Abstract
DEAD-box proteins comprise a large protein family with members from all kingdoms and play important roles in all types of processes in RNA metabolism. In this study, a rice gene OsBIRH1, which encodes a DEAD-box RNA helicase protein, was cloned and characterized. The predicted OsBIRH1 protein contains a DEAD domain and all conserved motifs that are common characteristics of DEAD-box RNA helicases. Recombinant OsBIRH1 protein purified from Escherichia coli was shown to have both RNA-dependent ATPase and ATP-dependent RNA helicase activities in vitro. Expression of OsBIRH1 was activated in rice seedling leaves after treatment with defence-related signal chemicals, for example benzothiadiazole, salicylic acid, l-aminocyclopropane-1-carboxylic acid, and jasmonic acid, and was also up-regulated in an incompatible interaction between a resistant rice genotype and the blast fungus, Magnaporthe grisea. Transgenic Arabidopsis plants that overexpress the OsBIRH1 gene were generated. Disease resistance phenotype assays revealed that the OsBIRH1-overexpressing transgenic plants showed an enhanced disease resistance against Alternaria brassicicola and Pseudomonas syringae pv. tomato DC3000. Meanwhile, defence-related genes, for example PR-1, PR-2, PR-5, and PDF1.2, showed an up-regulated expression in the transgenic plants. Moreover, the OsBIRH1 transgenic Arabidopsis plants also showed increased tolerance to oxidative stress and elevated expression levels of oxidative defence genes, AtApx1, AtApx2, and AtFSD1. The results suggest that OsBIRH1 encodes a functional DEAD-box RNA helicase and plays important roles in defence responses against biotic and abiotic stresses.
Keywords: DEAD-box RNA helicase, disease resistance, OsBIRH1, oxidative stress, rice (Oryza sativa L.)
Introduction
Plants, unlike animals, are sessile during their growth and development, and thus are unable to avoid unfavourable environmental conditions. To survive in stress environments, plants have developed a complex, precisely regulated defence network to cope with a variety of biotic and abiotic stresses. In particular, plant responses to invading pathogens involve a complex network of defence mechanisms, in which thousands of genes are coordinately activated and integrated upon recognition of pathogen-associated molecular patterns (Takken et al., 2007). These coordinately activated genes encode quite different species of proteins involved in numerous biological processes that are favourable for the defence response against pathogen attack. Recent studies have revealed that expression of many genes with regulatory functions such as transcription factors, protein kinases and phosphatases, RNA-binding proteins, calcium-binding proteins, etc. is altered during the defence response (Xiong et al., 2002; Zhu, 2002; Shinozaki et al., 2003; Bartels and Sunkar, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006).
RNA helicases, referred to as enzymes that use energy derived from the hydrolysis of a nucleotide triphosphate to unwind double-stranded RNAs, function as molecular motors that rearrange RNA secondary structure, potentially playing roles in many cellular process (de la Cruz et al., 1999). DEAD-box and related DEAH, DExH, and DExD families, which are commonly referred to as the DExD/H helicase family, are members of the SF2 family, and share nine highly conserved motifs (Gorbalenya and Koonin, 1993; Tanner and Linder, 2001; Caruthers and McKay, 2002). These closely related families can be distinguished by variations within their conserved motifs. The DEAD-box family is by far the largest one, and is characterized by the presence of nine conserved motifs that are involved in ATPase and helicase activities and in their regulation (Tanner et al., 2003). The conserved motifs of the DExD/H helicases are clustered in a central core region that spans 350–400 amino acids (Tanner and Linder, 2001; Caruthers and McKay, 2002). By contrast, the N- and C-terminal extensions are highly variable in size and composition (Caruthers and McKay, 2002).
The DEAD-box RNA helicases form a large family of proteins found in all eukaryotes, and most prokaryotes (Aubourg et al., 1999; de la Cruz et al., 1999; Rocak and Linder, 2004). For example, nearly 30 genes that encode DEAD-box RNA helicases were identified in genomes of Caenorhabditis elegans and Drosophila melanogaster (Boudet et al., 2001). In Arabidopsis, >50 members of DEAD-box RNA helicases have been identified (Aubourg et al., 1999; Boudet et al., 2001). Despite the involvement of DEAD-box RNA helicases in many diverse biological processes, their precise functions and regulation largely remain to be elucidated.
Increasing evidence suggests that the DEAD-box RNA helicases play important roles in plant growth and development processes, probably through regulation of RNA metabolism and gene expression (Itadani et al., 1994; Brander and Kuhlemeier, 1995; Okanami et al., 1998; Jacobsen et al., 1999; Li et al., 2001; Dalmay et al., 2001; Western et al., 2002; Gendra et al., 2004; Arciga-Reyes et al., 2006; Yoine et al., 2006a, b; Kobayashi et al., 2007; Matthes et al., 2007). ISE2, an Arabidopsis DEVH box RNA helicase, affects the structure of plasmodesmata in the embryo and genetic data, demonstrating that ISE2 is involved in determination of cell fate through a post-transcriptional gene silencing mechanism (Kobayashi et al., 2007). The Arabidopsis recessive mutant caf converts the floral meristems to an indeterminate state and thus CAF, a DExH/DEAD-box RNA helicase, appears to suppress cell division in floral meristems (Jacobsen et al., 1999). The tobacco VDL, a member of the DEAD-box RNA helicase family, controls early plastid differentiation and plant morphogenesis as its recessive mutant vdl showed variegated leaves and abnormal roots and flowers (Wang et al., 2000).
Involvement of the DEAD-box RNA helicases in plant stress responses has been evident recently in several studies in Arabidopsis. A cold stress-regulated DEAD-box RNA helicase, LOS4, was found to be an early regulator of CBF transcription factor expression in response to chilling (Gong et al., 2002, 2005). Genome-wide expression profiling has also identified additional Arabidopsis RNA helicase genes whose expression is cold-stress regulated (Seki et al., 2001, 2002; Kreps et al., 2002). STRS1 and STRS2, encoding DEAD-box RNA helicases, were also identified in a functional genomics screening. Loss of function of the STRS1 and STRS2 genes led to an increased expression of stress-responsive transcriptional activators and abiotic stress tolerance, and thus both STRS1 and STRS2 appear to function as an upstream negative regulator of ABA-dependent and ABA-independent abiotic stress signalling networks (Kant et al., 2007).
Recent studies have provided new insights into understanding the mechanism by which the DEAD-box RNA helicases exert their biological functions. The DEAD-box RNA helicases, CARPEL FACTORY/DICER-LIKE 1 (DCL1) and UPF1, are critical for the microRNA biogenesis and RNAi in Arabidopsis (Park et al., 2002; Reinhart et al., 2002). DCL1 and a dsRNA-binding protein, HYPONASTIC LEAVES 1 (HYL1), form a complex, which functions together in pri-miRNA processing in a distinct nuclear organelle (Kurihara et al., 2006). DCL1 is also involved in processing of the natural antisense siRNAs from transcripts of protein-encoding genes (Borsani et al., 2005). UPF1 is essential for the targeted destruction of mRNAs containing premature termination codons by nonsense-mediated mRNA decay, and upf1 mutants contain high levels of aberrant mRNAs and exhibit a range of unexpected vegetative and floral abnormalities (Arciga-Reyes et al., 2006; Yoine et al., 2006a, b).
In this study, the characterization of a rice gene, OsBIRH1, which encodes a DEAD-box RNA helicase with a DEVD motif, is reported. The OsBIRH1 protein has RNA-dependent ATPase and ATP-dependent RNA helicase activities in vitro. Expression of OsBIRH1 was activated by pathogen infection as well as by treatment with disease resistance-related signal molecules. Ectopic expression of OsBIRH1 in transgenic Arabidopsis led to increased expression of defence-related genes and enhanced disease resistance and oxidative stress tolerance. The results suggest that OsBIRH1 encodes a functional DEAD-box RNA helicase, which functions as a positive regulator in defence responses against biotic and abiotic stresses.
