Abstract
The regulation of ACC synthase (ACS) genes was studied in early (‘Early Golden’) and late (‘Shiro’) Japanese plum cultivars (Prunus salicina L.) in order to determine the role of this gene family in fruit ripening. Of the four Ps-ACS cDNAs isolated, two (Ps-ACS1 and -3) showed differential expression between the two cultivars. Ps-ACS1 accumulated during fruit ripening of ‘Early Golden’ (‘EG’) and ‘Shiro’ (‘SH’) in ethylene-dependent and -independent manners, respectively. Ps-ACS3a transcripts accumulated throughout fruit development and during ‘EG’ fruit ripening. Ps-ACS3b was detected only during ripening of ‘SH’ fruit. Furthermore, Ps-ACS3a transcript accumulation was negatively regulated by ethylene, whereas Ps-ACS3b was positively induced by the hormone. In both cultivars, the expression of Ps-ACS4 and -5 is under positive and negative feedback control by ethylene, respectively. Genetic analyses of ‘EG’ and ‘SH’ cultivars demonstrated that ‘EG’ is homozygous for Ps-ACS3a whereas ‘SH’ is heterozygous for Ps-ACS3 (a/b). The role of ethylene-overproducer 1-like in delaying fruit ripening by interacting with Ps-ACS proteins was also studied. The effect of the plant hormones, auxin, gibberellin, and cytokinin, in regulating ethylene production by promoting the induction of the different Ps-ACS mRNAs in plum was investigated. A model is presented in which differences in Ps-ACS alleles and gene expression between early and late plums are critical in determining the ripening behaviour of the cultivars.
Keywords: 1-Aminocyclopropane-1-carboxylic acid synthase (ACS), ACS alleles, auxin, cytokinin, double sigmoid curve growth pattern, ethylene-overproducer 1 (ETO1), gibberellin, plum fruit development and ripening, system 1 and system 2 ethylene production
Introduction
Ethylene, a gaseous phytohormone, regulates diverse developmental and physiological processes throughout the entire life cycle of plants, including seed germination, root initiation, flower and leaf senescence, abscission, wounding response, and disease resistance (Abeles et al., 1992). One of the most studied examples of ethylene regulation is the ripening of climacteric fruit. Climacteric fruit development typically includes a transition phase during which ethylene responsiveness and production levels are dramatically altered. While the outcome of these changes, the ripening process, has been extensively characterized in many plant species (Abeles et al., 1992; Fluhr and Mattoo, 1996), the developmental processes that lead up to and accompany the changes are less well understood (Giovannoni, 2001). Although tomato has generally been the model of choice in the study of climacteric fruit ripening (Alexander and Grierson, 2002), species such as plum have distinct features that are economically important and provide opportunities to dissect specific mechanisms.
Fruits can be classified as climacteric or non-climacteric depending on the presence or absence of post-harvest, ripening-associated rise in ethylene evolution and respiration (Biale and Young, 1981). Plum (Prunus salicina), like peach (P. persica) and apricot (P. armeniaca), is a climacteric fruit characterized by a large diversity for the date and rate of ripening. Some late plum cultivars such as ‘Shiro’ and ‘Golden Japan’ show a suppressed-climacteric pattern, while ‘Early Golden’ and ‘Santa Rosa’ behave as typical climacteric fruit (Abdi et al., 1997; Zuzunaga et al., 2001). However, treatment of such late cultivars with propylene showed that fruit displaying the suppressed-climacteric phenotype ripen as a typical climacteric fruit, so should still be placed in the climacteric class (Abdi et al., 1997).
The cumulative growth pattern of stone fruits (Prunus spp.), including plums, is portrayed by a typical double sigmoid curve during fruit development and ripening (Tonutti et al., 1997). Within this developmental period, four distinct stages (S1–S4) are clearly recognized. In the first stage (S1), of intense cell division and differentiation, growth is rapid. A long period of stasis follows (S2) during which the endocarp hardens to form a solid stone coincident with hardly any increase in fruit size. The third stage (S3) is accompanied by rapid cell division resulting in a significant increase in fruit size. Finally the fourth stage (S4), is when the fruit arrive at their fullsize and start to enhance ripening in an ethylene-dependent manner accompanied by changes in colour, texture, and aroma enrichment.
Ethylene is synthesized from S-adenosyl-L-methionine (SAM) via 1-aminocyclopropane-1-carboxylic acid (ACC). The conversion of SAM to ACC is catalysed by ACC synthase (ACS), and the subsequent oxidation of ACC to ethylene is catalysed by ACC oxidase (ACO) (Yang and Hoffman, 1984). Although ACS and ACO are the two key enzymes in ethylene biosynthesis, ACS is generally the rate-limiting enzyme in the biosynthetic pathway. In Arabidopsis, tomato, and other species, ACS and ACO proteins are encoded by small gene families whose members are differentially regulated at the transcriptional level by various developmental, environmental, and hormonal signals (Fluhr and Mattoo, 1996; Wang et al., 2002). Previous studies demonstrated that treatment of Arabidopsis seedlings with the phytohormones, auxin, gibberellin, or cytokinin, increase ethylene biosynthesis by activating ACS transcription only in the case of auxin and gibberellin (Abel et al., 1995; Kaneta et al., 1997), whereas cytokinin increases ethylene biosynthesis via a post-transcriptional mechanism (Vogel et al., 1998; Woeste et al., 1999).
Recent findings suggest that post-translational regulation is an important aspect of the control of ACS expression (Chae et al., 2003). Arabidopsis and tomato ACS proteins have been shown to be post-translationally phosphorylated by a wounding-inducible kinase activity within the Val and Ser residues found in the conserved C-terminal region (Tatsuki and Mori, 2001; Chae et al., 2003). Ethylene-overproducer 1 (ETO1) protein, a negative regulator of ethylene evolution, interacts strictly with this conserved C-terminal region, target of ETO1 (TOE) sequence, found uniquely in Type 2 ACS proteins, and degrades them via the ubiquitin–26S proteasome pathway (Chae et al., 2003; Chae and Kieber, 2005; Yoshida et al., 2005). Moreover, phosphorylation of the C-terminus of Type 1 ACS proteins by MPK6 suggests the involvement of the ACS C-terminal region in post-transcriptional regulation (Liu and Zhang, 2004; Chae and Kieber, 2005).
It has been proposed that specific members of ACS and ACO gene families control two systems of ethylene production in plants (Lelièvre et al., 1997). System 1 is the basal low rate of ethylene production that is detected in all tissues including non-climacteric fruits. System 2, or autocatalytic ethylene, operates during climacteric fruit ripening. In tomato fruit, ethylene biosynthesis is primarily regulated by three different Sl-ACS mRNAs, Sl-ACS2, -4, and -6 (Nakatsuka et al., 1998). Sl-ACS2 and -4 are responsible for system 2 autocatalytic ethylene production and Sl-ACS6 is responsible for the auto-inhibitory, low levels of system 1 ethylene (Barry et al., 2000).
The objective of this study is to understand the role of ACS gene family members in the capacity of the fruit to produce autocatalytic ethylene and ripen. The ethylene evolution and the accumulation pattern of four Ps-ACS mRNAs were studied during plum fruit development and ripening in early ‘Early Golden’ (‘EG’) and late ‘Shiro’ (‘SH’) cultivars. The effects of the phytohormones, auxin, gibberellin, and cytokinin, on the regulation of the different Ps-ACS mRNAs were investigated. The role of ETO1-like (EOL1) in controlling the date and rate of fruit ripening by suppressing ethylene production in plum was also studied. The results are discussed in terms of system 1 and system 2 ethylene synthesis, and a model of Ps-ACS gene regulation throughout the double sigmoid growth pattern is proposed.
Materials and methods
Plant material and post-harvest treatments
Fruits were harvested from two Japanese plum (Prunus salicina L.) cultivars ‘Early Golden’ (‘EG’) and ‘Shiro’ (‘SH’) grown at the experimental farm of the University of Guelph (Vineland Station, ON, Canada). These two varieties were chosen according to their maturity times, early and late, respectively. Plum fruit were treated as described previously (El-Sharkawy et al., 2007). Fruit tissues from five fruits exhibiting similar ethylene production were used for mRNA extraction and analysis. Since the seed is inseparable in S1 and S2, the whole fruit tissue was used for RNA extraction, while in S3 and S4 stages both fruit tissues (fruit pulp and seed) were carefully separated for RNA analysis. Other tissues such as shoot apex, axillary bud, young leaves, and flowers were collected from the ‘EG’ cultivar. All plant material was frozen in liquid nitrogen and stored at –80 °C.
Plant growth regulator (PGR) treatments and growth conditions
To minimize the effect of possible regulation of Ps-ACS mRNAs caused by the presence of endogenous hormones (ethylene, auxin, and/or gibberellin), leaves from the ‘EG’ cultivar were used for studying the effect of various PGRs. Leaves were thoroughly rinsed in tap water, surface sterilized for 10 min in 20% (v/v) commercial bleach, washed 3× with sterile distilled water, and the excess water was gently dried under aseptic conditions. The leaves were cultured in full-strength woody plant basal medium (Lloyd and McCown, 1981) (PhytoTechnology Laboratories, KS, USA) supplemented with 2% (w/v) sucrose, 0.7% (w/v) agar, and the respective PGR treatment as detailed below. The cultures were incubated in a controlled environmental condition (relative humidity 80%, 22 °C and 16 h light/8 h dark cycles) for 4 weeks. After that, the leaves were transferred to suspension cultures in 250 ml flasks containing 100 ml of liquid medium with appropriate PGRs as treated earlier. This ensures sufficient imbibition of PGRs by the leaves. The suspension cultures were incubated on a rotary shaker (100 rpm) and the leaves continued growing for 30 min at room temperature.
The PGR treatments include auxins, 2,4-D and picloram (0.5, 1, and 2 μM), IAA, IBA, and 1-NAA (0.4, 4, and 40 μM), gibberellins, GA3 and GA4+7 (0.4, 4, and 40 μM), and cytokinin-benzyladenine (0.4, 4, 20, and 40 μM). Leaves cultured in basal medium without any PGR were used as control. At the end of the suspension culture all the plant tissues were removed, briefly blotted dry, immediately frozen in liquid nitrogen, and stored at –80 °C for RNA analysis.
