transferase (GST)] have increased activity and increased gene expression upon As exposure. Requejo and Tena (2005), studying protein profiles, showed that 10% of the detectable proteins in maize roots were regulated (either up- or down-regulated) by As, and seven out of the 11 proteins whose identity was revealed were involved in cellular homeostasis for redox perturbation. Despite these studies, the molecular responses of plants to As have not been extensively characterized, yet a thorough analysis should provide insights into the nature of toxicity and the mechanisms of tolerance.
In mammals, inorganic As is metabolized to dimethyl arsenate [DMA(V)] which is then excreted in urine. This is achieved via repetitive reduction (from the +5 to the +3 valence state) and oxidative-methylation steps known as the Challenger pathway. Three enzymes are required, an arsenate reductase that converts As(V) to As(III), a methyltransferase that converts As(III) to monomethyl arsenate MMA(V) and converts monomethyl arsenite [MMA(III)] to DMA(V), and a monomethyl arsenate reductase which converts MMA(V) to MMA(III) (Aposhian et al., 2004; Thomas et al., 2004). While it has long been known that plants exposed to inorganic As accumulate both inorganic and organic forms (Meharg and Hartley-Whitaker, 2002), it has only recently been demonstrated that this is due to de novo synthesis of methylated species, rather than direct uptake of soil-microbially produced methylated species (Raab et al., 2005). The enzymology of methylation remains elusive in plants. The arsenate reductase from Holcus lanatus has been identified (the first in plants) (Bleeker et al., 2006), and more recently two rice genes (Os03g01770 and Os10g39860) encoding arsenate reductases have been cloned (Duan et al., 2007). The other enzymes in the pathway have not been characterized although it has been shown that plant extracts have the enzymatic ability to methylate As in vivo to MMA(V) and DMA(V) (Wu et al., 2002).
In animals, the enzymes of As(III) metabolism have recently been characterized. The methyltransferase has been cloned from a few species (Aposhian and Aposhian, 2006). It is classified as S-adenosine-L-methionine:As(III) methyltransferase, and can add a methyl group to As(III) to produce MMA(V) and to MMA(III) to make DMA(V). It has also been shown to have the ability to catalyse both intermediate reduction steps using thioredoxin or glutaredoxin as the reductant (Li et al., 2005). The gene is part of the glutathione transferase superfamily. Another gene in the pathway has been characterized in animals, described as monomethyl arsenate reductase (Aposhian et al., 2004), and it has been proposed that this enzyme can reduce all +5 valence state arsenicals [(As(V), MMA(V), and DMA(V)] to the +3 valence state [As(III), MMA(III), and DMA(III)]. In other words, it is also an arsenate reductase. This enzyme has an absolute requirement for reduced glutathione and it has been identified as an Omega class GST (Aposhian et al., 2004). This class of GST evolved in animals after their split with plants and do not, therefore, exist in plants, although plants do have two plant-specific classes of GST (the Phi and Tau classes with 16 and 40 genes each in rice), the functions of which are not known (Soranzo et al., 2004).
Recently, an alternative pathway for As metabolism has been suggested, with the +5 valence state as the end-product of the methylation reaction and involving conjugation of glutathione (Hayakawa et al., 2005). This suggests that the Challenger pathway might not be the only route for As methylation.
In this study, the alterations in gene expression of two varieties of rice when exposed to 13.3 μM sodium arsenate for 1 week were examined. The analysis focuses on genes that are commonly differentially regulated in both varieties. A large number of genes involved in glutathione synthesis, metabolism, and transport, and genes potentially involved in arsenate methylation suggests that glutathione conjugation and the Challenger-like pathway of arsenate methylation may be important biochemical responses to arsenate challenge.
Materials and methods
Hydroponic growth of rice plants
The two rice varieties Azucena (japonica) and Bala (indica) were germinated for 3 d at 37 °C and then floated on alkathene beads within 100 ml beakers filled with either phosphate-free nutrient solution containing 0.1 mM Mg2+ and SO42–, 0.2 mM Ca2+ and K+, and 0.6 mM NO3– (control), or the same nutrient solution supplemented with di-sodium hydrogen arsenate (treatment) at a concentration of 13.3 μM. The seedlings were grown in controlled conditions at 25 °C with 12 h day length (300 μmol m−2 s−1 PAR). After 1 week, the maximum length of the main root of plants was measured. The tolerance index was calculated as the percentage of root length in arsenate compared with control.
An experiment examining the root growth of Azucena and Bala when grown in arsenate was conducted using a similar methodology to that described above with the following amendments: half-strength Yoshida nutrient growth solution without phosphorus was used instead (Yoshida et al., 1976), and root lengths were measured on days 0, 2, 3, 5, and 7.
Microarray gene expression
For microarray analysis, the total root systems were harvested from control plants and plants treated with 13.3 μM sodium arsenate; growth conditions were as described above. A total of 100 mg of root material, pooled from four separate plants, was harvested for RNA extraction using the TRIzol method (Invitrogen) followed by further purification and DNase treatment using the RNeasy clean up system (Qiagen). RNA was processed for use with the Affymetrix GeneChip Rice Genome array according to the manufacturer's protocol. Briefly, 5 μg of total RNA was used in the one-cycle cDNA synthesis reaction. After the generation of double-stranded cDNA, an in vitro transcription reaction was performed to generate biotinylated cRNA. cRNA quality was assessed for fragment length using a bioanalyzer (Agilent 2100). Hybridization, washing, staining, and scanning procedures of the Affymetrix Test 3 arrays and Affymetrix GeneChip Rice Genome arrays were carried out by the Microarray Core Facility of the Institute of Medical Sciences, University of Aberdeen, UK, as described in the Affymetrix technical manual. Three independent replicated experiments were carried out for all four conditions (Azucena with/without sodium arsenate and Bala with/without sodium arsenate). Microarray data sets were deposited in the GEO public database, series accession number GSE4471.