Materials and methods
Plant growth and treatments
Rice cultivar Yuanfengzao (Oryza sativa L. subsp. indica) and a pair of isogenic lines, H8R and H8S, were used in this study. Cultivar Yuanfengzao is highly susceptible to rice blast fungus Magnaporthe grisea strain 85-14B1, belonging to race ZB1. H8S is susceptible, while H8R is resistant to M. grisea isolate 85-14B1. Seedlings were grown in soil in a growth chamber at 27 °C/22 °C with 14 h light/10 h dark (night/day). Three-week-old seedlings of cv. Yuanfengzao were used for most experiments unless indicated otherwise. Treatments with benzothiadiazole (BTH; Novartis, USA), salicylic acid (SA; Sigma-Aldrich, St Louis, MO, USA), jasmonic acid (JA; Sigma-Aldrich), and 1-aminocyclopropane-l-carboxlate (ACC; Sigma-Aldrich) were performed by foliar spraying with solutions of 0.3 mM BTH, 1 mM SA (pH 6.5), 100 μM JA, or 100 μM ACC, and with sterilized distilled water as a control. Inoculation with M. grisea was done as described previously (Luo et al., 2005a). Leaf samples were collected at different time points as indicated and stored at −80 °C until use.
Arabidopsis plants were grown in soil under fluorescent light (150 μE m2 s−1) at 22±3 °C with 60% RH and a 12 h light/12 h dark cycle. For axenic growth, seeds were sterilized and sown on a medium solidified with 1% agar that contained MS salts (Sigma-Aldrich) and 2% (w/v) sucrose. Conditions for axenic growth were 12 h of light of 60 μE m2 s−1.
Cloning of OsBIRH1
Differentially expressed clone BIHN-n5 was isolated from a suppression subtractive hybridization library and contained a 664 bp insert of cDNA, which showed a high level of similarity to plant DEAD-box RNA helicase proteins. The 5′ and 3′ ends flanking the sequence in the clone BIHN-n5 were amplified by PCR using phage DNA prepared from a rice cDNA library as template. Gene-specific primers used were BIHN-n5-1F (5′-ATG GAG GTG GCG GGT GCT CA-3′) and BIHN-n5-1R (5′-CTA GAC GCA TGG CAC ATC GT-3′). Primer BIHN-n5-1R and a vector primer T3-2 (5′-CCT GCA GGT CGA CAC TAG TG-3′) were used for amplification of the 5′ end, and the primer BIHN-n5-1F and the vector-specific primer T7-2 (5′-CCA CCC GGG TGG AAA ATC GA-3′) were used for amplification of the 3′ end. All PCR products were purified using the DNA Gel Purification Kit (Sangon, Shanghai, China) and cloned into pUCm-T-vector (Sangon, Shanghai, China) by T/A cloning. The plasmid containing the complete open reading frame (ORF) and the UTR sequences of both ends was designated as pUCm-BIHN-n5. The coding sequence was amplified using plasmid pUCm-BIHN-n5 as template, and a pair of specific primers BIHN-n5-11F (5′-CGC GGA TCC ATG GAG GTG GCG GGT GCT C-3′) and BIHN-n5-11R (5′-CCC AAG CTT CTA GAC GCA TGG CAC ATC G-3′), which contain a BamHI and a HindIII site (underlined), respectively. The entire ORF was cloned and confirmed by sequencing, yielding plasmid pUCm-OsBIRH1-1.
DNA sequencing and sequence analysis
DNA sequencing was performed on both strands on the MegaBACE 1000 DNA Analysis System (Amersham Biosciences, Piscataway, NJ, USA) at the Center of Analysis and Measurement in Zhejiang University. Similarity searches on nucleotide and amino acid sequences were carried out using BLAST at the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignments were conducted using CLUSTAL (http://www.ebi.ac.uk/clustalw/). A phylogenetic tree was constructed by the Neighbor–Joining method using Mega3.1 software (http://www.megasoftware.net/; Kumar et al., 2001).
Purification of recombinant OsBIRH1 protein
The OsBIRH1 coding region was released from pUCm-OsBIRH1-1 by digestion with BamHI/HindIII and cloned into pRSET-A vector (Invitrogen, Carlsbad, CA, USA), which was then introduced into Escherichia coli strain BL21 plus (DE3) cells. Total proteins were induced by 1 mM isopropyl-D-thiogalactoside at 37 °C for 4 h. Purification of OsBIRH1 fusion protein was performed using a His-Bind Kit (Nova-Gen, Madison, WI, USA) following the manufacturer's instructions. Protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA) following the recommended method.
Preparation of short double-stranded RNA substrates
RNA substrates used for helicase assays were prepared as described (Iost et al., 1999). Two double-stranded RNA molecules with different sizes of duplex were synthesized in vitro. Briefly, for L-dsRNA, two transcripts of 58 and 59 nucleotides (nt) were synthesized in vitro from pGEM-4Z (Promega, Madison, WI, USA) and the shorter one was labelled with [α-32P]dCTP. The nucleotide sequences of the two strands are: 58 nt strand, 5′-UGC AUG CCU GCA GGU CGA CUC UAG AGG AUC CCC GGG UAC CGA GCU CGA AUU CGU AUU C-3′; 59 nt strand, 5′-CCC UCU GUU CGA ACG UAC GGA CGU CCA GCU GAG AUC UCC UAG GGG CCC AUG GCU CGA GC-3′ (duplex regions underlined). For S-dsRNA, two transcripts of 45 nt and 34 nt were synthesized in vitro from pGEM-4Z and the shorter one was labelled. The sequence of the 45 nt strand is 5′-CCC UCU GUU CGA ACG UAC GGA CGU CCA GCU GAG AUC CUA GGG G-3′ and that of the 34 nt strand 5′-AGG AUC CCC GGG UAC CGA GCU CGA AUU CGU AUU C-3′ (with the duplex region underlined). The synthesized transcripts were annealed and the resulting partial duplex, which consists of a 47 bp and 9 bp region, respectively, was purified by native polyacrylamide gel electrophoresis.
Biochemical assays for OsBIRH1 activity
ATPase activity was monitored continuously by a coupled spectrophotometric method as described previously (Panuska and Goldthwait, 1980). Reactions were performed at 37 °C in a volume of 0.4 ml with buffer containing 20 mM TRIS–HCl, pH 8.0, 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, 300 mM NADH, 2 mM phosphoenolpyruvate, and 3 U ml−1 pyruvate kinase and lactate dehydrogenase. RNA and OsBIRH1 were added as indicated. The steady-state rate of ATP hydrolysis equals that of NADH oxidation, which was quantified using 6300 M−1 cm−1 for the extinction coefficient of NADH. RNA helicase activity was measured according to a previously reported method (Iost et al., 1999). Reaction mixtures of 15 μl in total contained 20 mM TRIS–HCl, pH 8.0, 70 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 15 U of RNasin, 45 fmol (3 nM) of partial duplex, and 2 mM ATP, where indicated. Reactions were initiated by addition of OsBIRH1. After incubation for 10 min at 37 °C, the reactions were stopped by addition of 4 μl of a solution containing 1.2% SDS, 10 mM EDTA, 40% glycerol, bromphenol blue, xylene cyanol, and 250 mg ml−1 proteinase K. For positive controls, duplex RNA was separated into monomers by heating at 95 °C for 2 min, followed by rapid cooling on ice. Samples were electrophoresed on a 10% polyacrylamide gel. Labelled RNAs were visualized by autoradiography.