RNA isolation
Total RNA from fruit samples was extracted using the methods described by Boss et al. (1996). For vegetative tissues and flowers, total RNA was extracted using the PureLink Plant RNA reagent (Invitrogen, Burlington, ON, Canada) as per the manufacturer's instructions. All RNA extracts were treated with DNase I (Promega, Madison, WI, USA), then cleaned up with an RNeasy mini kit (Qiagen, Mississauga, ON, Canada).
Isolation and in silico analysis of plum cDNA sequences
For the isolation of plum homologues of ACC synthase (ACS), first-strand cDNA synthesis was carried out using 20 μg of total DNase-treated RNA in a 50 μl aliquot. One microlitre of cDNA was used in a PCR with the appropriate degenerate primers. Several sets of primers (Jones and Woodson, 1999) were used to isolate plum Ps-ACS sequences that shared the structural characteristics associated with functional ACS proteins (Zarembinski and Theologis, 1994; Capitani et al., 1999). To isolate Ps-EOL1 cDNA, a PCR primer set was synthesized based on the conserved region of the nucleotide sequences of ETO1 (Wang et al., 2004; Yoshida et al., 2006) [Eol1-F 5′-ACCCAACACTCACATWCCCATAC-3′; Eol1-R 5′-CAAGATTGTTGAGGGCCTGACC-3′]. The isolated fragments were cloned in pGEM-T easy vector (Promega, Madison, WI, USA), sequenced and compared with database sequences using the BLAST program (Altschul et al., 1997). Extension of the partial cDNA clones was carried out using the 3′- and 5′-RACE kit (Invitrogen, Burlington, ON, Canada). Full-length amplification of cDNA sequences, designated Ps-ACS1, Ps-ACS3, Ps-ACS4, and Ps-ACS5, was carried out using Platinum Taq DNA Polymerase High Fidelity following the instructions provided by the manufacturer (Invitrogen). Alignments of the predicted protein sequences were performed with ClustalX (Jeanmougin et al., 1998) and GeneDoc (Nicholas and Nicholas, 1997). The Neighbor–Joining tree was constructed with PAUP* 4.0b3. Bootstrap values from 1000 replicates were obtained. The tree was visualized with the TreeView program (Page, 1996).
Real time quantitative RT-PCR
DNase-treated RNA (5 μg) was reverse transcribed in a total volume of 50 μl using SuperScript III Reverse Transcriptase (Invitrogen). Real-time quantitative PCR was performed using 20 ng of total RNA in a 20 μl reaction volume using SYBR GREEN PCR MasterMix (Qiagen) on a Mx4000® multiplex quantitative PCR system (Stratagene, La Jolla, CA, USA). Mx4000® v. 4.20 software (Stratagene) was used to design gene-specific primers (Table 1). For all the sequences studied here, optimal primer concentration was 200 nM. RT-PCR conditions were as described previously (El-Sharkawy et al., 2007). The PCR products were analysed further by a dissociation curve program at 95–60 °C (16 s). All RT-PCR experiments were run in triplicate with different cDNAs synthesized from three biological replicates. Each sample was run in three technical replicates on 96-well plates. For each sample, a Ct (threshold constant) value was calculated from the amplification curves by selecting the optimal ΔRn (emission of reporter dye over starting background fluorescence) in the exponential portion of the amplification plot. Relative fold differences were calculated based on the comparative Ct method using the β-actin as an internal standard (El-Sharkawy et al., 2007). To demonstrate that the efficiencies of the different gene primers were approximately equal, the absolute value of the slope of log input amount versus ΔCt was calculated for Ps-ACS1, Ps-ACS3, Ps-ACS4, Ps-ACS5, Ps-EOL1, and β-actin sequences, and was determined to be <0.1. To determine relative fold differences for each sample in each experiment, the Ct value for the five mRNAs studied was normalized to the Ct value for β-actin and was calculated relative to a calibrator using the formula 2–ΔΔCt. The calibrator is the sample that exhibited the minimum level of transcripts in the whole experiment (fruits 47 DAB for Ps-ACS3, fruits 52 DAB for Ps-ACS1, -4, and -5, and ‘EG’ fruits 82 DAB for Ps-EOL1; Figs 3–7) and control tissues (Figs 8–10).
Table 1.
Real-time quantitative PCR primers
| Name | Sequence |
| Ps-ACS1-F | 5′-ACCGCGTTCACGTTGTTT-3′ |
| Ps-ACS1-R | 5′-TGCCACAACCATGTCGTC-3′ |
| Ps-ACS3-F | 5′-GTGCAACTGCAGCCAATG-3′ |
| Ps-ACS3-R | 5′-GGTTGGCACAAGCAAAGC-3′ |
| Ps-ACS4-F | 5′-GCGTCGATGCTCTTGGAT-3′ |
| Ps-ACS4-R | 5′-AAAACCCCGTGCCTCTTC-3′ |
| Ps-ACS5-F | 5′-GATCTTGGATGGCGAACG-3′ |
| Ps-ACS5-R | 5′-AGCCTCCAAGGCTGCTCT-3′ |
| Ps-EOL1-F | 5′-TGATGCAGCAAAAGGTGTTC-3′ |
| Ps-EOL1-R | 5′-ACTTTTCATGCTCGCTGGAT-3′ |
| Ps-actin-F | 5′-CTGGACCTTGCTGGTCGT-3′ |
| Ps-actin-R | 5′-ATTTCCCGCTCAGCAGTG-3′ |
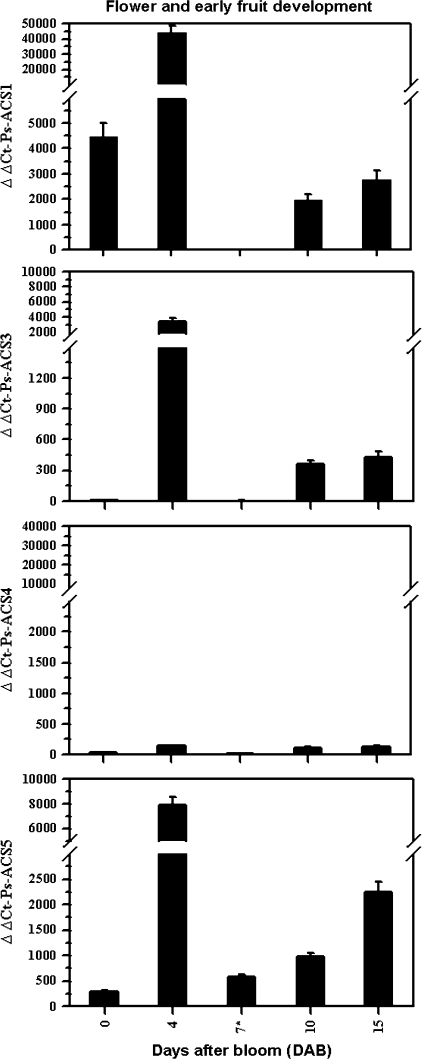
Fig. 3.
Steady-state transcript levels of Ps-ACS1, Ps-ACS3, Ps-ACS4, and Ps-ACS5 assessed by real-time quantitative PCR during flower and early fruit development of the ‘EG’ cultivar. The fertilized flower stage is marked with an asterisk. The experiments were carried out in triplicate. The x-axis represents the developmental stages indicated by number of days after bloom (DAB). ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression relative to fruits 47 DAB for Ps-ACS3 and fruits 52 DAB for Ps-ACS1, -4, and -5.
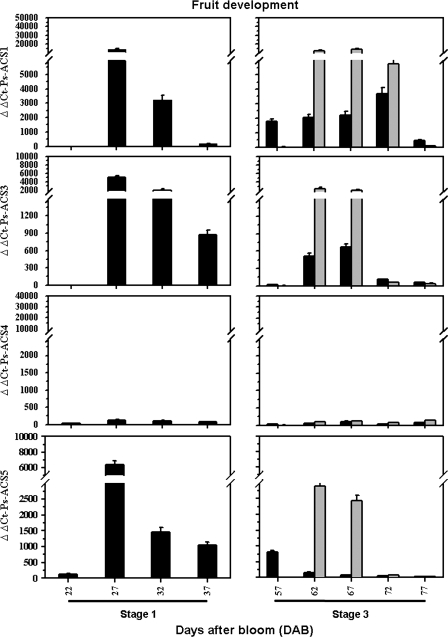
Fig. 4.
Steady-state transcript levels of Ps-ACS1, -3, -4, and -5 assessed by real-time quantitative PCR during S1 of fruit development using the ‘EG’ whole fruit. During S3 of fruit development the expression was studied in pulp (black-filled columns) and in seeds (grey-filled columns). The experiments were carried out in triplicate. The x-axis represents the developmental stage indicated by number of days after bloom (DAB) and by the name of the stage. ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression. Other details are as described in Fig. 3.
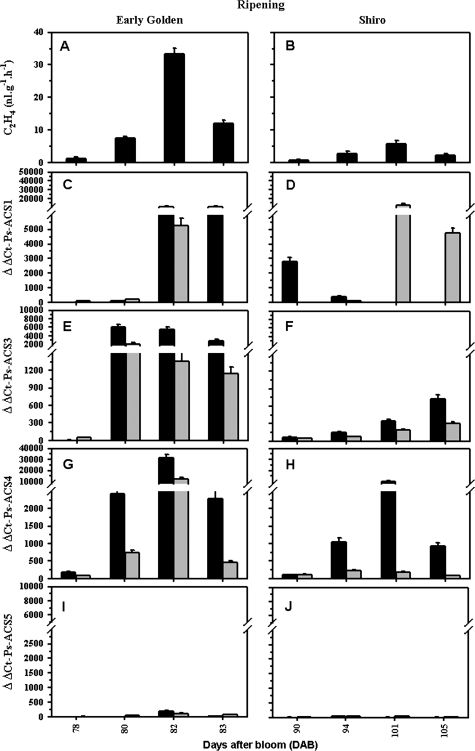
Fig. 5.
Ethylene evolution and steady-state transcript levels assessed by real-time quantitative PCR throughout fruit ripening: (A, B) ethylene production; (C, D) Ps-ACS1; (E, F) Ps-ACS3; (G, H) Ps-ACS4; and (I, J) Ps-ACS5 transcript levels during ripening of early ‘EG’ (left panel) and late ‘SH’ (right panel) plum fruits, respectively. The expression was studied in pulp (black-filled columns) and in seeds (grey-filled columns). The experiments were carried out in triplicate. The x-axis represents the developmental stage indicated by number of days after bloom (DAB). ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression. Other details are as described in Fig. 3.