Analysis of array data
Expression analysis was initially carried out using the Microarray analysis software 5.0 (MAS5.0) from the Affymetrix Gene Chip Operating software (GCOS, version 1.3). Further analysis was performed using the Bioconductor package (Gentleman et al., 2004) in R (Ihaka and Gentleman, 1996). The data were normalized using the modified robust multiarray average (RMA) (Irizarry et al., 2003) method GCRMA (Wu et al., 2004) which takes into account the GC content when performing RMA normalization. Differential gene expression was measured using the LIMMA linear model (Smyth, 2004) and a Benjamini and Hochberg (1995) correction of the P-value. Probe sets were only counted as differentially expressed if they met the following criteria: (i) there was a statistically significant differential expression (adjusted P-value ≤0.05); and (ii) the fold change in gene expression was ≥2.
Assignment of probe sets to genes
Probe sets were assigned to annotated genes using the web-based search tool on the National Science Foundation Rice Oligonucleotide Array Project website (www.ricearray.org). Annotation is based on the TIGR Pseudomolecule release 5 of 2007 (www.tigr.org).
Assignment of genes to biological processes
For the genes that were differentially regulated, the gene ontology classification provided in TIGR Pseudomolecule release 5 (GOSlim) was used to assign genes to a hierarchical biological process using the Web Gene Ontology Annotation Plotting tool (WEGO, http://wego.genomics.org.cn) (Ye et al., 2006). GOSlim information was available for 62% of the up-regulated genes and 68% of the down-regulated genes.
RT-PCR confirmation of Affymetrix array data
PCR primers were designed to amplify a number of genes to confirm the Affymetrix data. Primers were designed using Primer3, and purchased from Invitrogen. RNA was isolated as described above and reverse transcribed into cDNA from 2 μg of total RNA using the SuperScript™ Double-Stranded cDNA Synthesis Kit (Invitrogen). PCR was performed using 1 μl of cDNA reaction in 25 μl containing 16 mM (NH4)2SO4, 67 mM TRIS-HCl (pH 8.8), 0.01% Tween-20, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM of each primer, and 0.5 U of BioTaq polymerase (Bioline). Reactions were formed using the following cycle conditions, an initial 94 °C for 2 min, then 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by a final 5 min extension at 72 °C.
Quantitative PCR
Rice plants were grown as above except for the addition of 13.3 μM sodium arsenate either 168, 48, 24, 12, 5, 2, 1, 0.5, or 0 h before the roots were harvested.
RNA was isolated as described above and primers were designed for two genes, GSTU5 and GSTU19. The 18S gene was also amplified to normalize the RNA loading. cDNA was synthesized from 1 μg of total RNA using the ImProm-II cDNA synthesis kit (Promega) with random primers. Control reactions were performed without the reverse transcriptase enzyme to verify that no DNA contamination was present in the RNA samples. PCR amplification was performed in a 25 μl volume containing 1 μl of cDNA (for 18S reactions, cDNA was diluted 100-fold), 12.5 μl of 2× DyNAmo SYBR green (Finnzyme), and 0.25 μM of each primer. Analysis was performed in an Opticon2 (MJ research) with cycle conditions of an initial denaturing at 95 °C for 15 min, followed by 35 cycles (30 for 18S) of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s.
Results
Effect of 13.3 μM sodium arsenate on root length
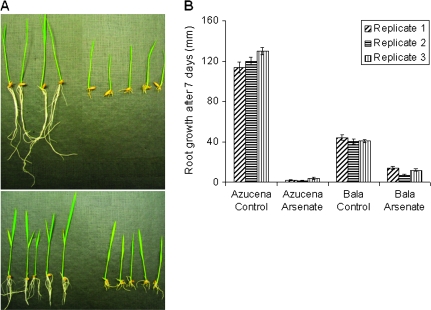
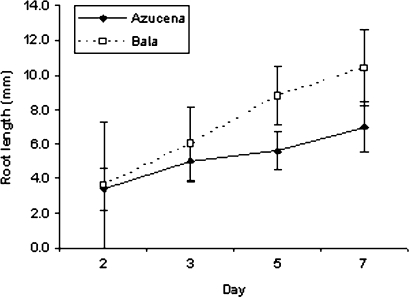
As(V) (13.3 μM) causes a dramatic decrease in root growth over a period of 7 d in both varieties (Fig. 1A). The root length measurements for the three replicate array experiments are presented in Fig. 1B. In the time-course experiments it is observed that root growth of arsenate-treated plants is linear (Fig. 2).
Fig. 1.
(A) Azucena control conditions (top left), Azucena grown in 13.3 μM sodium arsenate (top right), Bala control conditions (bottom left), and Bala grown in 13.3 μM sodium arsenate (bottom right) for 1 week showing marked inhibition of root growth. (B) Length of the roots for the three replicates used in the gene expression experiment. Bar = SE.
Fig. 2.
Root lengths of plants grown in 13.3 μM sodium arsenate. Maximum root length was measured on days 2, 3, 5, and 7. Bar = SE.
Genome-wide gene expression
Initial analysis of the microarray data indicated that an average of 43.9% of the probe sets were called present using the MAS5.0 software, indicating that 44% of genes were expressed in young hydroponically grown roots. A large number of probe sets (2304) were differentially expressed between Azucena and Bala in the control conditions. This suggests either a large degree of differential gene expression between these two rice varieties or differences in hybridization due to sequence polymorphisms in mRNA transcripts. A total of 1604 probe sets were significantly up-regulated ≥2-fold in Azucena in the presence of 13.3 μM sodium arsenate, whereas 1828 probe sets were down-regulated. In Bala, 909 probe sets were up-regulated and 935 probe sets were down-regulated. A full list of probe sets significantly differentially regulated can be found in Supplementary Tables S1–S4 at JXB online.