Generation of OsBIRH1-overexpressing transgenic Arabidopsis
The OsBIRH1 coding sequence was released from pUCm-OsBIRH1-1 by digestion with BamHI/HindIII and was then cloned into BamHI/HindIII sites of the binary vector pCAMBIA99-1 under control of the cauliflower mosaic virus (CaMV) 35S promoter in the sense orientation. The resulting plasmid, designated as pCAMBIA991-OsBIRH1, was introduced into Agrobacterium tumefaciens GV3101 by electroporation using a GENE PULSER II electroporation system (Bio-Rad Laboratories). Transformation of Arabidopsis was performed using the floral dip method as described previously (Clough and Bent, 1998). Transgenic plants were selected on MS media containing 50 mg l−1 hygromycin and allowed to grow to third generations. The wild-type plants and homozygous transgenic T3 lines were used in all experiments.
Genomic DNA of transgenic Arabidopsis plants was extracted by the CTAB protocol (Luo et al., 2005b). Approximately 100 ng of genomic DNA was used for detection of the transgene in transgenic plants by PCR with primers BIHN-n5-11F (5′-CGC GGA TCC ATG GAG GTG GCG GGT GCT C-3′) and BIHN-n5-11R (5′-CCC AAG CTT CTA GAC GCA TGG CAC ATC G-3′). Southern blotting analysis was performed in ULTRAhyb hybridization buffer and probed with a [α-32P]dCTP-labelled 856 bp fragment of OsBIRH1 cDNA. The membrane was blotted between waxfilm (Whatman International, Maidstone, UK) and autoradiographed by exposure to storage phosphor screens (Amersham BioScience) for 4 h.
Analysis of gene expression
Total RNA was extracted using an acid phenol–guanidine isothiocyanate–chloroform one-step method as described (Sambrook and Russell, 2001). Twenty micrograms of total RNA was used in northern blotting analysis as described above for Southern blot analysis.
Expression of defence-related genes in transgenic plants was analysed by reverse-transcriptase (RT)-PCR. Leaf samples were collected from 4-week-old plants and total RNA was extracted. Timers for the defence-related genes used in RT-PCR were as follows: AtPR1-F, 5′-TCGTCTTTGTAGCTCTTGTAGGTG-3′; AtPR1-R, 5′-TAGATTCTCGTAATCTCAGCTCT-3′; AtPR2-F, 5′-CGTTGTGGCTCTTTACAAACAACAAAAC-3′; AtPR2-R, 5′-GAAATTAACTTCATACTTAGACTGTCGAT-3′; AtPR5-F, 5′-ATGGCAAATATCTCCAGTATTCACA-3′; AtPR5-R, 5′-ATGTCGGGGCAAGCCGCGTTGAGG-3′; AtPDF1.2-F, 5′-GCTAAGTTTGCTTCCATCATCACCCTT-3′; AtPDF1.2-R, 5′-AACATGGGACGTAACAGATACACTTGTG-3′; AtApx1-1F, 5′-CTGTTGAGAAGTGCAGGAGGAAGC-3′; AtApx1-1R, 5′-CATGTGGGCCTCAGCGTAATCAGC-3′; AtApx2-1F, 5′-TGCTGTTGAGATCACTGGAGGAC-3′; AtApx2-1R, 5′-GATGAGCTTCCGTATAGTCTTCG-3′; AtFSD1-1F, 5′-AGTTCAATGCTGCTGCAGCCACTC-3′; AtFSD1-1R, 5′-GCAGAACTCACTGTCACTGAAGTC-3′; BIHN-n5-rt-1F, 5′-GATAATCGAGGAATCTCCTGTC-3′; BIHN-n5-rt-1R, 5′-TAGATGGTGCTATGTATGCTAG-3′; ATACTIN1-1F, 5′-GGCGATGAAGCTCAATCCAAACG-3′; ATACTIN1-1R, 5′-GGTCACGACCAGCAAGATCAAGACG-3′. AtACTIN1 was used as an internal control. One microgram of RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and gene-specific primers. Each PCR was carried out with specific primers for 30 cycles of 94 °C 15 s, 60 °C 15 s, and 72 °C 1 min, using Taq DNA polymerase (PrimerStar, Takara, Dalian, China).
Disease assays
Alternaria brassicicola was grown on potato dextrose agar, and spores were collected and suspended in distilled water for inoculation. Disease assay for A. brassicicola was performed on detached leaves (Mengiste et al., 2003). A single 5 μl spore suspension (1×106 spores ml−1) in water was dropped onto each detached leaf. Inoculated leaves were kept under a transparent cover to maintain high humidity and transferred to a growth chamber with 24 °C and a 12 h light/12 h dark cycle.
Leaves of 4-week-old plants were infiltrated with suspensions of Pseudomonas syringae pv. tomato DC3000 in 10 mM MgCl2 (OD600=0.001). To determine the bacterial growth curve, 10 leaves each were collected at 0, 2, and 4 d after inoculation. Leaf discs of the same size were made using a hole puncher, and bacterial titre per leaf area was determined as described previously (Mengiste et al., 2003).
Oxidative stress tolerance assays
Seeds were surface-sterilized and placed on 1/2 MS medium containing 1 μM or 100 μM methyl viologen (MV; Sigma-Aldrich) and were kept at 4 °C for 48 h to synchronize germination. Seed germination (emergence of radicals) was scored daily. Detached leaf assay was used to determine the sensitivity of plants to oxidative stress induced by MV. Leaves from soil-grown plants were placed on 1/2 MS medium without sucrose, and a single drop of a 2 μl solution with different concentrations of MV (0, 25, and 50 μM) was placed on each leaf. Chlorophyll content was measured spectrophotometrically after treatments (Veronese et al., 2003). Chlorophyll content was calculated according to the formula Chl (a+b) = 5.24A664+22.24A648, where Chl is the chlorophyll concentration in micrograms per millilitre and A is the absorption (Lichtenthaler, 1987).
Results
Cloning of OsBIRH1, a rice gene encoding a DEAD-box RNA helicase
In previous studies aimed at elucidating the molecular biology of disease resistance in rice, differentially expressed cDNA clones associated with BTH-induced defence response were isolated and identified (Song and Goodman, 2002). Among these differentially expressed cDNAs, BLAST similarity searches revealed that the 664 bp insert in clone BIHN-n5 (GenBank accession no. BI118654) has a high level of similarity to genes encoding DEAD-box RNA helicases. Full-length cDNA of this putative DEAD-box RNA helicase gene was obtained by amplification of the 5′ and 3′ ends flanking the sequence in the clone BIHN-n5, and this gene was designated as OsBIRH1 for Oryza sativa BTH-induced RNA helicase 1. The full-length cDNA of OsBIRH1 was 2167 bp with a predicted 1926 bp ORF and has three nucleotide differences compared with the full-length cDNA AK065776 from O. sativa ssp. japonica but with no difference in predicted proteins. The OsBIRH1 gene, the same as the predicted locus Os03g01830 in rice genome sequence annotation, is localized on chromosome 3 of the rice genome and consists of eight exons and seven introns.