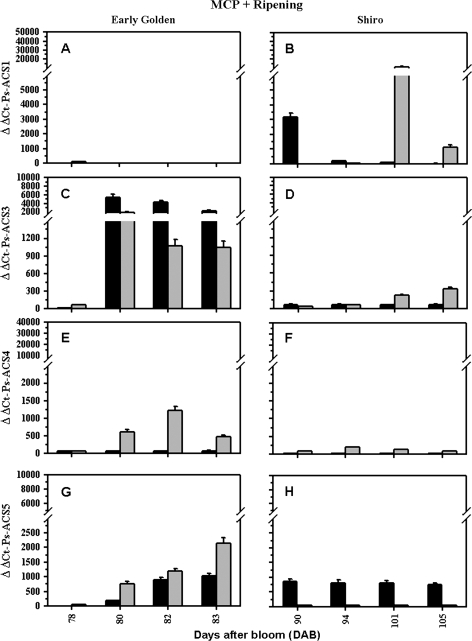
Fig. 6.
Ethylene evolution and steady-state transcript levels assessed by real-time quantitative PCR during ripening of 1-MCP-pre-treated fruit: (A, B) Ps-ACS1; (C, D) Ps-ACS3; (E, F) Ps-ACS4; and (G, H) Ps-ACS5 transcript levels during ripening of early ‘EG’ (left panel) and late ‘SH’ (right panel) plum fruits, respectively. The expression was studied in pulp (black-filled columns) and in seeds (grey-filled columns). Fruits were exposed overnight immediately at harvest (76 DAB and 88 DAB for ‘EG’ and ‘SH’, respectively) to 1-MCP (1 μl l-1) before the onset of endogenous ethylene. The experiments were carried out in triplicate. The x-axis represents the developmental stage indicated by number of days after bloom (DAB). ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression. Other details are as described in Fig. 3.
Fig. 7.
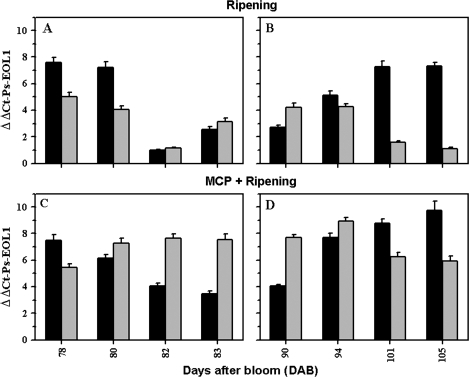
Ps-EOL1 (EU176813) gene expression in early ‘EG’ (left panel) and late ‘SH’ (right panel) plum fruits. The steady-state transcript levels were assessed by real-time quantitative PCR during ripening of ‘EG’ (A) and ‘SH’ (B) fruits, and in MCP-treated ‘EG’ (C) and ‘SH’ (D) fruits. The experiments were carried out in triplicate. The x-axis represents the developmental stage indicated by number of days after bloom (DAB). ΔΔCt on the y-axis of each figure refers to the fold difference in Ps-EOL1 gene expression relative to fruits 82 DAB.
Fig. 8.
Steady-state transcript levels of Ps-ACS1, -3, -4, and -5 assessed by real-time quantitative PCR in control ‘EG’ leaves and ‘EG’ leaves treated with cytokinin using the indicated concentrations. The experiments were carried out in triplicate. The x-axis represents the type and concentration (μM) of cytokinin used in this study. ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression relative to the control.
Fig. 9.
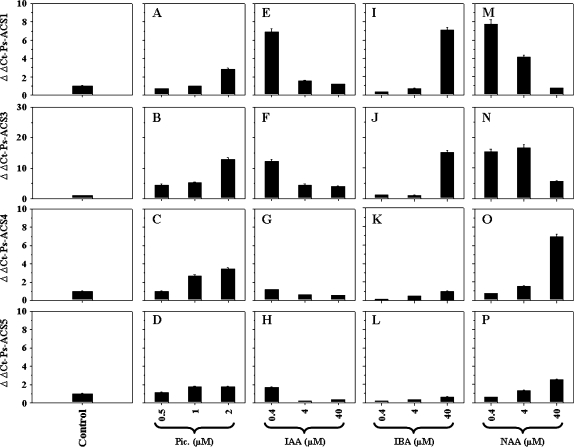
Steady-state transcript levels of Ps-ACS1, -3, -4, and -5 assessed by real-time quantitative PCR in control ‘EG’ leaves and ‘EG’ leaves treated with different types of auxins (picloram, IAA, IBA, and NAA) using the indicated concentrations. The experiments were carried out in triplicate. The x-axis represents the type and concentration (μM) of auxin used in this study. ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression relative to the control.
Fig. 10.
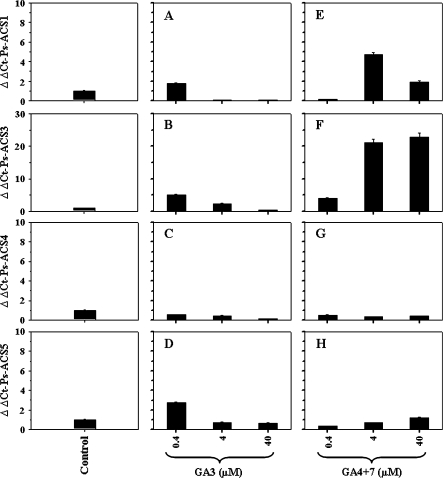
Steady-state transcript levels of Ps-ACS1, -3, -4, and -5 assessed by real-time quantitative PCR in control ‘EG’ leaves and ‘EG’ leaves treated with different types of gibberellins (GA3 and GA4+7) using the indicated concentrations. The experiments were carried out in triplicate. The x-axis represents the type and concentration (μM) of gibberellin used in this study. ΔΔCt on the y-axis of each figure refers to the fold difference in gene expression relative to the control.
Promoter isolation
Genomic DNA was extracted from immature leaves according to the DNeasy Plant Maxi Kit (Qiagen). Promoters of Ps-ACS3 [Ps-DACS3a (EU034651), Ps-DACS3b (EU034652)] were isolated from ‘EG’ and ‘SH’ genotypes using the Universal Genome Walker Kit (Clontech, Palo Alto, CA, USA). In order to identify the presence of the different Ps-ACS3 promoters in ‘EG’ and ‘SH’ cultivars, four specific primers for Ps-DACS3a and b, ACS3a-F [–1078] (5′-GCGTAATTTATGAATACAATATCCCTAC-3′), ACS3a-R (5′- CCCCAGTTGGAAGCTTCTGAGTGCTCTTCC-3′) [331], ACS3b-F [–1456] (5′-ATCTACTGATTCCAAGTCGTTAATAC-3′), and ACS3b-R [–810] (5′-GAGAATTTACGTATATAAGGTAATAGAG-3′), were designed from each promoter sequence. PCR contained ∼150 ng of template genomic DNA and were carried out using an Advantage Genomic PCR Kit (Clontech).
Results
Protein structure and organization
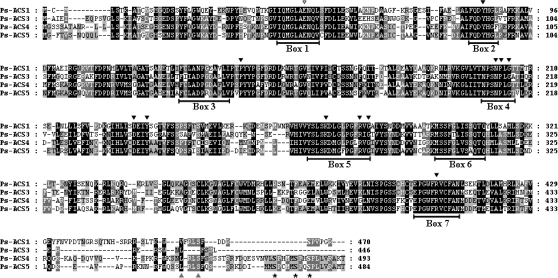
ACS cDNAs and proteins have been isolated from a wide variety of plant species. Although sequence identity can be as low as 45% in ACS proteins, there are highly conserved signature elements. Four novel Ps-ACS sequences were isolated from two plum cultivars using an RT-PCR approach in order to investigate the involvement of ACS proteins in fruit ripening (Fig. 1). All Ps-ACS isozymes contain the seven conserved boxes found in ACS from Arabidopsis, tomato, and other plant species. Eleven out of 12 amino acid residues conserved in aminotransferase and ACS proteins, as well as the glutamate (E) residue in box 1 that is responsible for substrate specificity (McCarthy et al., 2001), are present in all of the plum sequences (Mehta et al., 1989; Yamagami et al., 2003). All Ps-ACS predicted proteins, except Ps-ACS3, have the Ser residue that is phosphorylated by a calcium-dependent protein kinase, CDPK (Tatsuki and Mori, 2001), whereas only Ps-ACS4 and -5 predicted proteins have the three Ser residues that are targets of the protein kinase MPK6 phosphorylation (Liu and Zhang, 2004) (Fig. 1). Interestingly, Ps-ACS1 predicted protein has only one out of the three Ser residues (S465) important for phosphorylation by MPK6 (Fig. 1).
Fig. 1.
Amino acid sequence alignment of Ps-ACS1 (EU034649), Ps-ACS3 (EU034650), Ps-ACS4 (EU034653), and Ps-ACS5 (EU034654) using the ClustalX program. Conserved residues are shaded in black. Dark grey shading indicates similar residues in three out of four of the sequences and light grey shading indicates similar residues in two out of four of the sequences. The 11 black arrows designate the residues that represent the conserved amino acids in aminotransferases. The conserved glutamate residue (E) marked with an open arrow is involved in substrate specificity (McCarthy et al., 2001). The two grey arrows indicated the Val and Ser residues important for interaction with ETO1 and CDPK phosphorylation (Tatsuki and Mori, 2001; Yoshida et al., 2005). The Ser residues that are targets of the protein kinase MPK6 phosphorylation (Liu and Zhang, 2004) are marked with asterisks.
The relationships between the predicted amino acid sequences, as indicated by percentage similarity over the whole sequence, are presented in Table 2. Although there was considerable divergence among the Ps-ACS mRNAs, there were also highly homologous sequences putatively coding for closely related isozymes. Sequences related to the isolated plum cDNAs have been identified previously in pear (El-Sharkawy et al., 2004). Strong sequence similarity between specific plum and pear sequences (89–93%; Table 2) indicates that these sequences are likely to be orthologous.
Table 2.