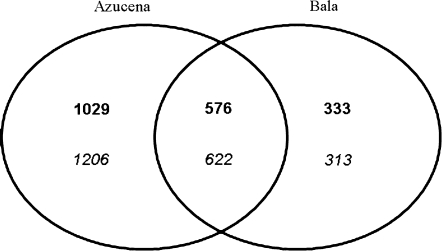
A proportion of the probe sets either up- or down-regulated were common in both Azucena and Bala (Fig. 3). A total of 576 probe sets were significantly up-regulated ≥2-fold in both Azucena and Bala, and 622 sets were significantly down-regulated at least 2-fold in both Azucena and Bala.
Fig. 3.
Venn diagram of the number of probe sets up- and down-regulated in response to arsenate in Azucena and Bala. The number of probe sets which were up-regulated (bold) and the number of probe sets which were down-regulated (italic) with an adjusted P-value of ≤0.05 and a ≥2-fold change in gene expression.
There were three probe sets which were significantly regulated in both varieties, but in opposing directions. The probe set Os.9396.1.S1_at (Os03g48810, permease 1 gene) is up-regulated in Bala whereas in Azucena it is down-regulated in response to As(V). The probe sets Os.16305.1.S1_at (Os01g03330, Bowman–Birk-type bran trypsin inhibitor precursor) and Os.19472.1.S1_at (Os04g10160 cytochrome P450 CYP99A1) are down-regulated in Bala whereas they are up-regulated in the Azucena response to As(V).
Confirmation of the array data was carried out using primers designed for probe sets that were expressed in at least one of the conditions and absent in other conditions. A fourth replicate of the arsenate test was performed and cDNA synthesized from the harvested RNA was used as templates for confirmatory PCR. For all nine probe sets tested in this way, the amplification in the PCRs exactly matched the present/absent score from the array data (Supplementary Fig. S1 at JXB online).
Gene annotation
An analysis of those genes that respond to As(V) in both varieties is conducted below. Some genes have more than one probe set and, therefore, the number of responsive probe sets is greater than the number of responsive genes. Taking this into account, the number of genes significantly up-regulated ≥2-fold was 460, while 523 genes were down-regulated. The annotation of the rice genome does not give a putative function to all pseudomolecules so a large number of genes are still annotated as either ‘expressed’ or ‘hypothetical’. The ‘expressed’ genes accounted for 92 of the genes down-regulated and 80 of those up-regulated, while genes annotated as ‘hypothetical’ accounted for 18 down-regulated and eight up-regulated. A number of the probe sets on the genome array do not match or have a low match to the TIGR Pseudomolecule release 5; 59 of the up-regulated and 40 down-regulated probe sets fell into this category.
A summary of the biological processes (as assigned by WEGO) in which the numbers of genes in a category that are significantly different between up- and down-regulation is given in Table 1. This reveals that As(V)-responsive genes that are involved in heat and toxin responses, the metabolism of toxins, sulphur, amines, and organic acids, cellular and macromolecule catabolism, and nitrogen biosynthesis are predominantly up-regulated. In contrast, the As(V)-responsive genes that are involved in transport, regulation of metabolism and cell size, metabolism of phosphorus, phenylpropanoids, and aromatic compounds, cellular morphogenesis, cell growth, and responses to auxin are predominantly down-regulated.
Table 1.
Biological process categorization (from GOSlim data, using WEGO), showing only categories with a significant difference between up- and down-regulated genes
| Up-regulated | Down-regulated | Biological process |
| 27 | 48 | Transport |
| 16 | 55 | Regulation of cellular metabolism |
| 4 | 17 | Phenylpropanoid metabolism |
| 8 | 20 | Aromatic compound metabolism |
| 5 | 22 | Phosphorus metabolism |
| 10 | 1 | Sulphur metabolism |
| 15 | 6 | Amine metabolism |
| 15 | 1 | Toxin metabolism |
| 23 | 12 | Organic acid metabolism |
| 34 | 10 | Cellular catabolism |
| 14 | 5 | Macromolecule catabolism |
| 14 | 2 | Nitrogen compound biosynthesis |
| 2 | 14 | Cellular morphogenesis |
| 1 | 12 | Regulation of cell size |
| 1 | 10 | Unidimensional cell growth |
| 3 | 11 | Response to auxin stimulus |
| 22 | 6 | Response to heat |
| 20 | 2 | Response to toxin |
In order to provide a better insight into the nature of the genes that respond to As(V), the following section describes a number of notable differentially regulated genes. The largest category of responsive genes was transcription factors (25 up-regulated and 64 down-regulated) followed by protein kinases (seven up-regulated and 23 down-regulated), but with the present state of gene annotation for rice it is not yet straightforward to draw useful conclusions about these genes. A total of eight genes annotated as UDP-glucuronosyl and UDP-glucosyl transferase were up-regulated, and three were down-regulated. Gene ontology classification indicates that this class of gene is involved in xenobotic metabolism and response to toxins.
Transporter genes
The phosphate:H+ symporter gene OsPT2 (03g05640) was down-regulated. This gene was previously found to be highly expressed under phosphate starvation but expression decreased with elevated phosphate (Paszkowski et al., 2002; Güimil et al., 2005). In the present study, 13.3 μM arsenate dramatically reduced the expression of OsPT2 transcripts in rice compared with control plants, which were probably phosphate starved given that phosphate was omitted from the nutrient solution. A gene annotated as an inorganic phosphate transporter (Os10g30770) was also down-regulated. Five sulphate transporters were up-regulated (Os03g09940, Os03g09970, Os09g06499, Os08g31410, and Os09g06510). Transporters of chloride (Os02g35190 and Os08g20570), ammonium (Os01g61550), and nitrate (Os02g38230) were down-regulated. Also down-regulated were genes involved in sugar (Os02g36414 and Os04g37990), amino acid (Os04g39489), peptide (Os01g04950), and oligopeptide (Os02g46850 and Os08g38400) transport.