OsBIRH1 is a member of DEAD-box RNA helicases
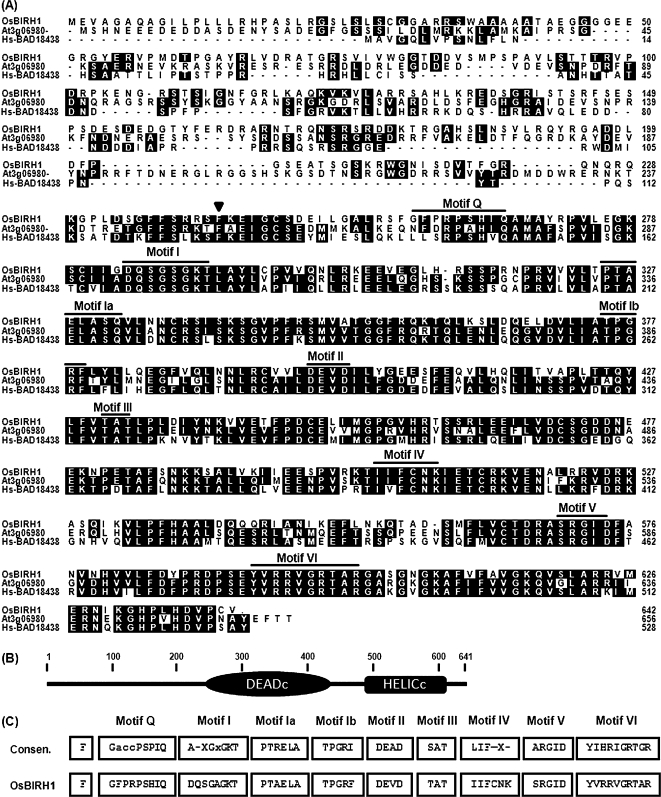
OsBIRH1 is predicted to encode a putative protein consisting of 641 amino acid residues with a calculated molecular weight of 71 kDa and isoelectric point of 9.1. Searching for conserved domains revealed that amino acids 242–455 and 473–613 in the OsBIRH1 protein represent two typical conserved domains, DEADc (PF00270) and HELICs (helicase superfamily C-terminal domain, PF00271), which are characteristic of the DEAD-box protein family (Fig. 1A, B). OsBIRH1 also possesses all nine highly conserved amino acid motifs characterizing the DEAD-box proteins (Fig. 1A). However, the sequences of these conserved motifs in OsBIRH1 are not identical to those of the consensus sequences (Fig. 1C). Notably, the sequence of the motif II in OsBIRH1 is DEVD, instead of the well-characterized motif sequences, for example, DEAD and DEAH, and thus OsBIRH1 should belong to the DExD group (Cordin et al., 2006). OsBIRH1 has a relatively long N-terminal extension of 241 amino acids, counting upstream from the conserved phenylalanine, but it has a short C-terminal extension of 39 amino acids, counting downstream from the conserved motif VI.
Fig. 1.
Structure of OsBIRH1 protein. (A) Alignment of OsBIRH1 with Arabidopsis AtRH50 (At3g06980) and human BAD18438. Conserved motifs are indicated by short thick lines and the conserved phenylalanine residue is indicated by an inverted filled triangle. (B) Domain organization of the OsBIRH1 protein. The amino acid positions are indicated. (C) Comparison of the conserved motifs in OsBIRH1 with consensus from other DEAD-box RNA helicase proteins.
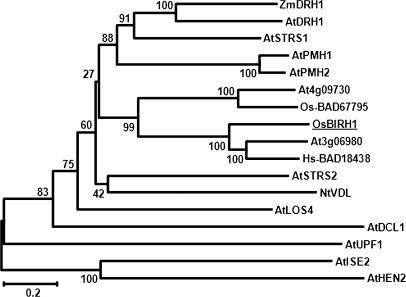
Alignment and phylogenetic tree analyses revealed that OsBIRH1 has a high level of identity to Arabidopsis AtRH50 (At3g06980) and a human protein BAD18438, showing identities of 53.6% and 46.7%, respectively (Fig. 1A). OsBIRH1, AtRH50, and human BAD18438 form a group of proteins, which all contain the conserved DEVD motif (Fig. 2). OsBIRH1 shows less identity to other known DEAD-box proteins identified so far. Sequence identity/similarity of OsBIRH1 to other DEAD-box proteins resides in the conserved motifs and the C-terminal extension, whereas there is no significant sequence similarity in the N-terminal extension.
Fig. 2.
Phylogenetic tree analysis of OsBIRH1 with DEAD-box RNA helicase protein from other organisms. Phylogenetic trees were constructed using the Neighbor–Joining method and genetic distances were calculated using the Kimura two-parameter model. Bootstrap values from 1000 replicates were used to assess the robustness of the trees. The DEAD-box RNA helicase proteins used were: Arabidopsis thaliana At3g06980 (AAO00880, AY113064), At4g09730 (BAD43116, AK175435), AtLOS4 (At3g53110, BT002444), AtSTRS1 (At1g31970, AY080680), AtSTRS2 (At5g08620, AY035114), AtISE2 (At1g70070, AF387007), AtUPF1 (At5g47010, AF484122), AtDCL1 (At1g01040, AF292940), AtPMH1 (At3g22310, AY091091), AtPMH2 (At3g22330, AY062502), AtHEN2 (At2g06990, AY080791), and AtDRH1 (At3g01540, AY062591); Oryza sativa Os01g0184500 (BAD67795); Homo sapiens Hs-BAD18438 (BAD18438); Zea mays ZmDRH1 (AAR29370, AY466159); and Nicotinana tabacum NtVDL (AAG34873, AF261032). The OsBIRH1 protein is underlined.
OsBIRH1 has RNA-dependent ATPase and ATP-dependent RNA helicase activity
Generally, as reported previously, the DEAD-box RNA helicases have both RNA-dependent RNA helicase activity and ATP-dependent RNA helicase activity (Cordin et al., 2006). To determine whether OsBIRH1 encodes a functional DEAD-box RNA helicase, ATPase and RNA helicase activity of recombinant OsBIRH1 protein were analysed. Combinant OsBIRH1 was purified from E. coli and the ATPase activity of OsBIRH1 was first measured using a continuous spectrophotometric method, in which ATP hydrolysis is coupled to NADH oxidation allowing monitoring of any change by measurement of the absorbance at 338 nm (A338). In the absence of RNA, no change of A338 was detected, indicating that OsBIRH1 has no ATPase activity by itself. By contrast, in the presence of total rice RNA, the A338 value decreased linearly over time during the experimental period (Fig. 3A). Addition of RNase A to the reaction abolished the activity, confirming that it was dependent on the added RNA (data not shown). These results confirm that OsBIRH1 has an RNA-dependent ATPase activity.
Fig. 3.
RNA-dependent ATPase and ATP-dependent RNA helicase activities of OsBIRH1. (A) ATPase activity was measured by a spectrophotometric assay in the presence or absence of total rice RNA. Oxidation of NADH, which is directly proportional to the rate of ATP hydrolysis, was continuously monitored by measuring the absorbance at 338 nm. Diamonds, OsBIRH1 (150 nM) without RNA; squares, OsBIRH1 (150 nM) with RNA (50 mg ml−1). (B) Scheme of the partial RNA duplexes used for unwinding activity assays. (C) RNA unwinding activity of OsBIRH1. Activity was measured by the ability of the protein to dissociate the partial RNA duplex. Fifty femtomol of labelled substrate was incubated with increasing concentrations of OsBIRH1 in the presence or absence of 2 mM ATP. The products of the reaction were separated by a 10% native PAGE and visualized by autoradiography. ss probe, Single strand probe as positive control; Boiled ds probe, the duplex was boiled before loading on the gel.
To examine RNA helicase activity of OsBIRH1, two different dsRNA substrates with partial duplexes of 47 bp and 9 bp, respectively, were synthesized. L-dsRNA was prepared by annealing a 59 nt ssRNA to a radiolabelled 58 nt ssRNA to form a 47 bp duplex; while S-dsRNA was made by annealing a 34 nt ssRNA to a longer radiolabelled 45 nt RNA to form a 9 bp duplex (Fig. 3B). As shown in Fig. 3C, the L-dsRNA and S-dsRNA duplexes migrated much slower than the corresponding ssRNAs (Fig. 3C, lanes 1 and 3). When the L-dsRNA was included as substrate, no duplex dissociation was detected in the presence or absence of ATP and even at higher concentrations of OsBIRH1 protein in the reactions (Fig. 3C). Similarly, when the S-dsRNA was included as substrate, no duplex dissociation was detected in reactions without ATP (Fig. 3C). In the presence of ATP, duplex dissociation was detected when a higher concentration of OsBIRH1 protein was included in the reactions (Fig. 3C, lower panel, lane 8). However, no duplex dissociation was observed when a low concentration of OsBIRH1 protein was used (Fig. 3C, lower panel, lanes 6 and 7). These results clearly indicate that OsBIRH1 has an ATP-dependent RNA helicase activity and that OsBIRH1 requires a high concentration for its RNA helicase activity.