Amino acid sequence comparison between the predicted full-length plum and pear ACC synthase cDNAs
| Amino acid similarity percentage | |||||
| Protein size (no. of aa) | Ps-ACS1 | Ps-ACS3 | Ps-ACS4 | Ps-ACS5 | |
| Prunus salicina | |||||
| Ps-ACS1 | 470 | 100 | |||
| Ps-ACS3 | 446 | 68 | 100 | ||
| Ps-ACS4 | 493 | 64 | 65 | 100 | |
| Ps-ACS5 | 484 | 65 | 65 | 73 | 100 |
| Pyrus communis | |||||
| Pc-ACS1 | 473 | 93 | 68 | 62 | 65 |
| Pc-ACS2 | 446 | 67 | 89 | 63 | 63 |
| Pc-ACS3 | 446 | 68 | 91 | 64 | 64 |
| Pc-ACS4 | 495 | 64 | 65 | 91 | 72 |
| Pc-ACS5 | 487 | 64 | 64 | 72 | 93 |
Arabidopsis and tomato ACS proteins have been shown to be post-transcriptionally and/or post-translationally regulated by the phosphorylation of target residues found in the conserved C-terminal region (Tatsuki and Mori, 2001; Chae et al., 2003; Liu and Zhang, 2004; Yoshida et al., 2005). A classification of ACC synthase based on the similarity of DNA sequences and the C-terminal consensus motif has been reported (Yoshida et al., 2005). Predicted proteins of both Ps-ACS4 and -5 are members of Type 1 ACC synthases that have the Ser residue in the ‘RLSF’ motif, necessary for CDPK phosphorylation (Tatsuki and Mori, 2001), followed by a long C-terminal tail (27–33 aa) containing the three conserved Ser residues proved to be the targets of MPK6 phosphorylation (Liu and Zhang, 2004) but lack the Val in the ‘WVF’ motif (Fig. S1 in Supplementary data available at JXB online). In both of these predicted proteins the ‘WVF’ motif is degenerated to ‘WKSNL’ and ‘WQSSL’, respectively (Fig. 1). Ps-ACS1 predicted protein belongs to Type 2 ACS isozymes that possess the conserved TOE sequence (Yoshida et al., 2006), which includes the ‘WVF’ sequence just before the ‘RLSF’ motif, followed by a short tail (nine amino acids) rich in basic and acidic amino acids (Supplementary Fig. S1). The predicted Ps-ACS3 protein is a member of Type 3 isozymes, where the C-terminus is truncated by 24–47 amino acids and consequently lacks all the residues important for phosphorylation and the ‘WVF’ motif degenerates to ‘WNLF’ (Fig. 1).
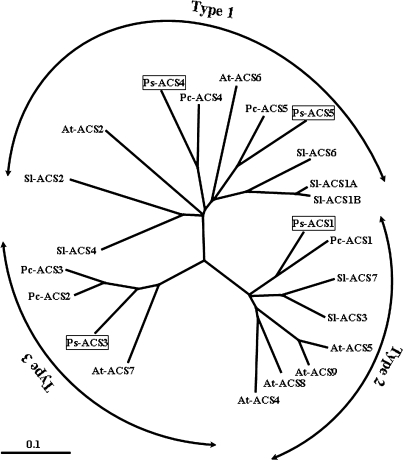
The phylogenetic tree presented in Fig. 2 was obtained by comparing the C-terminal amino acid sequences of 23 ACS proteins from four species–plum, pear, Arabidopsis, and tomato. It indicates that ACS proteins can be divided into three main subfamilies (1, 2, and 3). Sequences corresponding to the first subfamily, Type 1, comprising all the At-ACS proteins, have been shown to be post-transcriptionally regulated by MPK6 phosphorylation (Liu and Zhang, 2004). The second subfamily, Type 2, isozymes containing all the Arabidopsis and tomato ACS proteins, have been shown to confer a strong ability to bind ETO1 and to be post-translationally regulated by CDPK phosphorylation (Vogel et al., 1998; Chae et al., 2003; Yoshida et al., 2005). The third subfamily, Type 3, contains the ACS proteins that have a long C-terminal (Sl-ACS4) or a short C-terminal tail (Ps-ACS3, Pc-ACS2, Pc-ACS3, and At-ACS7) but in both cases lack all the residues important for post-transcriptional and/or post-translational regulation.
Fig. 2.
Phylogenetic relationships between Prunus salicina (Ps-ACS1, Ps-ACS3, Ps-ACS4, Ps-ACS5), Arabidopsis thaliana [At-ACS2 (Q06402), At-ACS4 (NP179866), At-ACS5 (AAG50098), At-ACS6 (T13019), At-ACS7 (AAG48754), At-ACS8 (AAG50090), At-ACS9 (AAG48755)], Solanum lycopersicon [Sl-ACS1A (AAF97614), Sl-ACS1B (AAF97615), Sl-ACS2 (CAA41855), Sl-ACS3 (AAB48945), Sl-ACS4 (AAA03164), Sl-ACS6 (BAA34923), Sl-ACS7 (AAC32317),], and Pyrus communis [Pc-ACS1 (X87112), Pc-ACS2 (AF386519), Pc-ACS3 (AY388988), Pc-ACS4 (AF386518), Pc-ACS5 (AF386523)] based on amino acid sequences. Type 1, Type 2, and Type 3 show the different ACS protein subfamilies.
A plum homologue of the Arabidopsis ethylene-overproducer 1-like (At-ETO1) has also been isolated. The partial Ps-EOL1 sequence encoded a putative protein of 245 amino acid residues. The predicted protein displayed strong homology (92% identity, 97% similarity) to the tomato Sl-EOL1 sequence (Yoshida et al., 2006). Analysis of the partial Ps-EOL1 amino acid sequence revealed the presence of four out of six TPR (tetratricopeptide repeat) motifs which are involved in diverse protein–protein interactions and can serve as a scaffold for assembly of multi-protein complexes (D'Andrea and Regan, 2003).
Ethylene production during fruit development and ripening
Low levels of ethylene production, ranging from 0.8 to 2 nl g−1 h−1 ±0.2, have been detected during S1 of fruit development (22–37 DAB). In both studied cultivars, ‘EG’ and ‘SH’, the whole fruit produced ethylene throughout S4 fruit ripening (Fig. 5A, B). No ethylene emission was detected from the separated S3 and S4 seeds (data not shown). ‘EG’ fruit displayed an early, rapid ripening, and a short and rapid (maximal 5 d) ethylene production profile (Fig. 5A). ‘SH’ fruit exhibited a suppressed climacteric pattern. The fruit ripened more slowly and later than ‘EG’, with ethylene production in ‘SH’ fruit reaching a maximum at ∼11 d after the onset of ethylene emission (Fig. 5B). 1-Methylcyclopropene (1-MCP) treatment immediately at harvest before the onset of endogenous ethylene production completely abolished the ethylene burst and ripening in the pre-treated fruits (data not shown).
The expression of the Ps-ACS cDNAs during fruit development and ripening
To investigate the possible role of ACC synthase in controlling ethylene production and subsequently in the capacity of plum fruit to ripen, four Ps-ACS mRNAs were identified. Real-time PCR analysis of steady-state mRNA levels was carried out to determine their mRNA accumulation patterns throughout fruit development and ripening.
Transcripts of Ps-ACS1, -3, and -5 accumulated at constant levels in vegetative tissues, while those of Ps-ACS4 mRNA remained low or undetectable in such tissues (data not shown). Figures 3 and 4 show the expression pattern of the four cDNAs during flower and early fruit development (0–15 DAB), and throughout fruit development (22–77 DAB), respectively. Ps-ACS4 transcripts were expressed at a basal low level or were almost undetectable during these periods of fruit development at 0–77 DAB (Figs 3, 4). Ps-ACS1, -3, and -5 mRNAs accumulated in the same pattern during the periods mentioned (Figs 3, 4). The three Ps-ACS transcripts peaked in opened flowers (∼4 DAB), followed by strong inhibition in their transcript levels after fertilization, ∼7 DAB (Fig. 3). During early fruit development (7–15 DAB), transcript levels for Ps-ACS1, -3, and -5 increased slightly (Fig. 3).
Throughout S1 of fruit development (∼27 DAB), another peak of the three transcripts was detected (Fig. 4), declining slightly thereafter (32–37 DAB) to reach their basal levels, and becoming almost undetectable during S2 of fruit development at 42–52 DAB (data not shown). During S3 (57–77 DAB) of fruit development and when the pulp was separated from the seed (endocarp+embryo), high transcript levels of Ps-ACS1, -3, and -5 were only detected in the fruit between 62 and 72 DAB (Fig. 4). Markedly higher accumulation levels of the three mRNAs were detected in the seed than in the pulp. The expression levels of Ps-ACS1, -3, and -5 were ∼4, 150, and 110 times higher, respectively, in the seed than in the pulp (Fig. 4). Only Ps-ACS1 and -3 transcript levels showed a steady increase in the pulp tissue, whereas those of Ps-ACS5 decreased in abundance. The three transcripts reached a basal level in the whole fruit at the end of S3 at ∼77 DAB.
‘EG’ fruit showed a dramatic increase in the transcript levels of Ps-ACS1, -3, and -4 mRNAs throughout S4 at fruit ripening (78–83 DAB). Transcripts of the three cDNAs were low or almost undetectable in the non-climacteric stage of ‘EG’ fruit at ∼78 DAB (Fig. 5A, C, E, G). As ripening progressed and higher levels of autocatalytic ethylene were produced (Fig. 5A), the expression levels of Ps-ACS1, -3, and -4 mRNAs increased in abundance. Ps-ACS1 transcripts markedly increased in whole fruit but late, during the climacteric stage (∼82 DAB), and strongly declined thereafter but only in the seed (Fig. 5C). Ps-ACS3 transcript levels increased to a peak ∼80 DAB and decreased slightly thereafter (Fig. 5E), whereas those for the Ps-ACS4 mRNA accumulation peak correlated well with the increase in ethylene production at ∼82 DAB, (Fig. 5A, G).