A total of seven multidrug and toxic compound extrusion (MATE) transporters were up-regulated (Os03g37490, Os05g48040, Os08g37430, Os08g37432, Os10g20450, Os10g20470, and Os12g03260) and two down-regulated (Os04g48290 and Os09g29284). MATE transporters cover a diverse range of functions including the transport of lipophilic cations and related compounds; they have also been linked with growth and development (Debeaujon et al., 2001). While MATE transporters do not transport glutathione-conjugated molecules, a number of proteins that do also responded to As(V). A single gene with an annotated putative function of a glutathione conjugate transporter (Os04g13210) was up-regulated. A gene annotated as a multidrug resistance-associated protein MRP2 (Os01g07870) was also up-regulated. MRPs are a subclass of ATP-binding cassette (ABC) transporters, which are involved in the transport of glutathione-conjugated compounds into the vacuole of plants for subsequent detoxification (Rea et al., 1998; Theodoulou, 2000). Another three genes with the predicted putative function of an ABC transporter family protein (Os04g49890, Os04g52900, and Os11g05700) were up-regulated. Phylogenetic analysis of these ABC transporter gene sequences revealed that they are also MRP transporters.
A number of metal transporters responded to As(V). Two genes annotated as Nramp1, which are broad range membrane-bound metal transporters (Bereczky et al., 2003), were differentially regulated; Os07g15460 was >50-fold up-regulated whereas Os07g15370 was down-regulated. In addition, two potassium (Os01g45990 and Os07g47350), one zinc (Os01g74110), and a ZIP zinc/iron transporter (Os05g10940) were down-regulated.
Two aquaporin genes were down-regulated (Os05g14240 and Os12g10280); neither exactly matches any of the previously described aquaporins (Sakurai et al., 2005). Phylogenetic analysis of the protein sequence revealed that Os05g14240 is a member of the tonoplast intrinsic protein subclass and Os12g10280 is a member of the Nod26-like intrinsic protein subclass of aquaporins. One final class of transporters that was up-regulated by As(V) were five major facilitator superfamily proteins (Os01g16260, Os03g58080, Os11g04020, Os11g05390, and Os12g03899).
Stress proteins
A number of genes involved in molecular chaperoning responded to As(V). This included 13 up-regulated heat shock proteins (Os01g04360, Os01g04370, Os01g04380, Os02g32590, Os02g54140, Os03g14180, Os03g16020, Os03g16030, Os03g60620, Os04g01740, Os06g11610, Os09g31486, and Os11g13980) and two down-regulated proteins (Os07g33350 and Os10g30180). A related class of proteins are the chaperones, of which five were up-regulated (Os03g04970, Os03g31300, Os05g46290, Os09g11250, and Os10g32550). Three other genes in the same class were up-regulated, a late embryogenesis abundant protein (Os05g46480) and two dehydrin genes (Os11g26750 and Os11g26790).
Oxidative stress
Members of two large gene families that are implicated in oxidative stress responded to As(V). A total of 12 peroxidases were down-regulated and one up-regulated, while cytochrome P450-type genes were also differentially expressed (eight up-regulated and 12 down-regulated). Two genes annotated as chloroplastic quinone-oxidoreductase (Os04g30420 and Os09g32570) were also up-regulated. This enzyme protects cells against free radicals and toxic oxygen metabolites generated by the one-electron reductions of cytochrome P450s and other enzymes, as well as exogenous free radicals and other electrophiles (Winner et al., 1997).
Glutathione S-transferases
A total of 15 GST genes (Os01g49710, Os01g49720, Os01g72150, Os01g72160, Os06g12290, Os09g20220, Os10g34020, Os10g38140, Os10g38340, Os10g38360, Os10g38470, Os10g38489, Os10g38590, Os10g38610, and Os10g38740) were up-regulated. Twelve of these genes belong to the 40-member Tau class GSTs described by Soranzo et al. (2004). While the other three have not been categorized into a GST subclass by Soranzo et al. (2004), phylogenetic analysis confirmed they cluster with the Tau class of GSTs. Two up-regulated genes are annotated as IN2-1 proteins (Os03g17470 and Os03g17480). This category of gene has been described as a Lambda class GST by Dixon et al. (2002). One GST (Os01g27390) was down-regulated, and this was a Phi class GST.
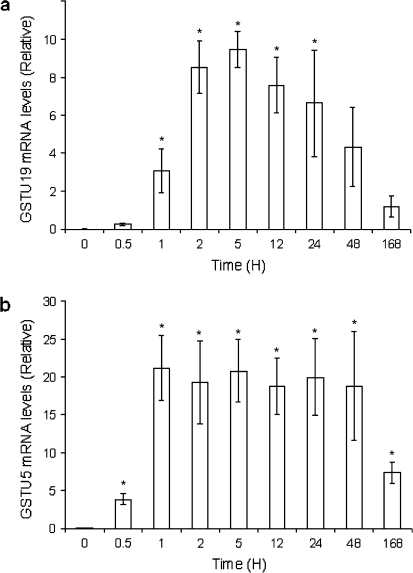
Two of the Tau class GSTs (Os09g20220 and Os10g38340) displayed a >30-fold increase in expression from the array data and these were further studied using real-time PCR (Fig. 4). Both genes were significantly up-regulated within 0.5 h, and their expression peaked at 5 h, with an increase of >250-fold for Os09g20220 and >600-fold for Os10g38340 compared with controls. For both genes the expression dropped between 2 d and 7 d, but was still substantially elevated over controls.
Fig. 4.
Gene expression of GSTU5 (A) and GSTU19 (B) in response to 13.3 μM sodium arsenate over time as detected by quantitative real-time PCR (relative to actin expression). Bar = SE.