Expression of OsBIRH1 is induced in rice defence responses
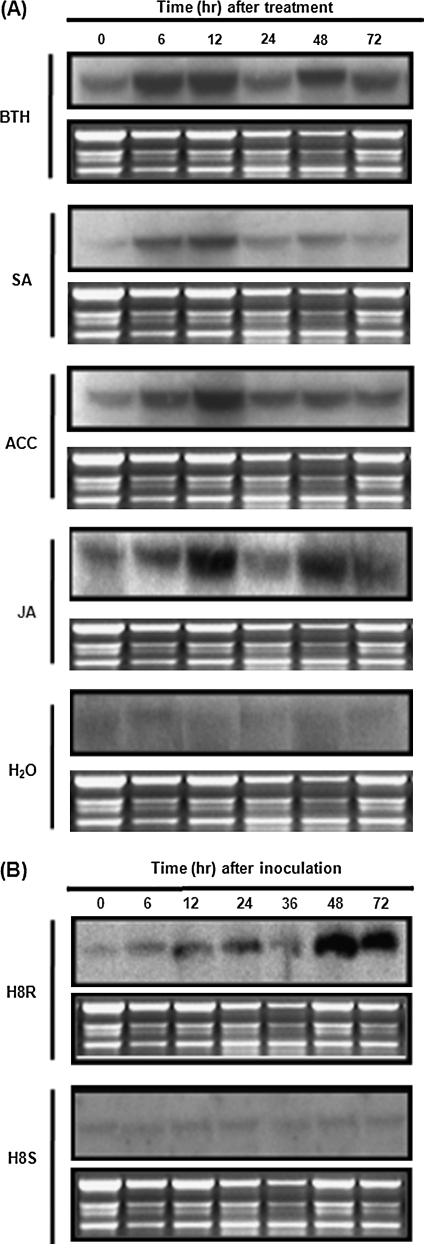
Originally, the differentially expressed cDNA clone BIHN-n5 was identified from a suppression subtractive hybridization library constructed by subtracting cDNAs from BTH-induced and M. grisea-inoculated rice leaf tissue with cDNAs from uninoculated control leaf tissue, implying that this gene was activated in rice defence responses (Song and Goodman, 2002). Therefore, an analysis was carried out to see if expression of OsBIRH1 in rice seedlings was induced by well-known disease resistance-related signal molecules such as SA, BTH, JA, and ACC. Transcripts corresponding to OsBIRH1 could be detected in water-treated healthy rice seedlings, but BTH treatment did induce expression of OsBIRH1 (Fig. 4A). Induced expression of OsBIRH1 was observed as early as 6 h after treatment, peaked at 12 h and maintained a relatively high level for 12–72 h. SA treatment also led to a rapidly induced expression of OsBIRH1 within 6 h which was maintained at a relatively higher level for 12–72 h after treatment (Fig. 4A). It is likely that expression of OsBIRH1 was also induced by JA and ACC, with kinetics similar to those by BTH and SA (Fig. 4A).
Fig. 4.
Expression patterns of OsBIRH1 in rice seedlings after treatment with different resistance signal chemicals and in interactions between rice and Magnaporthe grisea. (A) Expression of OsBIRH1 by disease resistance-related signal molecules. Rice seedlings were treated by spraying with solutions of 0.3 mM BTH, 1 mM SA, 100 μM JA, 100 μM ACC, or water. (B) Induced expression of OsBIRH1 in rice and the blast fungus interactions. Three-week-old rice seedlings of H8R and H8S were inoculated with M. grisea. Rice leaf samples were collected at each time point (hours) as indicated after treatment or inoculation. Twenty micrograms of total RNA were fractionated on a 1.2% agarose formaldehyde gel and hybridized with the α-32P-labelled 856 bp fragment of OsBIRH1 as a probe. The corresponding ethidium bromide gel images show equal loading of total samples.
Expression of OsBIRH1 in the disease resistance response was further examined using a pair of near-isogenic lines, H8R and H8S, which show incompatible and compatible interaction, respectively, with the tested strain of M. grisea. Similar to the kinetics after BTH treatment, expression of OsBIRH1 in H8R leaves was induced as early as 6 h after inoculation with M. grisea and the expression levels increased progressively within 72 h after inoculation (Fig. 4B). By contrast, no significant induced expression was detected in H8S leaves after inoculation with M. grisea (Fig. 4B). These results suggest that expression of OsBIRH1 is induced by multiple disease resistance-related signal molecules and pathogen infection, and indicate that induced expression of OsBIRH1 only occurs in an incompatible interaction between rice and the blast fungus.
Generation of OsBIRH1-overexpressing transgenic Arabidopsis plants
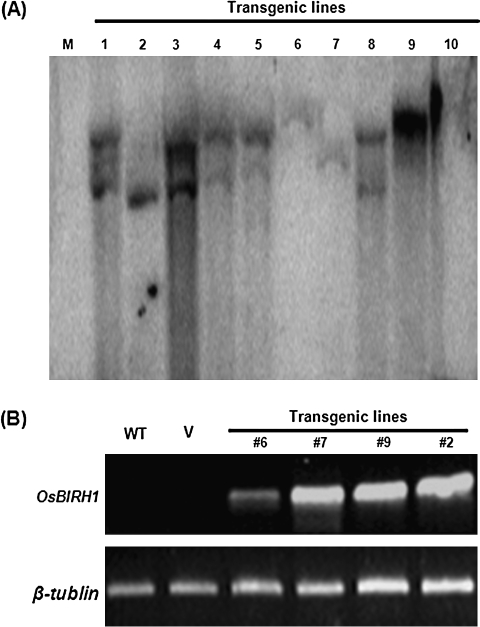
To gain further information on the biological function of OsBIRH1, transgenic Arabidopsis lines that ectopically express OsBIRH1 under the control of the CaMV 35S promoter were generated. A total of 15 independent transgenic lines were obtained through the floral dip transformation method and antibiotic screening. Southern blot analysis of genomic DNA from the transgenic plants confirmed that OsBIRH1 was integrated into the genome of transgenic plants and that more than half of the transgenic lines contain two or more copies of the transgene (Fig. 5A). The transgenic lines with a single copy of the OsBIRH1 gene were selected to grow for three generations to obtain homozygous lines for further studies. RT-PCR analysis was performed to examine expression levels of OsBIRH1 in T3 plants of the transgenic lines (#2, #6, #7, and #9), and a high level of OsBIRH1 expression was detected in these lines (Fig. 5B). During the experiments with these transgenic lines, no obvious differences in morphology, growth, and development under normal growth condition were observed (data not shown).
Fig. 5.
Southern blot and RT-PCR analysis of the OsBIRH1 transgenic plants. (A) Southern blot analysis of the transgenic lines. M, Non-transgenic plant as a negative control; 1–10, individual OsBIRH1 transgenic plants. (B) RT-PCR analysis of OsBIRH1 expression in homozygous transgenic lines. WT, Non-transgenic plant as a negative control (CK-); V, pCAMBIA 99-1 transgenic plants; #6, #7, #9, and #2, individual OsBIRH1 transgenic plants carrying a single copy of the OsBIRH1 gene.