Throughout ripening of late ‘SH’ fruit (90–105 DAB), Ps-ACS1 decreased in abundance in the pulp but steadily increased in the seeds (Fig. 5D). Ps-ACS3 transcripts slightly accumulated in both fruit tissues to reach their maximum in the post-climacteric stage at ∼105 DAB (Fig. 5F). The expression of Ps-ACS4 in ‘SH’ fruit has the same pattern as in ‘EG’ fruit, except that its accumulation in the seed remained constant and low throughout ‘SH’ fruit ripening (Fig. 5H). In both plum cultivars, Ps-ACS5 transcripts were absent or expressed in very low levels in the whole fruit throughout ripening (Fig. 5I, J).
Effect of 1-MCP on the expression of the four Ps-ACS genes during fruit ripening
To determine the role of ethylene in regulating Ps-ACS gene expression throughout early and late fruit ripening, the expression of the four cDNAs was studied in 1-MCP-pre-treated fruit during ripening.
The increases in Ps-ACS3 transcripts during early ‘EG’ fruit ripening were not found to be ethylene dependent, but seemed to be ripening related, since no significant changes in its accumulation level or pattern were observed in the MCP-treated fruits (Fig. 6C). 1-MCP completely inhibited the ethylene-associated transcription of Ps-ACS1 and -4 in the whole fruit (Fig. 6A, E). The effect of 1-MCP in the inhibition of Ps-ACS4 mRNA accumulation was stronger in the pulp than that in the seed (Fig. 6E).
In late ‘SH’ fruit, the Ps-ACS1 accumulation level or pattern was not significantly affected by MCP treatment (Fig. 6B). 1-MCP treatment strongly reduced the ripening-related increase in Ps-ACS4 expression and, in contrast to the situation in ‘EG’, also for Ps-ACS3 (Fig. 6D, F). Accumulation of both transcripts did not significantly respond to 1-MCP treatment in ‘SH’ seeds (Fig. 6D, F).
Contrary to the situation during fruit ripening, 1-MCP treatment enhanced the accumulation of Ps-ACS5 transcripts in both cultivars (Fig. 6G, H). The level of Ps-ACS5 mRNA was higher in the seed than in the pulp of ‘EG’ MCP-treated fruit (Fig. 6G); whereas 1-MCP treatment did not affect the transcript level of Ps-ACS5 in ‘SH’ MCP-treated seed (Fig. 6H).
The expression of the Ps-EOL1 during fruit ripening
To investigate if the differences between the two plum cultivars in the date and rate of ripening are related to the differences in Ps-EOL1 mRNA accumulation pattern and/or level, the expression of Ps-EOL1 mRNA during ripening of early and late plum cultivars was studied.
Throughout ripening of early ‘EG’ fruit, high levels of Ps-EOL1 transcripts accumulated in the non-climacteric stage at ∼78 DAB (Fig. 7A). As ripening progressed and higher levels of ethylene were produced, the expression levels of Ps-EOL1 mRNAs decreased in abundance to reach their basal level at the climacteric stage (∼82 DAB). In the post-climacteric stage (∼83 DAB), and when climacteric ethylene evolution starts to decline, Ps-EOL1 transcripts start to increase again (Fig. 7A). Unlike ‘EG’ fruit, the expression levels of Ps-EOL1 mRNA increased in abundance during late ‘SH’ fruit ripening to reach a maximum at the post-climacteric stage (Fig. 7B). In both cultivars, 1-MCP treatment enhanced the accumulation of Ps-EOL1 transcripts in the whole fruit (Fig. 7C, D).
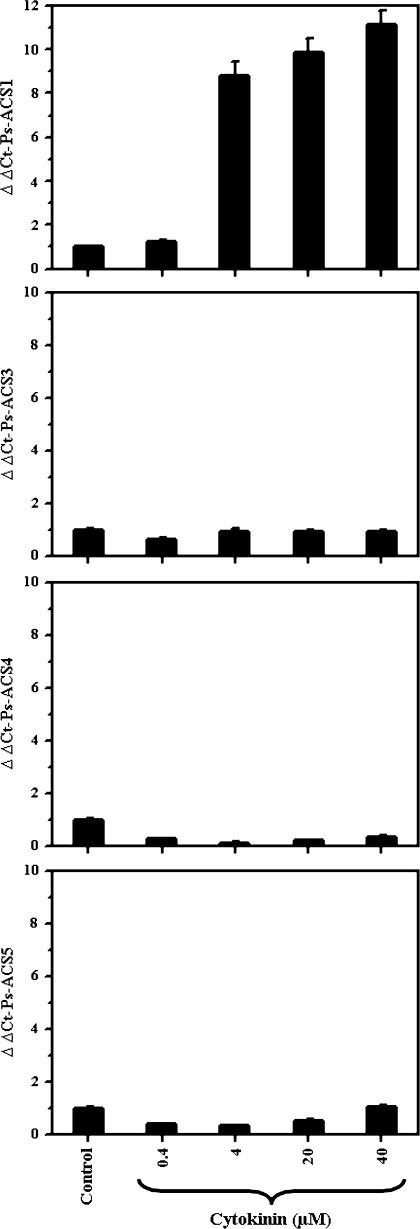
Effect of cytokinin on the expression of the four Ps-ACS cDNAs
Cytokinin increases ethylene biosynthesis by increasing uniquely Type 2 ACS isozyme activity via a post-transcriptional mechanism (Vogel et al., 1998; Woeste et al., 1999). To explore this effect, the expression level of the four Ps-ACS transcripts was studied in leaves treated with different concentrations of cytokinin. From the four Ps-ACS mRNAs isolated, only Ps-ACS1 mRNA showed a steady increase in response to cytokinin treatment (Fig. 8). Ps-ACS1 mRNA levels did not respond significantly to low cytokinin concentration (0.4 μM; Fig. 8). A dramatic increase in Ps-ACS1 transcript levels was detected with increasing cytokinin concentrations (4–40 μM; Fig. 8).
Effect of auxins and gibberellins (GAs) on the expression of the four Ps-ACS mRNAs
High levels of different Ps-ACS transcripts accumulated in the seed during fruit development and ripening. To investigate whether any of the Ps-ACS mRNAs under study were induced by auxin and/or gibberellin that strongly occurred during seed development, the expression of the four cDNAs was studied in leaves treated with different types and concentrations of synthetic and natural auxins and gibberellins.
None of the Ps-ACS transcripts significantly responded to 2,4-D treatment, at least under the conditions used in this study (data not shown). The transcript levels of the four cDNAs increased along with increasing picloram concentrations (Fig. 9A–D).
Plant tissues treated with IAA, IBA, and NAA exhibited dramatic increases in Ps-ACS1, -3, and -5 mRNA levels. IAA- and NAA-treated leaves strongly accumulated Ps-ACS1 and -3 mRNAs but only with low concentrations of hormones (0.4 μM and/or 4 μM) and declined thereafter with higher concentrations (Fig. 9E, F, M, N). While the Ps-ACS5 accumulation pattern was similar to those of Ps-ACS1 and -3 in response to IAA treatment (Fig. 9H), in NAA-treated tissues Ps-ACS5 mRNA expression levels significantly increased only at high NAA concentrations (Fig. 9P).
Ps-ACS1 and -3 mRNAs were mostly detected only in leaves treated with elevated IBA concentrations (Fig. 9I, J), whereas Ps-ACS5 expression levels did not significantly respond to IBA treatment (Fig. 9L). The expression of Ps-ACS4 was not affected by IAA and IBA treatments, but was markedly induced by NAA at high concentrations (40 μM; Fig. 9G, K, O).
Ps-ACS4 mRNA levels did not significantly respond to any of the gibberellin treatments used in this study (Fig. 10C, G). Ps-ACS1, -3, and -5 expression levels were slightly induced by low GA3 concentration (0.4 μM) and declined thereafter to reach their basal level with higher concentrations (Fig. 10A, B, D). The effect of GA3 treatment on the induction of the different Ps-ACS mRNAs was significantly higher for Ps-ACS5 transcripts than those of Ps-ACS1 and -3.
Transcript levels of the three Ps-ACS mRNAs increased after treatment with GA4+7 (Fig. 10E, F, H). The patterns of transcript accumulation for the three cDNAs were, however, quite different. The levels of Ps-ACS1 mRNA increased to reach their maximum with 4 μM GA4+7 concentrations and declined thereafter at the highest concentration (40 μM) (Fig. 10E). Ps-ACS3 transcript levels increased dramatically along with increasing GA4+7 concentrations (Fig. 10F). Ps-ACS5 mRNA accumulated in GA4+7-treated leaves in the same manner as Ps-ACS3 but at much lower levels (Fig. 10H).
Alleles identified for the Ps-ACS3 promoter region
Since Ps-ACS3 mRNA showed differential expression between early and late plum cultivars, further genomic characterization was undertaken in order to determine possible genetic linkages with the level of autocatalytic ethylene and the capacity of fruit to ripen.
Two alleles of the 5′-non-coding regions of the Ps-ACS3 [Ps-DACS3a (–1349 bp) and Ps-DACS3b (–1496 bp)] gene were identified from the ‘EG’ and ‘SH’ cultivars. The percentage similarity between the isolated sections of the 5′-non-coding regions was 57% (Fig. S2 in Supplementary data available at JXB online). Interestingly, the analysis of both promoter sequences revealed the presence of a sequence motif box only in the Ps-DACS3b promoter sequence (Supplementary Fig. S2) which shows 87.5% identity with an ERE (ethylene responsive element) regulatory element found in the promoter of tomato E4 gene (Montgomery et al., 1993).
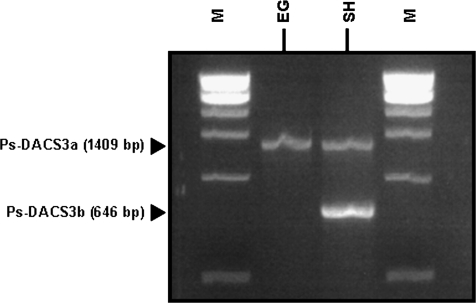
PCR was performed to determine the presence of the Ps-DACS3a/b alleles in the ‘EG’ and ‘SH’ genomes. While the early ‘EG’ genotype was homozygous for Ps-DACS3a, the late ‘SH’ genotype was found to be Ps-DACS3a/b heterozygous (Fig. 11).
Fig. 11.
Diagnosis of the two Ps-DACS3a/b alleles in two plum cultivars, ‘EG’ and ‘SH’. DNA from both cultivars was extracted from leaves. Specific primers were used to amplify each 5′-flanking region and the PCR products were viewed on a 1.2% agarose gel. The sizes of the fragments are given on the left side. Bands of 1409 bp and 646 bp correspond to Ps-DACS3a (EU034651) and Ps-DACS3b (EU034652), respectively.