Cell growth
A number of genes involved in cell growth and the cell cycle were down-regulated; these included two expansin genes (Os01g14660 and Os04g46650), two tubulin genes (Os03g45920 and Os03g56810), an actin gene (Os01g64630), and two microtubule genes (Os03g13460 and Os09g27700).
Methyltransferases
A gene annotated as a methyltransferase (Os02g51030) was up-regulated. This gene contains a UbiE/Coq5 family protein motif, and it has been reported that genes from bacteria and archaea with this motif are involved in As detoxification (Qin et al., 2006). A second class of methyltransferase was up-regulated (Os03g12110 and Os10g28630), both of which are annotated as homocysteine S-methyltransferase, which catalyses the formation of S-adenosyl-L-homocysteine and L-methionine from S-adenosyl-L-methionine and L-homocysteine. This enzyme is involved in S-methylmethionine synthesis (Ranocha et al., 2000), and this pathway has been proposed for sustaining a pool of soluble methionine (Mudd and Datko, 1990).
Discussion
As previously reported, 13.3 μM As(V) caused a decrease in root growth over a 7 d period in both the varieties tested in this study (Dasgupta et al., 2004). These two rice varieties have previously been shown to differ in their tolerance to As(V) (Dasgupta et al., 2004), and a companion paper (Norton et al., 2008) focuses on a three-gene model for tolerance and potential candidate genes within the tolerance loci.
The effect of As(V) at the concentration used had a constant effect on root growth (Fig. 2). The observation that root growth is constantly affected rather than an initial inhibition of root growth justifies the testing of plants exposed for 7 d rather than a shorter period of time. It is also more relevant to consider long-term exposure if the ultimate aim is to reveal mechanisms of adaptation because natural exposure to arsenic will always be prolonged.
The presented analysis of global gene expression focuses on the gene expression responses that are common in both rice varieties. To obtain the most meaningful comparison between tolerant and sensitive varieties, an additional treatment would be required in Azucena such that its root growth was inhibited by a similar amount to the inhibition observed in Bala grown in 13.3 μM arsenate. This is because the large number of genes that are uniquely responsive in one variety compared with the other may be due to differences in the level of intercellular exposure if transport of arsenicals is different between varieties, or differences in modified arsenicals if the two varieties have different biochemistry. An additional drawback of comparison is that there are such major differences in expression between varieties in control treatment (Fig. 2), which emphasizes the large degree of genetic diversity between them.
A total of 1198 probe sets responded significantly to As(V) treatment across both varieties, which represents 2.09% of probe sets present on the array (not including control probe sets). This number is not surprising, given that Requejo and Tena (2005) observed that 10% of proteins in maize exposed to As(V) at 300 μM for 24 h showed changed intensities. Despite the much lower concentration used here, the exposure time was much longer, and it has been demonstrated that longer exposure times (up to several days) generally increase the number of gene responses (Gadjev et al., 2006).
It has previously been demonstrated that As(V) presents the plant with an oxidative stress that results in responses of catalases, superoxide dismutase, and GSTs (Mylona et al., 1998). While a small number of genes involved in oxidative stress responses were among the list of up- and down-regulated genes here, the genes classically used to link stress to reactive oxygen species such as catalases, superoxide dismutase, glutathione reductases, or ascorbate peroxidase did not respond. The only agreement with Mylona et al. (1998) were the GSTs, where some of the Tau class GSTs showed remarkable changes in expression. Gadjev et al. (2006) revealed that four GSTs commonly respond to different oxidative stresses, and the present phylogenetic analysis indicates that these are all Tau class GSTs, which might indicate evidence of an oxidative component to As(V) phytotoxicity as shown by Hartley-Whitaker et al. (2001). However, it is worth noting that in animals the enzyme involved in the reduction steps of the Challenger pathway that methylates As(III) to organic MMA and DMA is a GST, but of the animal-specific Omega class (Tanaka-Kagawa et al., 2003; Schmuck et al., 2005; Zakharyan et al., 2005). Two other up-regulated GSTs identified here are of the Lambda class GSTs, which have a similar active site motif to Omega class GSTs (Board et al., 2000; Dixon et al., 2002).
The Challenger pathway that requires reduction of mono-methylated As(V) using Omega GSTs also requires a methyltransferase (Aposhian et al., 2004; Thomas et al., 2004). It may be significant, therefore, that the As(V)-responsive methyltransferase (Os02g51030) has the UbiE/Coq5 motif that is homologous with As methyltransferases from prokaryotes (Qin et al., 2006), and has some motif homology with the animal S-adenosylmethionine:arsenic(III) methyltransferase (Lin et al., 2002). The Challenger pathway has not been demonstrated in plants, but it has been shown that extracts from Agrostis capillaris produce DMA and MMA when treated with As(V) (Wu et al., 2002).
Another known direct mechanism by which GSTs combat xenobiotic stresses is the conjugation of glutathione, and it has been suggested (Liu et al., 2001) that arsenical–glutathione conjugates are produced by GSTs in rat liver epithelial cells. Liu et al. (2001) also suggest that acquired tolerance in rat epithelial cells to As(III) is afforded by induction of a GST, MRPs, and a multidrug resistance gene, with the latter two types of gene being considered to function as transporters of glutathione conjugates. It is noteworthy, therefore, that in the present array results, a total of four MRPs and one glutathione conjugate transporter were up-regulated.