Overexpression of OsBIRH1 in Arabidopsis confers enhanced disease resistance
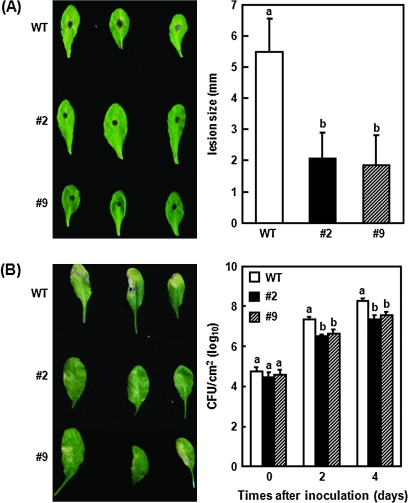
First, the disease resistance phenotype of OsBIRH1-overexpressing plants was examined against A. brassicicola, a necrotrophic fungal pathogen causing early blight disease in cruciferous plants, using a detached leaf assay (Mengiste et al., 2003). In wild-type plant leaves, typical disease symptoms were seen around 3 d after inoculation with spores of the fungus, and the lesion enlarged to form a yellow chlorotic area at 5 d after inoculation (Fig. 6A). By contrast, disease symptoms developed much more slowly and lesions were mainly restricted to a small area at the inoculation sites with less chlorosis at 5 d after inoculation (Fig. 6A). Further measurement of the disease lesions also revealed that lesion sizes in both of the transgenic lines tested were significantly reduced compared with the lesion size in the wild-type plants (Fig. 6A). These results indicate that overexpression of OsBIRH1 in transgenic Arabidopsis enhances disease resistance against A. brassicicola.
Fig. 6.
Enhanced disease resistance of OsBIRH1-overexpressing Arabidopsis plants against Pseudomonas syringae pv. tomato and Alternaria brassicicola. (A) Representative leaves showing disease symptoms (left panel) and lesion size (right panel) after inoculation with Alternaria brassicicola. Leaves from wild-type (Wt) and OsBIRH1 transgenic plants were inoculated with a 4 μl droplet of spores (106 spores ml−1) and photographs were taken at 5 d after inoculation. Lesion size was measured 3 d after inoculation. Data presented are means ±SD from a minimum of 40 lesions. (B) Representative leaves showing disease symptoms (left panel) and bacterial growth (right panel) after infiltration with Pseudomonas syringae pv. tomato DC3000. Bacterial growth [colony-forming units (CFU) cm−2 leaf area] in leaves at 0, 2, and 4 d after inoculation was measured. Photographs were taken 3 d after inoculation. Data presented are means ±SD from three independent experiments. Different letters above the columns indicate significant differences at P=0.05. WT, Wild type; #2 and #9, OsBIRH1-overexpressing plants.
The disease resistance phenotype of OsBIRH1-overexpressing plants against P. syrinage pv. tomato DC3000 was also examined. Leaves of wild-type and transgenic plants were infiltration-inoculated with Pseudomonas syringae pv. tomato suspension, and disease development was observed over 1 week after inoculation. Chlorosis symptoms were visible 3 d after inoculation under the experimental conditions and developed progressively on the infected leaves of the wild-type plants (Fig. 6B). Tissue collapse was also observed in some of the inoculated leaves of wild-type plants. By contrast, development of chlorosis symptoms in inoculated leaves of the transgenic plants was slower, typically seen 4–5 d after inoculation, and lesion size was also significantly reduced (Fig. 6B). Measurement of bacterial titres in inoculated leaves also indicated that the bacterial population in transgenic plant leaves was reduced by at least 10-fold compared with that in the wild-plant leaves (Fig. 6B). These results suggest that ectopic expression of OsBIRH1 in transgenic Arabidopsis confers disease resistance against P. syringae pv. tomato.
OsBIRH1-overexpressing plants have up-regulated PR gene expression
To gain further information on the increased disease resistance phenotype observed in OsBIRH1-overexpressing Arabidopsis plants, the expression levels of some defence-related genes were analysed by RT-PCR. In the present experiments, induced expression of the selected defence-related genes in wild-type plants was not detected, suggesting that the growth regime did not cause significant stress. Results from RT-PCR experiments showed that the expression levels of PR-1, PR-2, PR-5, and PDF1.2 were constitutively up-regulated in the OsBIRH1-overexpressing plants in the absence of the pathogen as compared with those in the wild-type plants (Fig. 7). PR-2 and AtPR-5 were up-regulated in all transgenic lines, while PR-1 and PDF1.2 were up-regulated in most of the transgenic lines tested.
Fig. 7.
Expression of defence-related genes in the OsBIRH1-overexpressing plants. Two-week-old seedlings grown on MS medium were collected for analysis of gene expression by RT-PCR. WT, Non-transgenic plant as a negative control (CK-); V, pCAMBIA 99-1 transgenic plants; #2, #6, #7 and #9, individual transgenic plants carrying single copy of OsBIRH1 gene.
Increased tolerance of transgenic plants to oxidative stress
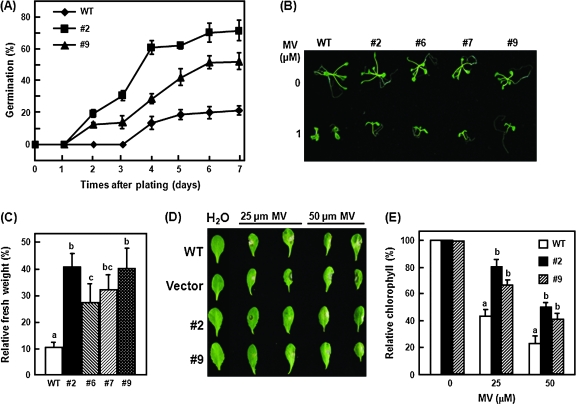
The possible function of OsBIRH1 in the abiotic stress response was also examined by analysing tolerance of OsBIRH1-overexpressing plants to oxidative stress. Tolerance levels of the transgenic plants were tested at different stages, for example seed germination, seedling growth, and mature leaves of 4-week-old plants, using MV as the oxidative stress-generating chemical. In MS medium containing 100 μM MV, seeds from the transgenic lines germinated much better than the wild-type seeds (Fig. 8A). At 7 d after treatment, ∼70% and ∼50% of seeds from transgenic lines #2 and # 9, respectively, germinated, but the germination rate of the wild-type seeds was only about 20%. No difference in seed germination was observed between the transgenic line and wild-type plants on MS medium without MV (data not shown). Seedlings of the wild-type and the transgenic lines grown on MV-containing MS medium showed growth retardation, but oxidative stress-caused growth retardation in transgenic lines was less severe than in the wild-type as indicated by the seedling fresh weight (Fig. 8B, C). The fresh weight of the transgenic seedlings grown on MS medium containing 1 μM MV was 2.5- to 4.0-fold higher than that of the wild-type seedlings (Fig. 8C). Tolerance of OsBIRH1-overexpressing Arabidopsis plants was further examined in mature leaves of plants. Detached leaves from wild-type plants showed significant necrosis and bleaching after treatment with 20 μM or 50 μM MV, while less necrosis and bleaching was observed in detached leaves from the transgenic plants (Fig. 8D). Total chlorophyll content in leaves of the wild-type plants decreased to 45% and 20% of the normal levels in untreated leaves after treatment with 20 μM or 50 μM MV, respectively, whereas the chlorophyll content in leaves of the transgenic plants was maintained at >60% and 40% after MV treatment (Fig. 8E). These results suggest that overexpression of OsBIRH1 in transgenic Arabidopsis plants confers enhanced tolerance to oxidative stress in all developmental stages.
Fig. 8.