Discussion
In general Japanese plum fruits are climacteric, characterized by producing a burst of autocatalytic ethylene that triggers and maintains many aspects of the ripening process (Abdi et al., 1997). However, within these plums there are marked differences that distinguish them from the other climacteric fruits. Prunus spp., including plum, exhibit a typical double sigmoid growth pattern during fruit development and ripening (Tonutti et al., 1997). Within this developmental period, four distinct stages are clearly recognized.
There are big differences in ripening behaviour and in ethylene production and responses in the plum cultivars used in this study. ‘EG’ fruits showed ripening patterns typical of climacteric fruits, while the late cultivar (‘SH’) exhibited a suppressed-climacteric pattern with only a slight increase in autocatalytic ethylene production related to ripening.
Some ethylene responses, including ACO1 gene expression and enzyme activity, can be induced in immature green plum fruits and seeds, which remain unable to produce autocatalytic ethylene and to ripen (Fernández-Otero et al., 2006). This indicates that other factors control the ethylene response and ripening, and consequently control the ethylene levels during later stages of fruit ripening. In tomato, ACS has been shown to control the onset and maintenance of the ripening-related autocatalytic ethylene (Nakatsuka et al., 1998).
Four ethylene biosynthetic mRNAs, putatively encoding ACS isozymes, were isolated and characterized from two plum cultivars in order to determine their role in fruit ripening in terms of the double sigmoid growth pattern. The predicted amino acid sequences had the traits generally associated with ACS post-transcriptional and/or post-translational regulation and related aminotransferase activity (Yamagami et al., 2003; Yoshida et al., 2005).
In the dendrogram, a number of well-defined branches have both Arabidopsis and tomato sequences but lack plum sequences, suggesting that there are likely to be as yet unidentified ACS genes within the plum genome. In species where multiple ACS genes have been characterized, each mRNA appears to have a unique mode of transcriptional and/or post-translational regulation (Wang et al., 2002).
Post-translational regulation has been identified in a number of Arabidopsis and tomato ACS proteins (Chae et al., 2003; Yoshida et al., 2005). Ps-ACS4 and -5 predicted proteins are closely related to At-ACS2 and -6 (Type 1), respectively, that were characterized as lacking the Val residue in the conserved C-terminal ‘WVF’ motif but have the Ser target of CDPK phosphorylation in the ‘RLSF’ motif (Tatsuki and Mori, 2001) followed by an extended C-terminal region containing the three conserved Ser residues that are the target for MPK6 phosphorylation (Liu and Zhang, 2004). Replacement of the conserved Val residue with a Leu in all ACS proteins belonging to the Type 1 subfamily (Fig. S1 in Supplementary data available at JXB online) suggests that these proteins have special post-translational behaviour. The ethylene-overproducing phenotype of the eto3 mutation is a result of replacing the Val residue in the ‘WVF’ motif, which is essential for the regulation of At-ACS9 activity, with aspartic acid (Chae et al., 2003), suggesting an important role for this small motif in protein stability. ACS isozymes from Type 2 have been shown to possess the TOE sequence that includes the ‘WVF’, ‘RLSF’, and R/D/E-rich motifs (Yoshida et al., 2006). Additionally, all ACS isoforms that have been shown to be post-translationally regulated by CDPK phosphorylation fall into this group (Chae et al., 2003; Sebastià et al., 2004). Ps-ACS1 as At-ACS4, -5, -8, and -9 and Sl-ACS3 has the corresponding Val and Ser residues necessary for ETO1 interaction and CDPK phosphorylation (Tatsuki and Mori, 2001; Yoshida et al., 2005). None of the ACS isoforms from Type 2 has the three conserved Ser residues found in Type 1 ACC synthase, and MPK6 does not phosphorylate At-ACS5 in vitro (Liu and Zhang, 2004), indicating a restricted specific regulation of Type 1 ACS isozymes by MPK6. Interestingly, the Ps-ACS1 predicted protein has one putative phosphorylation site (S465) where the Ser residue was followed by Pro (Fig. S1 in Supplementary data available at JXB online), which provides the ability of Ps-ACS1 protein to serve as an MPK6 substrate. Liu and Zhang (2004) suggested that the phosphorylation of the three Ser residues by MPK6 is independent of each other, but the level of phosphorylation is related to the number of phosphorylation sites. Thus, at least two parallel signalling pathways, containing either a CDPK, for Type 2 ACS isozymes, and/or MPK6 for Type 1 ACS isozymes, might be involved in modulating ACS function and activity. Ps-ACS3 (Type 3) has a short C-terminal region compared with the other Ps-ACS sequences and consequently lacks all the residues important for protein phosphorylation. It seems that Ps-ACS3 protein may play an important role in stabilizing ACC levels in plums. Yoshida et al. (2005) showed that ETO1 specifically interacts with Type 2 of ACC synthase (At-ACS5 and Sl-ACS3), but not with Type 1 (Sl-ACS2) or Type 3 (Sl-ACS4) in the yeast two-hybrid system. Mutation of the Sl-ACS2 C-terminus, by adding the ‘WVF’ motif just before the ‘RLSF’ motif to mimic Sl-ACS3 protein, was sufficient to recover the strong interaction with ETO1 to a level comparable with Sl-ACS3 (Yoshida et al., 2005).
Ps-ACS1, -3, and -5 are expressed in very high levels in opened flowers and rapidly decline after fertilization. The complete inhibition of the different Ps-ACS transcripts, post-fertilization, correlated well with a large accumulation of free polyamines that occur strongly at fertilization (Pérez-Amador et al., 1995; Antognoni et al., 2002; Aziz, 2003; DeDios et al., 2006). Ethylene and polyamines share SAM as a common intermediate. It was shown that ethylene biosynthesis may compete with the biosynthesis of polyamines (Kushad and Dumbroff, 1991; Escribano and Merodio, 1994), and it cannot be ruled out that SAM may be alternatively channelled towards ethylene or the polyamine pathway, constituting a control mechanism for certain physiological processes in higher plants (Walden et al., 1997). The gradual increase in abundance of the three transcripts throughout early fruit development may reflect the important roles of ethylene in petal wilt, flower abscission, and embryo development.
Ps-ACS1, -3, and -5 increased, again, to a peak in the young fruit at ∼27 DAB. The increase of the three transcripts corresponds to low levels of ethylene production, and high levels of IAA and cytokinin contents, important for cell division during embryogenesis, which occurred throughout S1 of fruit development (Miller et al., 1987; Hartmann et al., 2002; this study). Ethylene production in the S1 stage is perhaps needed for regulating the abundant cell division and embryo development characterizing this stage of fruit development. It was found that the low ethylene emission throughout this stage of fruit development coincided with high ACC and N-malonyl-ACC (MACC) contents detected in the non-lignified endocarp (DeDios et al., 2006). The absence of all the studied Ps-ACS transcripts throughout S2 of fruit development coincided with strong down-regulation of four different ethylene perception and signal transduction component elements (El-Sharkawy et al., 2007), suggesting only a minor role for ethylene during this stage.
Although there was hardly any ethylene detected throughout S3 of fruit development, high transcript levels of Ps-ACS1, -3, and -5 were detected in the seed from this stage. During S3, the endocarp is almost fully developed to form the highly lignified seed, which could not be involved in any synthesis of ethylene for many reasons: (i) any accumulation of MACC and ACC coincided with very high levels of free polyamines in the lignified seed; (ii) the seed, which develops much later than the fruit, does not have in vivo ACO activity (DeDios et al., 2006). The present results showed that the strong accumulation of Ps-ACS1, -3, and -5 in the seed coincides with the high auxin and gibberellin content detected in the seed during S3 (Jackson, 1968; Miller et al., 1987). The role of seeds in stone fruit development was determined by Jackson (1968) and Miller et al. (1987) who provided evidence that seeds stimulate fruit growth and ripening by providing auxins and gibberellins. The accumulation of Ps-ACS1 and -3 transcripts in the fruit pulp was ethylene independent but seemed to be auxin and/or gibberellin dependent. Early induction of both transcripts suggests their importance for the fruit to ripen.
Differences in the ripening behaviour between the cultivars reflect an altered capacity to produce and respond to ethylene. Mature ‘EG’ fruit are capable of producing autocatalytic ethylene, and ripen, ∼82 DAB, as a typical climacteric fruit. Late ‘SH’ fruit exhibited a very low ethylene emission and ripened more slowly and later than ‘EG’ fruit. However, fruits pre-treated with propylene could recover the climacteric pattern and ripen as rapidly as early cultivars (Abdi et al., 1997).
From the different ACS isozymes studied, Ps-ACS1 and -3, and partially Ps-ACS4 cDNAs showed a cultivar-specific expression pattern and/or level throughout fruit ripening. In both cultivars studied, Ps-ACS1, -3, and -4 expression levels dramatically increased in abundance during the ripening process (S4). Transcripts of the three mRNAs in ‘SH’ fruits were at least four times lower than in ‘EG’ fruits.
During ‘EG’ and ‘SH’ fruit ripening, Ps-ACS1 mRNA accumulated in ethylene-dependent and -independent manners, respectively. Isolation of the Ps-ACS1 promoter sequence (∼1.8 kb) from both cultivars showed no significant differences in their homology (data not shown).
In ‘EG’ fruits, Ps-ACS3 transcript levels increased early during the onset of ripening in an ethylene-independent manner. By contrast, the induction of Ps-ACS3 transcripts, throughout ripening of ‘SH’ fruit, was dependent on ethylene action. Both plum cultivars share a common allele of the Ps-DACS3 promoter associated with Ps-DACS3a, whereas the second allele, associated with Ps-DACS3b, was found only in the late ‘SH’ genotype. It is clear that the Ps-ACS3 isoform that functions during fruit development and throughout ‘EG’ fruit ripening is Ps-ACS3a, while the Ps-ACS3b isoform is only functional during ‘SH’ fruit ripening. Interestingly, the promoter of Ps-DACS3b has two boxes of motif sequences that share high sequence identity with a regulatory element found in the promoter of Arabidopsis ERF1 (Solano et al., 1998). This sequence has also been found in the promoter regions of many genes which are up-regulated by ethylene (Itzhaki and Woodson, 1993; Montgomery et al., 1993). The presence of the two boxes in the promoter is necessary for full promoter activity (Tapia et al., 2005). In ‘SH’ fruit, the low transcript levels of Ps-ACS3 (Ps-ACS3b) expressed during ripening may be related to the low quantities of autocatalytic ethylene produced.