If GSTs have a role in As detoxification, they can be expected to utilize reduced glutathione that is biosynthesized by two sequential ATP-dependent reactions. There is evidence from the gene list presented in Supplementary Tables S1–S4 at JXB online that enzymes involved in glutathione synthesis are differentially regulated. The first step is the synthesis of γ-glutamylcysteine from glutamate and cysteine by the enzyme γ-glutamylcysteine synthetase which is also known as glutamate–cysteine ligase. The second step is the addition of glycine to the C-terminal end by the enzyme glutathione synthase (Noctor et al., 1998). A probe set (Os.12568.2.S1_at) that was significantly up-regulated has homology to two glutamate–cysteine ligases (Os05g03820 and Os07g27790), and the former of these two genes has a unique probe set that is up-regulated. No genes annotated as glutathione synthase passed the criteria for reporting used here because there was insufficient match between probe sets and the matching pseudomolecule. However, of the three genes in the TIGR Pseudomolecule release 5 which are annotated as gluthathione synthetases, the array data suggest that two are differentially regulated. For Os12g16200 (for which only seven of the 11 Affymetrix probes matched the predicted pseudomolecule, which is why this gene is not included in the results reported), the probe set (Os.7101.1.S1_at) was 25-fold down-regulated, but for Os12g34380 (for which only five of the 11 Affymetrix probes matched the predicted pseudomolecule), the probe set (Os.12023.1.S1_at) was 3-fold up-regulated. The Os12g16200 gene is a chloroplastic isozyme, while Os12g34380 is probably cytosolic, suggesting a shift in the site of glutathione synthesis in response to As(V). This would be interesting to investigate further.
Evidence that glutathione synthesis is indeed increased under As(V) exposure is provided by the up-regulation of a number of genes involved in sulphate transport (as reported in the Results), a gene annotated as a bifunctional 3′-phosphoadenosine 5′-phosphosulphate synthetase 2 (Os04g02050) (that is involved in sulphur assimilation), and a gene annotated as a cysteine synthase (Os04g08350). In other stresses, such observations have been linked to an increased demand for glutathione (Gratao et al., 2005).
Another class of responsive genes that is involved in glutathione biochemistry is the glutaredoxins. Two glutaredoxin genes (Os01g13950 and Os02g40500) were up-regulated in As(V). These small proteins are in the same superfamily as GSTs, glutathione peroxidases, and thioredoxins (Lemaire and Miginiac-Maslow, 2004). They are involved in redox homeostasis via gluthathione (Holmgren, 1989; Porras et al., 2002).
Glutathione is a precursor in phytochelatin synthesis (Cobbett, 2000; Inouhe, 2005) and these compounds are implicated in responses to several metals and metalloids including As (Raab et al., 2005). No phytochelatin synthases or synthetase (of which there are five in rice, three of which are expressed and the other two marginally expressed in all four conditions) were up-regulated. This suggests that at this level of As(V) exposure phytochelatin synthases and synthetases are constitutively expressed and are not involved in adaptive tolerance. This is in agreement with a number of previous studies which show that exposure to heavy metals does not alter expression of phytochelatin synthases (Ha et al., 1999; Ouziad et al., 2005) and overexpression of an Arabidopsis phytochelatin synthase led to hypersensitivity to both cadmium and zinc (Lee et al., 2003). Contradicting these observations, however, a number of studies have shown that these genes are induced by heavy metals (He et al., 2005; Zhang et al., 2005; Li et al., 2006). Also, phytochelatins have been shown to be involved in genetic differences in arsenate tolerance in H. lanatus (Bleeker et al., 2006).
Two arsenate reductase genes have recently been identified in rice (OsArs1, Os10g39860; and OsArs2, Os03g01770) (Duan et al., 2007). Neither of the probe sets for these genes is differentially regulated by As(V) in this study.
Conclusions
One week of arsenate treatment caused altered gene expression in a large number of genes, but not classic oxidative stress, phytochelatin synthases/synthetases, or arsenate reductase genes that might have been expected. However, the large number of responses from genes involved in glutathione synthesis, metabolism, and transport as well as genes potentially involved in arsenate methylation suggests that glutathione conjugation and the Challenger-like pathway of arsenate methylation may be important biochemical responses to arsenate challenge. The results of this transcriptomic study are employed in a companion paper (Norton et al., 2008) aiming to identify candidate genes for three epistatic loci that have been shown to determine the difference in arsenate tolerance observed between the two rice varieties used here.
Supplementary data
Supplementary data (Fig. S1 and Tables S1–S4) can be found at JXB online.
Fig. S1. RT-PCR confirmation of array results for actin and eight genes chosen to represent contrasting patterns of differential expression from the array data.
Table S1. List of probe sets significantly up-regulated 2-fold in Azucena in response to 13.3 μM sodium arsenate.
Table S2. List of probe sets significantly down-regulated 2-fold in Azucena in response to 13.3 μM sodium arsenate.
Table S3. List of probe sets significantly up-regulated 2-fold in Bala in response to 13.3 μM sodium arsenate.
Table S4. List of probe sets significantly down-regulated 2-fold in Bala in response to 13.3 μM sodium arsenate.
Supplementary Material
Acknowledgments
GJN was employed on a BBSRC grant (BB/C509931/1) which funded a proportion of the array analysis. DL-H is funded by a BBSRC studentship. The authors acknowledge the assistance of Dr Elaina Collie-Duguid at the University of Aberdeen Affymetrix core facility.