Enhanced tolerance to oxidative stress of OsBIRH1-overexpressing plants. (A) Percentage of seed germination of the wild-type (WT) and transgenic Arabidopsis lines on 1/2 MS agar medium containing 100 μM MV. Germination was scored when the radical tip had fully emerged from the seed coat. (B) Enhanced tolerance to MV of the transgenic plants at the seedling stage. Representative seedlings 14 d after MV treatment are shown. (C) Relative fresh weight of the WT or transgenic lines grown in the absence or presence of MV. Seedlings grown without MV are given a value of 100%. (D) Representative leaves from WT and transgenic plants 5 d after MV treatment are shown. (E) Chlorophyll concentration of the leaf tissues of the WT or transgenic lines was measured in the presence or absence of different MV concentrations. Data presented are means ±SD from three independent experiments. Different letters above the columns indicate significant differences at P=0.05.
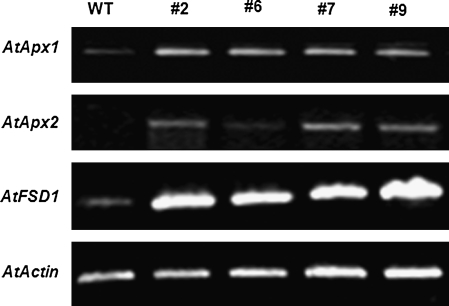
To confirm further the oxidative stress tolerance phenotype in OsBIRH1-overexpressing transgenic plants, expression of some known oxidative stress defence genes was analysed (Mittler et al., 2004). As shown in Fig. 9, the transgenic plants showed up-regulated expression of three different oxidative stress defence genes, AtApx1 (ascorbate peroxidase 1), AtApx2 (ascorbate peroxidase 1), and AtFSD1 (Fe-superoxide dismutase 1), under normal growth conditions. Therefore, the enhanced tolerance observed in transgenic plants might be due to an increased capacity of anti-oxidative stress to be modulated by OsBIRH1.
Fig. 9.
Expression of oxidative stress defence genes in the OsBIRH1-overexpressing plants. Two-week-old seedlings grown on MS medium were collected for analysis of gene expression by RT-PCR. WT, Non-transgenic plant as a negative control (CK-); #2, #6, #7, and #9, individual transgenic plants carrying single copy of the OsBIRH1 gene.
Discussion
The involvement and significance of RNA helicases in response to biotic and abiotic stress have only recently begun to emerge (Owttrim, 2006; Vashisht and Tuteja, 2006). In the present study, the characterization of the rice OsBIRH1 gene, which encodes a DEAD-box RNA helicase with RNA-dependent ATPase and ATP-dependent RNA helicase activities, is reported. Evidence that OsBIRH1 plays important roles in disease resistance response and oxidative stress tolerance, as revealed by its inducible expression by pathogen infection and multiple defence signal molecules as well as by phenotypes of the OsBIRH1-overexpressing Arabidopsis plants, was also presented. The findings not only extend knowledge of the biological function of the DEAD-box RNA helicases, but also provide new clues to explore further the significance of RNA helicase-mediated RNA metabolism in regulation of plant defence responses.
The OsBIRH1 protein contains all conserved domains and motifs that are characteristic of the DEAD-box proteins. However, sequences of the conserved motifs in OsBIRH1 are different from those of the consensus sequences; in particular, motif II of OsBIRH1 is DEVD, instead of the well-characterized motif sequences, for example DEAD and DEAH, indicating that OsBIRH1 is most likely to belong to the DExD group (Cordin et al., 2006). In the phylogenetic tree analysis, OsBIRH1, Arabidopsis AtRH50 (At3g06980), and human BAD18438 form a small related subgroup. These three proteins not only contain the same conserved DEVD motif but also have almost identical sequences in the conserved motifs I–VI. However, neither AtRH50 or human BAD18438 has been characterized in terms of their function so far. The present results from the OsBIRH1 studies might provide preliminary information on the biological function of this group.
Although a number of DEAD-box proteins or genes have been identified from various different plants, most of the proteins were not analysed for their biochemical activity in vitro. The Arabidopsis LOS4 was shown to have RNA-dependent ATPase activity in vitro, and AtDRH1 was found to possess both ATP-dependent RNA helicase and RNA-dependent ATPase activities (Okanami et al., 1998; Gong et al., 2005). In the present study, the recombinant OsBIRH1 protein showed specific RNA-dependent ATPase and ATP-dependent RNA helicase activities in vitro, suggesting that OsBIRH1 is a functional DEAD-box RNA helicase. It is considered that unwinding of RNA duplexes by DEAD-box RNA helicases occurs in a processive manner (Jankowsky et al., 2000, 2005; Eoff and Raney, 2005; Cordin et al., 2006). However, the DEAD-box proteins are feeble helicases in nature, and are thus supposed to act at key steps of RNA metabolism with short duplexes (<10 bp) preferred as their RNA substrates (Cordin et al., 2006; Linder and Lasko, 2006). This better explains the observations that OsBIRH1 was unable to unwind long RNA duplex substrates and its unwinding activity on the short RNA duplex required a high concentration of protein in vitro.
Previous studies have demonstrated that a number of DEAD-box RNA helicases are involved in regulating several key steps in development of plant embryos, plastids, and floral meristems (Jacobsen et al., 1999; Wang et al., 2000; Kobayashi et al., 2007). A relatively high level of basal expression of the OsBIRH1 gene under normal growth conditions may imply a function for OsBIRH1 in growth and/or development processes, although ectopic expression in transgenic Arabidopsis does not cause any visible growth and development changes. Thus, knockouts or knockdown mutants of rice OsBIRH1 and Arabidopsis AtRH50 (At3g06980) should be used to check whether these RNA helicases play a role in growth and development.
Evidence is accumulating that expression of RNA helicase genes or RNA helicase activity is regulated in response to changes in environmental conditions (Vashisht and Tuteja, 2006). Several DEAD-box RNA helicase genes were indeed identified from genome-wide expression profiling analysis, suggesting that these genes might be stress regulated (Seki et al., 2001, 2002; Kreps et al., 2002; Kant et al., 2007). STRS1 and STRS2 were shown to be down-regulated by multiple abiotic stresses, for example salt, drought, and heat stress, but they both attenuated abiotic stress tolerance (Kant et al., 2007). Although OsBIRH1 has a relatively high level of basal expression, its expression was induced not only by pathogen infection, but also by several disease resistance-related signalling molecules, for example SA, JA, and ACC, suggesting a role for OsBIRH1 in disease resistance signalling pathways. This is further supported by the observation that overexpression of OsBIRH1 in transgenic Arabidopsis plants enhanced disease resistance against infection by A. brassicicola and P. syringae pv. tomato. Studies with knockout or knockdown mutants of OsBIRH1 will further provide direct genetic evidence to demonstrate its function in disease resistance signalling pathways.
Alternaria brassicicola and P. syringae represent two different parasitic modes, necrotrophic and biotrophic, respectively. Correspondingly, plant defence responses against necrotrophic and biotrophic pathogens and the signalling pathways involved are also distinct in nature (Glazebrook, 2005). Generally, expression of PR genes and the SA-dependent signalling pathway are involved in the regulation of defence responses against biotrophic pathogens, while the PDF1.2 gene and the JA/ET signalling pathway are involved in modulating defence responses against necrotrophic pathogens (Gupta et al., 2000; Kunkel and Brooks, 2002; Spoel et al., 2003). The results indicate that OsBIRH1 might be involved in both of these two different signalling pathways. This is supported by several lines of evidence obtained from this study; for example, induction of OsBIRH1 expression by SA and JA/ACC, representing the SA signalling pathway and the JA/ET signalling pathway, respectively, enhanced disease resistance against A. brassicicola and P. syringae pv. tomato in OsBIRH1-overexpressing transgenic Arabidopsis, and constitutively up-regulated expression of both PR genes and PDF1.2 in transgenic plants. Such constitutively up-regulated expression of defence-related genes in transgenic plants indicates that ectopically expressed OsBIRH1 activates endogenous signalling pathway(s) resulting in enhanced defence responses. However, the mechanisms linking the OsBIRH1-mediated signalling pathway and expression of defence-related genes remain unknown.