In both cultivars, Ps-ACS4 mRNA levels increased linearly with the ethylene levels. 1-MCP treatment completely repressed Ps-ACS4 expression. The transcript levels of Ps-ACS1, -3, and -4 appear to play an important role in determining the level of autocatalytic ethylene production and the capacity of the fruit to ripen. Ps-ACS5 mRNA was negatively regulated by ethylene during ripening of both cultivars studied, since its transcripts only accumulated in non-autocatalytic ethylene-produced tissues such as vegetative tissues, flowers, immature fruit, and MCP-treated fruits.
The high transcript levels of Ps-ACS1, -3, and -4 detected in the seeds during ripening have no role in the synthesis of autocatalytic ethylene produced throughout this stage of fruit development, since the endocarp is already lignified at this stage and, as in other seeds (Rodrìguez-Gacio and Matilla, 2001), the activity of the last step of ethylene synthesis is inhibited. By contrast, the greatest amount of ethylene produced and ACC and MACC content have been observed in the ripening mesocarp and epicarp (DeDios et al., 2006).
In both plum cultivars, the fruit development process seems to progress in parallel. Once the ripening stage (S4) has started, the ripening process in the late ‘SH’ cultivar seems to progress much more slowly than in the early ‘EG’ cultivar. As mentioned previously, there are large differences in ripening behaviour and in ethylene production and responses in the plum cultivars used in this study. ETO1-like protein (EOL1) is a negative regulator of ethylene evolution and interacts with Type 2 ACS proteins to degrade them via the ubiquitin–26-S proteasome pathway (Chae et al., 2003; Chae and Kieber, 2005; Yoshida et al., 2005).
The expression of Ps-EOL1 throughout ‘EG’ and ‘SH’ fruit ripening was studied to explore the possible role of EOL1 protein in suppressing ethylene production and delaying ripening in plum fruit. Ps-EOL1 transcript levels decreased in abundance along with increasing ethylene evolution. Contrarily, ‘SH’ fruits significantly accumulated Ps-EOL1 mRNA during ripening. Interestingly, Ps-EOL1 transcripts were negatively regulated by ethylene in both the cultivars studied, since 1-MCP treatment increased its transcript accumulation. It seems that late ‘SH’ fruit produces insufficient amounts of ethylene to inhibit Ps-EOL1 mRNA induction. However, treatment of ‘SH’ fruit with propylene restored the climacteric pattern and the fruit ripens as a typical climacteric fruit (Abdi et al., 1997). Recent studies suggest that cytokinin increased ethylene biosynthesis by inhibiting the interaction between Type 2 ACS isozymes and ETO1 protein, which resulted in reducing ACS protein degradation by a proteasome-dependent pathway (Wang et al., 2004; Chae and Kieber, 2005). Cytokinin activity tends to be high in developing fruits but decreases thereafter and becomes difficult to detect in mature fruits (Hartmann et al., 2002). This indicates that cytokinin does not have any role in enhancing ethylene production in plum fruit through the inhibition of EOL1 and Type 2 ACC synthesis interaction (Chae and Kieber, 2005; this study). Taken together, the data suggest that there is a minimum requirement for ethylene quantity necessary to inhibit the negative ethylene regulator, Ps-EOL1, transcript levels, but once this minimum requirement is met and Ps-EOL1 transcripts are down-regulated, the developmental changes required for ripening will be accelerated.
As mentioned before, cytokinin increases ethylene biosynthesis via a post-transcriptional mechanism (Vogel et al., 1998; Woeste et al., 1999) by blocking the interaction of the Type 2 ACS isozymes with ETO1 protein that leads to an increase in ACS protein stability (Chae and Kieber, 2005), whereas the application of other plant hormones such as auxin and gibberellin can affect ethylene levels through increased ACS transcription levels (Abel et al., 1995; Kaneta et al., 1997). Ethylene can also affect its own biosynthesis, either increasing (autocatalysis) or decreasing (autoinhibition) its rate of production (Nakatsuka et al., 1998). All these plant hormones play an important role in the differential accumulation of the various Ps-ACS mRNAs in flowers, and throughout fruit development and ripening. Among the four Ps-ACS mRNAs characterized, only Ps-ACS1 is up-regulated by cytokinin. Ps-ACS1 and -3 transcripts were found to be strongly up-regulated by ethylene, auxin, and gibberellin. Despite the strong accumulation of Ps-ACS4 mRNA by the synthetic auxins, picloram and NAA, none of the natural auxins or gibberellins induced its expression. It seems that Ps-ACS4 transcript abundance strongly increased in response to ethylene evolution. Unlike Ps-ACS4, Ps-ACS5 transcripts were completely inhibited by ethylene; however, auxin and gibberellin significantly induced its mRNA accumulation. Thus the various Ps-ACS isozymes seem to regulate distinct developmental processes and environmental responses differentially.
In tomato, it has been proposed that there are two systems of ethylene production. System 1 ethylene is autoinhibitory and operates in immature fruit and vegetative tissues. System 2, or autocatalytic ethylene, is produced during climacteric fruit ripening (Lelièvre et al., 1997; Barry et al., 2000). In tomato and other climacteric fruits, ACS genes play a critical role in the transition from system 1 to system 2.
Throughout plum fruit development, Ps-ACS1, -3, and -5 transcripts are present well above their basal levels. In both the cultivars studied, autocatalytic ethylene resulted in complete inhibition of Ps-ACS5 transcripts. Ps-ACS1 and -3 transcripts were induced in ethylene-independent and/or -dependent manners during ripening of both cultivars. Together, these results suggest that Ps-ACS5 and, to some extent Ps-ACS1 and -3, are important for system 1 ethylene production and offer further evidence for the existence of a dual system of ethylene production in plum.
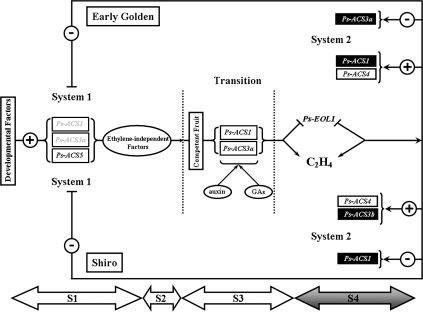
The model presented in Fig. 12 illustrates our current understanding of the control of ethylene production in plum fruit within the two-systems approach. In immature plum fruit, system 1 ethylene relies on the expression of Ps-ACS5 and partially on Ps-ACS1 and -3a. System 1 ethylene synthesis continues throughout fruit development (0–62 DAB) until the fruit attains a state of physiological maturity, at which point a transition occurs (62–77 DAB). In mature plum fruit, the transition from system 1 to system 2 is induced throughout the induction and/or repression of unknown ethylene-independent factors so that the fruit gains the competence to ripen. These factors could be the increase of plant hormones, auxin, gibberellin, and, in slightly later stages, ethylene, which coincided with the repression of Ps-EOL1 transcripts. Ps-ACS1 and -3a mRNAs are expressed during this transition period in an ethylene-independent manner but in auxin- and/or gibberellin-dependent manner. This resulted in ethylene production that activates the expression of Ps-ACS1, -3a, and -4 in ‘EG’ fruit, and Ps-ACS1, -3b, and -4 in ‘SH’ fruit. The elevated levels of ethylene produced in system 2 resulted in negative feedback of the system 1 developmental pathway and consequently reduced Ps-ACS5 expression. In tomato, transition from system 1 to system 2 is associated with the expression of Sl-ACS4 which triggers the expression of Sl-ACS2 and inhibits the expression of Sl-ACS6 (Alexander and Grierson, 2002).
Fig. 12.
Model for the regulation of Ps-ACS gene expression during fruit development and throughout ripening of early (Early Golden) and late (Shiro) plum cultivars. The plus (+) and minus (–) symbols refer to the positive and negative actions of ethylene, respectively, during system 1 and system 2 ethylene synthesis that leads to the activation (+) or repression (–) of Ps-ACS gene expression. Filled black rectangles correspond to mRNAs that are differentially regulated in early and late plum cultivars. Throughout the transition stage both hormones, auxin and gibberellin, from the seed play a major role in initiating the ripening process through increasing Ps-ACS1 and -3a transcript levels, which result in increasing endogenous ethylene levels. High levels of ethylene repress Ps-EOL1 transcripts and accelerate the autocatalytic ethylene production in the fruit (system 2). The four stages of plum fruit development throughout the double sigmoid growth pattern are indicated by S1 to S4.
The results presented here suggest that differences in Ps-ACS gene expression between early and late plums are important determinators in the ripening behaviour of the cultivars. However, in addition to Ps-ACS expression, it is likely that Ps-EOL1 is also involved in determining the suppressed-climacteric pattern in late plum cultivars. In tomato, the rin mutant fails to produce autocatalytic ethylene and to ripen (Giovannoni, 2001). This non-ripening phenotype is caused by mutations in transcription factors that control the expression of ripening-related genes. Sl-ACS4 and Sl-ACS2 transcript accumulation, critical for the onset of system 2 ethylene, does not occur in rin mutant fruit. Partially in ‘SH’ fruit, an ethylene-independent developmental process which operates to reduce Ps-ACS mRNA expression resulted in this late phenotype, ‘SH’, which appears to result from an inability of the fruit to produce sufficient quantities of ethylene to coordinate the transition from system 1 to system 2 ethylene production and ripening.
Supplementary data
Figure S1: Classification based on C-termini of ACC synthases of plum, tomato and Arabidopsis.
Figure S2: Nucleotide sequence alignment of the promoter regions of Ps-ACS3a and Ps-ACS3b genes using ClustalX program.
Acknowledgments
We thank Dr Dennis Murr (University of Guelph) for providing generous use of the GC equipment, Dr John Cline for helpful discussions, and Peter Alm for technical assistance. Financial assistance by the Canadian Foundation for Innovation, Ontario Innovation Trust and Ontario Tender Fruit Marketing Board (SJ), and the NSERC-Fellowship (IE-S) is also gratefully acknowledged.