References
- Abedin J, Cresser MS, Meharg AA, Feldmann J, Cotter-Howells J. Arsenic accumulation and metabolism in rice (Oryza sativa L.) Environmental Science and Technology. 2002;36:962–968. doi: 10.1021/es0101678. [DOI] [PubMed] [Google Scholar]
- Aposhian HV, Aposhian MM. Arsenic toxicology: five questions. Chemical Research in Toxicology. 2006;19:1–15. doi: 10.1021/tx050106d. [DOI] [PubMed] [Google Scholar]
- Aposhian VH, Zakharyan RA, Avram MD, Sampayo-Reyes A, Wollenberg ML. A review of the enzymology of arsenic metabolism and a new potential role of hydrogen peroxide in the detoxication of the trivalent arsenic species. Toxicology and Applied Pharmacology. 2004;198:327–335. doi: 10.1016/j.taap.2003.10.027. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Bereczky Z, Wang H-, Schubert V, Ganal M, Bauer P. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. Journal of Biological Chemistry. 2003;278:24697–24704. doi: 10.1074/jbc.M301365200. [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Hakvoort HWJ, Bliek M, Souer E, Schat H. Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. The Plant Journal. 2006;45:917–929. doi: 10.1111/j.1365-313X.2005.02651.x. [DOI] [PubMed] [Google Scholar]
- Board PG, Coggan M, Chelvanayagam G, et al. Identification, characterization, and crystal structure of the Omega class glutathione transferases. Journal of Biological Chemistry. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- Cobbett CS. Phytochelatins and their roles in heavy metal detoxification. Plant Physiology. 2000;123:825–832. doi: 10.1104/pp.123.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Hossain SA, Meharg AA, Price AH. An arsenate tolerance gene on chromosome 6 of rice. New Phytologist. 2004;163:45–49. doi: 10.1111/j.1469-8137.2004.01109.x. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. The transparent testa12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. The Plant Cell. 2001;13:853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Davis BG, Edwards R. Functional divergence in the glutathione transferase superfamily in plants: identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. Journal of Biological Chemistry. 2002;277:30859–30869. doi: 10.1074/jbc.M202919200. [DOI] [PubMed] [Google Scholar]
- Duan G, Zhou Y, Tong Y, Mukhopadhyay R, Rosen BP, Zhu Y. A CDC25 homologue from rice functions as an arsenate reductase. New Phytologist. 2007;174:311–321. doi: 10.1111/j.1469-8137.2007.02009.x. [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiology. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratão PL, Polle A, Lea PJ, Azevedo RA. Making the life of heavy metal-stressed plants a little easier. Functional Plant Biology. 2005;32:481–494. doi: 10.1071/FP05016. [DOI] [PubMed] [Google Scholar]
- Güimil S, Chang HS, Zhu T, et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proceedings of the National Academy of Sciences, USA. 2005;102:8066–8070. doi: 10.1073/pnas.0502999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Smith AP, Howden R, Dietrich WM, Bugg S, O'Connell MJ, Goldsbrough PB, Cobbett CS. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. The Plant Cell. 1999;11:1153–1163. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley-Whitaker J, Ainsworth G, Meharg AA. Copper- and arsenate-induced oxidative stress in Holcus lanatus L. clones with differential sensitivity. Plant, Cell and Environment. 2001;24:713–722. [Google Scholar]
- Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: arsenic–glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Archives of Toxicology. 2005;79:183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- He Z, Li J, Zhang H, Ma M. Different effects of calcium and lanthanum on the expression of phytochelatin synthase gene and cadmium absorption in Lactuca sativa. Plant Science. 2005;168:309–318. [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. Journal of Biological Chemistry. 1989;264:13963–13966. [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- Inouhe M. Phytochelatins. Brazilian Journal of Plant Physiology. 2005;17:65–78. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Lee S, Moon JS, Ko T, Petros D, Goldsbrough PB, Korban SS. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiology. 2003;131:656–663. doi: 10.1104/pp.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Miginiac-Maslow M. The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynthesis Research. 2004;82:203–220. doi: 10.1007/s11120-004-1091-x. [DOI] [PubMed] [Google Scholar]
- Li J, Guo J, Xu W, Ma M. Enhanced cadmium accumulation in transgenic tobacco expressing the phytochelatin synthase gene of Cynodon dactylon L. Journal of Integrative Plant Biology. 2006;48:928–937. [Google Scholar]
- Li M, Huang W, Yang Q, Liu X, Wu Q. Expression and oxidative stress tolerance studies of glutaredoxin from cyanobacterium Synechocystis sp. PCC 6803 in Escherichia coli. Protein Expression and Purification. 2005;42:85–91. doi: 10.1016/j.pep.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. Journal of Biological Chemistry. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen H, Miller DS, Saavedra JE, Keefer LK, Johnson DR, Klaassen CD, Waalkes MP. Overexpression of glutathione S-transferase II and multidrug resistance transport proteins is associated with acquired tolerance to inorganic arsenic. Molecular Pharmacology. 2001;60:302–309. doi: 10.1124/mol.60.2.302. [DOI] [PubMed] [Google Scholar]
- Meharg AA, Hartley-Whitaker J. Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytologist. 2002;154:29–43. [Google Scholar]
- Meharg AA, Jardine L. Arsenite transport into paddy rice (Oryza sativa) roots. New Phytologist. 2003;157:39–44. doi: 10.1046/j.1469-8137.2003.00655.x. [DOI] [PubMed] [Google Scholar]
- Mudd SH, Datko AH. The S-methylmethionine cycle in Lemna paucicostata. Plant Physiology. 1990;93:623–630. doi: 10.1104/pp.93.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona PV, Polidoros AN, Scandalios JG. Modulation of antioxidant responses by arsenic in maize. Free Radical Biology and Medicine. 1998;25:576–585. doi: 10.1016/s0891-5849(98)00090-2. [DOI] [PubMed] [Google Scholar]
- Noctor G, Gomez L, Vanacker H, Foyer CH. Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. Journal of Experimental Botany. 2002;53:1283–1304. doi: 10.1093/jexbot/53.372.1283. [DOI] [PubMed] [Google Scholar]
- Norton GJ, Nigar M, Williams PN, Dasgupta T, Meharg A, Price AH. Rice–arsenate interactions in hydroponics: a three-gene model for tolerance. Journal of Experimental Botany. 2008;59 doi: 10.1093/jxb/ern098. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouziad F, Hildebrandt U, Schmelzer E, Bothe H. Differential gene expressions in arbuscular mycorrhizal-colonized tomato grown under heavy metal stress. Journal of Plant Physiology. 2005;162:634–649. doi: 10.1016/j.jplph.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences, USA. 2002;99:13324–13329. doi: 10.1073/pnas.202474599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras P, Pedrajas JR, Martínez-Galisteo E, Padilla CA, Johansson C, Holmgren A, Bárcena JA. Glutaredoxins catalyze the reduction of glutathione by dihydrolipoamide with high efficiency. Biochemical and Biophysical Research Communications. 2002;295:1046–1051. doi: 10.1016/s0006-291x(02)00771-4. [DOI] [PubMed] [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proceedings of the National Academy of Sciences, USA. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaghebeur M, Rengel Z. The distribution of arsenate and arsenite in shoots and roots of Holcus lanatus is influenced by arsenic tolerance and arsenate and phosphate supply. Plant Physiology. 2003;132:1600–1609. doi: 10.1104/pp.103.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab A, Schat H, Meharg AA, Feldmann J. Uptake, translocation and transformation of arsenate and arsenite in sunflower (Helianthus annuus): formation of arsenic–phytochelatin complexes during exposure to high arsenic concentrations. New Phytologist. 2005;168:551–558. doi: 10.1111/j.1469-8137.2005.01519.x. [DOI] [PubMed] [Google Scholar]
- Ranocha P, Bourgis F, Ziemak MJ, Rhodes D, Gage DA, Hanson AD. Characterization and functional expression of cDNAs encoding methionine-sensitive and -insensitive homocysteine S-methyltransferases from Arabidopsis. Journal of Biological Chemistry. 2000;275:15962–15968. doi: 10.1074/jbc.M001116200. [DOI] [PubMed] [Google Scholar]
- Rea PA, Li Z, Lu Y, Drozdowicz YM, Martinoia E. From vacuolar GS-X pumps to multispecific ABC transporters. Annual Review of Plant Biology. 1998;49:727–760. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- Requejo R, Tena M. Proteome analysis of maize roots reveals that oxidative stress is a main contributing factor to plant arsenic toxicity. Phytochemistry. 2005;66:1519–1528. doi: 10.1016/j.phytochem.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant and Cell Physiology. 2005;46:1568–1577. doi: 10.1093/pcp/pci172. [DOI] [PubMed] [Google Scholar]
- Schmuck EM, Board PG, Whitbread AK, Tetlow N, Cavanaugh JA, Blackburn AC, Masoumi A. Characterization of the monomethylarsonate reductase and dehydroascorbate reductase activities of Omega class glutathione transferase variants: implications for arsenic metabolism and the age-at-onset of Alzheimer's and Parkinson's diseases. Pharmacogenetics and Genomics. 2005;15:493–501. doi: 10.1097/01.fpc.0000165725.81559.e3. [DOI] [PubMed] [Google Scholar]
- Scott N, Hatlelid KM, MacKenzie NE, Carter DE. Reactions of arsenic(III) and arsenic(V) species with glutathione. Chemical Research in Toxicology. 1993;6:102–106. doi: 10.1021/tx00031a016. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Sari Gorla M, Mizzi L, De Toma G, Frova C. Organisation and structural evolution of the rice glutathione S-transferase gene family. Molecular Genetics and Genomics. 2004;271:511–521. doi: 10.1007/s00438-004-1006-8. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004 doi: 10.2202/1544-6115.1027. 3. [DOI] [PubMed] [Google Scholar]
- Takamatsu T, Aoki H, Yoshida T. Determination of arsenate, arsenite, monomethylarsonate, and dimethylarsinate in soil polluted with arsenic. Soil Science. 1982;133:239–246. [Google Scholar]
- Tanaka-Kagawa T, Jinno H, Hasegawa T, Makino Y, Seko Y, Hanioka N, Ando M. Functional characterization of two variant human GSTO 1-1s (Ala140Asp and Thr217Asn) Biochemical and Biophysical Research Communications. 2003;301:516–520. doi: 10.1016/s0006-291x(02)03066-8. [DOI] [PubMed] [Google Scholar]
- Theodoulou FL. Plant ABC transporters. Biochimica et Biophysica Acta. 2000;1465:79–103. doi: 10.1016/s0005-2736(00)00132-2. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Waters SB, Styblo M. Elucidating the pathway for arsenic methylation. Toxicology and Applied Pharmacology. 2004;198:319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Ullrich-Eberius CI, Sanz A, Novacky AJ. Evaluation of arsenate- and vanadate-associated changes of electrical membrane potential and phosphate transport in Lemna gibba G1. Journal of Experimental Botany. 1989;40:119–128. [Google Scholar]
- Winner EJ, Prough RA, Brennan MD. Human NAD(P)H:quinone oxidoreductase induction in human hepatoma cells after exposure to industrial acrylates, phenolics, and metals. Drug Metabolism and Disposition. 1997;25:175–181. [PubMed] [Google Scholar]
- Wu J, Zhang R, Lilley RM. Methylation of arsenic in vitro by cell extracts from bentgrass (Agrostis tenuis): effect of acute exposure of plants to arsenate. Functional Plant Biology. 2002;29:73–80. doi: 10.1071/PP01022. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- Ye J, Fang L, Zheng H, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Research. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Lock JH, Gomez KA. A laboratory manual for the physiological studies of rice. Manilla: IRRI; 1976. pp. 61–66. [Google Scholar]
- Zakharyan RA, Tsaprailis G, Chowdhury UK, Hernandez A, Aposhian HV. Interactions of sodium selenite, glutathione, arsenic species, and Omega class human glutathione transferase. Chemical Research in Toxicology. 2005;18:1287–1295. doi: 10.1021/tx0500530. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu W, Guo J, He Z, Ma M. Coordinated responses of phytochelatins and metallothioneins to heavy metals in garlic seedlings. Plant Science. 2005;169:1059–1065. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.