Glossary
Abbreviations
- ACC
1-aminocyclopropane-1-carboxylic acid
- ACO
1-aminocyclopropane-1-carboxylic acid oxidase
- ACS
1-aminocyclopropane-1-carboxylic acid synthase
- DAB
days after bloom
- ‘EG’
‘Early Golden’
- ETO1
ethylene-overproducer 1
- EOL1
ETO1-like
- GA
gibberellin
- PGR
plant growth regulator
- 1-MCP
1-methylcyclopropene
- SAM
S-adenosyl-L-methionine
- ‘SH’
‘Shiro’
- TOE
target of ETO1
References
- Abdi N, Holford P, McGlasson WB, Mizrahi Y. Ripening behavior and responses to propylene in four cultivars of Japanese type plums. Postharvest Biology and Technology. 1997;12:21–34. [Google Scholar]
- Abel S, Nguyen MD, Chow W, Theologis A. ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana: structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. Journal of Biological Chemistry. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- Abeles FB, Morgan PW, Saltveit ME. Ethylene in plant biology. 2nd edn. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. Journal of Experimental Botany. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antognoni F, Ghetti F, Mazzucato A, Franceschetti M, Bangi N. Polyamine pattern during flower development in the parthenocarpic fruit (pat) mutant of tomato. Physiologia Plantarum. 2002;116:539–547. [Google Scholar]
- Aziz A. Spermidine and related-metabolic inhibitors modulate sugar and amino acid levels in Vitis vinifera L.: possible relationships with initial fruitlet abscission. Journal of Experimental Botany. 2003;54:355–363. doi: 10.1093/jxb/erg029. [DOI] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiology. 2000;123:979–986. doi: 10.1104/pp.123.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biale JB, Young RE. Respiration and ripening in fruits – retrospect and prospect. In: Friend J, Rhodes MJC, editors. Recent Advances in the Biochemistry of Fruits and Vegetables. London: Academic Press; 1981. pp. 1–39. [Google Scholar]
- Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera cv. Shiraz grape berries and the implications for pathway regulation. Plant Physiology. 1996;111:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitani G, Hohenester E, Feng L, Storici P, Kirsch JF, Jansonius JN. Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. Journal of Molecular Biology. 1999;294:745–756. doi: 10.1006/jmbi.1999.3255. [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. The eto1, eto2 and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. The Plant Cell. 2003;15:545–559. doi: 10.1105/tpc.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Kieber JJ. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends in Plant Science. 2005;10:291–296. doi: 10.1016/j.tplants.2005.04.006. [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends in Biochemical Sciences. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- DeDios P, Matilla AJ, Gallardo M. Flower fertilization and fruit development prompt changes in free polyamines and ethylene in damson plum (Prunus insititia L.) Journal of Plant Physiology. 2006;163:86–97. doi: 10.1016/j.jplph.2005.03.007. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy I, Jones B, Gentzbittel L, Lelièvre JM, Pech JC, Latché A. Differential regulation of ACC synthase genes in cold-dependent and -independent ripening in pear fruit. Plant, Cell and Environment. 2004;27:1197–1210. [Google Scholar]
- El-Sharkawy I, Kim WS, El-Kereamy A, Jayasankar S, Svircev AM, Brown DCW. Isolation and characterization of four ethylene signal transduction elements in plums (Prunus salicina L.) Journal of Experimental Botany. 2007;58:3631–3643. doi: 10.1093/jxb/erm213. [DOI] [PubMed] [Google Scholar]
- Escribano MI, Merodio C. Relevance of polyamine levels in cherimoya (Annona cherimola Mill.) fruit ripening. Physiologia Plantarum. 1994;73:201–205. [Google Scholar]
- Ferández-Otero C, Matilla AJ, Rasori A, Ramina A, Bonghi C. Regulation of ethylene biosynthesis in reproductive organs of damson plum (Prunus domestica L. subsp. Syriaca) Plant Science. 2006;171:74–83. [Google Scholar]
- Fluhr R, Mattoo AK. Ethylene-biosynthesis and perception. Critical Reviews in Plant Sciences. 1996;15:479–524. [Google Scholar]
- Giovannoni J. Molecular biology of fruit maturation and ripening. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- Hartmann HT, Kester DE, Davies FT, Geneve RL. Plant propagation. 7th edn. Upper Saddle River, NJ: Pearson Education; 2002. [Google Scholar]
- Itzhaki H, Woodson WR. Characterization of an ethylene-responsive glutathione S-transferase gene cluster in carnation. Plant Molecular Biology. 1993;22:43–58. doi: 10.1007/BF00038994. [DOI] [PubMed] [Google Scholar]
- Jackson DI. Gibberellin in the growth of peach and apricot fruits. Australian Journal of Biological Science. 1968;21:209–215. [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends in Biochemical Science. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jones ML, Woodson WR. Differential expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in carnation. Plant Physiology. 1999;119:755–764. doi: 10.1104/pp.119.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneta T, Kakimoto T, Shibaoka H. Gibberellin A3 causes a decrease in the accumulation of mRNA for ACC oxidase and in the activity of the enzyme in azuki bean (Vigna angularis) epicotyls. Plant and Cell Physiology. 1997;38:1135–1141. doi: 10.1093/oxfordjournals.pcp.a029098. [DOI] [PubMed] [Google Scholar]
- Kushad MM, Dumbroff EB. Metabolic and physiological relationship between the polyamine and ethylene biosynthetic pathways. In: Slocum RD, Flores HE, editors. Biochemistry and physiology of polyamines in plants. Boca Raton, FL: CRC Press; 1991. pp. 77–92. [Google Scholar]
- Lelièvre JM, Latché A, Jones B, Bouzayen M, Pech JC. Ethylene and fruit ripening. Physiologia Plantarum. 1997;101:727–739. [Google Scholar]
- Liu Y, Zhang S. Phosphorylation of ACC synthase by MPK, a stress-responsive MAPK, induces ethylene biosynthesis in Arabidopsis. The Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd G, McCown BH. Commercially-feasible micropropagation of Mountain Laurel, Kalmia latifolia, by shoot tip culture. Proceeding of the International Plant Propagators’ Society. 1981;30:421–427. [Google Scholar]
- McCarthy DL, Capitani G, Feng L, Gruetter MG, Kirsch JF. Glutamate 47 in 1-aminocyclopropane-1-carboxylate synthase is a major specificity determinant. Biochemistry. 2001;40:12276–12284. doi: 10.1021/bi011050z. [DOI] [PubMed] [Google Scholar]
- Mehta PK, Hale TI, Christen P. Evolutionary relationships among aminotransferases: tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase are homologous proteins. European Journal of Biochemistry. 1989;186:249–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- Miller AN, Walsh CS, Cohen JD. Measurement of indole-3-acetic acid in peach fruits (Prunus persica L. Batsch cv Redhaven) during development. Plant Physiology. 1987;84:491–494. doi: 10.1104/pp.84.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proceedings of the National Academy of Sciences, USA. 1993;90:5939–5943. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, Inaba A. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiology. 1998;118:1295–1305. doi: 10.1104/pp.118.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HBJ. GeneDoc: a tool for editing and annotating multiple sequence alignments, distributed by the authors. 1997 [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pérez-Amador MA, Carbonell J, Granell A. Expression of arginine decarboxylase is induced during early fruit development and in young tissues of Pisum sativum (L.) Plant Molecular Biology. 1995;28:997–1009. doi: 10.1007/BF00032662. [DOI] [PubMed] [Google Scholar]
- Rodrìguez-Gacio MC, Matilla AJ. The last step of the ethylene-biosynthesis pathway in turnip tops (Brassica rapa) seeds: alterations related to development and germination and its inhibition during desiccation. Physiologia Plantarum. 2001;111:273–279. doi: 10.1034/j.1399-3054.2001.1120216.x. [DOI] [PubMed] [Google Scholar]
- Sebastià HC, Hardin SC, Clouse SD, Kieber JJ, Huber SC. Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Archives of Biochemistry and Biophysics. 2004;428:81–91. doi: 10.1016/j.abb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by Ethylene-Insensitive3 and Etylene-Response-Factor1. Genes and Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia G, Verdugo I, Yañez M, Ahumada I, Theoduloz C, Cordero C, Poblete F, González E, Ruiz-Lara S. Involvement of ethylene in stress-induced expression of the TLC1.1 retrotransposon from Lycopersicon chilense Dun. Plant Physiology. 2005;138:2075–2086. doi: 10.1104/pp.105.059766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Mori H. Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase LE-ACS2, at the C-terminal region. Journal of Biological Chemistry. 2001;276:28051–28057. doi: 10.1074/jbc.M101543200. [DOI] [PubMed] [Google Scholar]
- Tonutti P, Bonghi C, Ruperti B, Tornielli GB, Ramina A. Ethylene evolution and 1-aminocyclopropane-1-carboxylate oxidase gene expression during early development and ripening of peach fruit. Journal of the American Society of Horticultural Science. 1997;122:642–647. [Google Scholar]
- Vogel JP, Woeste KW, Theologis A, Kieber JJ. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proceedings of the National Academy of Sciences, USA. 1998;95:4766–4771. doi: 10.1073/pnas.95.8.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden R, Cordeiro A, Tiburcio AF. Polyamines: small molecules triggering pathways in plant growth and development. Plant Physiology. 1997;113:1009–1013. doi: 10.1104/pp.113.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. The Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428:945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- Woeste K, Ye C, Kieber JJ. Two Arabidopsis mutants that overproduce ethylene are affected in the post-transcriptional regulation of ACC synthase. Plant Physiology. 1999;119:521–530. doi: 10.1104/pp.119.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology. 1984;35:155–189. [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. Biochemical diversity among the 1-aminocyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. Journal of Biological Chemistry. 2003;278:49102–49111. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KLC, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biology. 2005;10:5–14. doi: 10.1186/1471-2229-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Wang KLC, Chang CM, Mori K, Uchida E, Ecker JR. The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Molecular Biology. 2006;62:427–437. doi: 10.1007/s11103-006-9029-7. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Molecular Biology. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- Zuzunaga M, Serrano M, Martínez-Romero D, Valero D, Riquelme F. Comparative study of two plum (Prunus salicina Lindl.) cultivars during growth and ripening. Food Science and Technology International. 2001;7:123–130. [Google Scholar]