Some of the DEAD-box RNA helicases have been shown to integrate into multiple signalling pathways, leading to participation in regulation of different aspects of growth/development and stress responses. The cold stress-regulated LOS4 was found to play an important role in responses to cold and heat stress (Gong et al., 2002, 2005), while STRS1 and STRS2 were also shown to be negative regulator nodes in the multiple abiotic stress signalling subnetwork (Kant et al., 2007). In the present studies, it was found that the ectopic expression of OsBIRH1 in transgenic Arabidopsis plants led to enhanced tolerance of abiotic stresses, including oxidative stress and drought (D Li and F Song, unpublished data). It was also found that ectopic expression of OsBIRH1 results in the enhanced expression of AtApx1, AtApx2, and AtFSD1 in the transgenic plants under normal growth conditions. This is in agreement with previous observations that enhanced oxidative stress tolerance is associated with high levels of expression of genes that are involved in oxidative stress responses in plants (Mittler et al., 2004, 2006). Furthermore, the preliminary results also revealed that the OsBIRH1-overexpressing transgenic Arabidopsis plants showed an increased sensitivity to ABA (D Li and F Song, unpublished data). Taken together, like Arabidopsis STRS1 and STRS2, OsBIRH1 might also participate in regulation of different signalling pathways leading to defence responses against biotic and abiotic stresses.
In summary, the present study has demonstrated that OsBIRH1 is a functional DEVD-box RNA helicase belonging to the DEAD-box protein superfamily and that OsBIRH1 plays important roles in regulating different defence signalling pathways against both biotic and abiotic stresses. However, many questions regarding the OsBIRH1 function and its action mechanisms remain to be solved in the future; for example, how OsBIRH1 protein regulates expression of defence-related genes in plants, what role OsBIRH1 may play in plant growth and development, how OsBIRH1 operates different stress signalling pathways, etc. On the other hand, DEAD-box RNA helicases belong to a large protein family, but only a few of them have been identified recently for their biological functions in plant growth/development and stress responses (Park et al., 2002; Reinhart et al., 2002; Gendra et al., 2004; Gong et al., 2005; Arciga-Reyes et al., 2006; Yoine et al., 2006a, b; Kant et al., 2007; Kobayashi et al., 2007; Matthes et al., 2007). Definitely, additional extensive studies with knockout or knockdown mutants in model species, for example Arabidopsis and rice, will be very helpful in elucidating both biochemical and biological functions of each member of this huge protein family in plants.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grants no. 30571209), the National Key Basic Research and Development Program (2006CB101903), the Fund for the New Century Talent Program from MOE of China, and the Excellent Young Teachers Program of MOE. The authors thank Dr Zuhua He, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Science, for the rice near-isogenic lines H8R and H8S, and Mr Rongyao Chai, Zhejiang Academy of Agricultural Science, for the Magnaporthe grisea isolate 85-14B1.
References
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. The Plant Journal. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Research. 1999;27:628–636. doi: 10.1093/nar/27.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. CRC Critical Reviews in Plant Sciences. 2005;24:23–58. [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Reseach. 2001;11:2101–2114. doi: 10.1101/gr.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brander KA, Kuhlemeier C. A pollen-specific DEAD-box protein related to translation initiation factor eIF-4A from tobacco. Plant Molecular Biology. 1995;27:637–649. doi: 10.1007/BF00020219. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, McKay DB. Helicase structure and mechanism. Current Opinion in Structural Biology. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO Journal. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD box proteins and related families. Trends in Biochemical Sciences. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Eoff RL, Raney KD. Helicase-catalysed translocation and strand separation. Biochemical Society Transactions. 2005;33:1474–1478. doi: 10.1042/BST0331474. [DOI] [PubMed] [Google Scholar]
- Gendra E, Moreno A, Alba MM, Pages M. Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. The Plant Journal. 2004;38:875–886. doi: 10.1111/j.1365-313X.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. The Plant Cell. 2005;17:256–267. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proceedings of the National Academy of Sciences, USA. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Current Opinion in Structural Biology. 1993;3:419–429. [Google Scholar]
- Gupta V, Willits MG, Glazebrook J. Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Molecular Plant–Microbe Interactions. 2000;13:503–511. doi: 10.1094/MPMI.2000.13.5.503. [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. Journal of Biological Chemistry. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Itadani H, Sugita M, Sugiura M. Structure and expression of a cDNA encoding an RNA helicase-like protein in tobacco. Plant Molecular Biology. 1994;24:249–252. doi: 10.1007/BF00040593. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Fairman ME, Yang Q. RNA helicases: versatile ATP-driven nanomotors. Journal of Nanoscience and Nanotechnology. 2005;5:1983–1989. doi: 10.1166/jnn.2005.508. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Kant P, Kant S, Gordon M, Shaked R, Barak S. STRS1 and STRS2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiology. 2007;145:814–830. doi: 10.1104/pp.107.099895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Otegui MS, Krishnakumar S, Mindrinos M, Zambryski P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. The Plant Cell. 2007;19:1885–1897. doi: 10.1105/tpc.106.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. Arizona State University: Tempe; 2001. MEGA2: Molecular Evolutionary Genetics Analysis Software. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Chung MC, Chen CS. Cloning and characterization of a DEAD box RNA helicase from the viable seedlings of aged mung bean. Plant Molecular Biology. 2001;47:761–770. doi: 10.1023/a:1013687412020. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology. 1987;148:350–382. [Google Scholar]
- Linder P, Lasko P. Bent out of shape: RNA unwinding by the DEAD-box helicase Vasa. Cell. 2006;125:219–221. doi: 10.1016/j.cell.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Luo H, Song F, Goodman RM, Zheng Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biology. 2005a;7:459–68. doi: 10.1055/s-2005-865851. [DOI] [PubMed] [Google Scholar]
- Luo H, Song F, Zheng Z. Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. Journal of Experimental Botany. 2005b;56:2673–2682. doi: 10.1093/jxb/eri260. [DOI] [PubMed] [Google Scholar]
- Matthes A, Schmidt-Gattung S, Kohler D, Forner J, Wildum S, Raabe M, Urlaub H, Binder S. Two DEAD-box proteins may be part of RNA-dependent high molecular weight protein complexes in Arabidopsis thaliana mitochondria. Plant Physiology. 2007 doi: 10.1104/pp.107.108076. DOI: 10.1104/pp.107.108076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. The Plant Cell. 2003;15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Letters. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. The reactive oxygen gene network of plants. Trends in Plant Sciences. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Okanami M, Meshi T, Iwabuchi M. Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana. Nucleic Acids Research. 1998;26:2638–2643. doi: 10.1093/nar/26.11.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Research. 2006;34:3220–3230. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panuska JR, Goldthwait DA. A DNA-dependent ATPase from T4-infected Escherichia coli: purification and properties of a 63 000-dalton enzyme and its conversion to a 22 000-dalton form. Journal of Biological Chemistry. 1980;255:5208–5214. [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biology. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes and Development. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Linder P. DEAD-box proteins: the driving force behind RNA metabolism. Nature Reviews Molecular Cell Biology. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. The Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Song F, Goodman RM. OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta. 2002;215:997–1005. doi: 10.1007/s00425-002-0794-5. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. The Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI. Resistance proteins: molecular switches of plant defence. Current Opinion in Plant Biology. 2007 doi: 10.1016/j.pbi.2006.05.009. DOI: 10.1016/j.pbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Molecular Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Molecular Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Tuteja N. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. Journal of Photochemistry and Photobiology B. 2006;84:150–160. doi: 10.1016/j.jphotobiol.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Veronese P, Narasimhan ML, Stevenson RA, Zhu JK, Weller SC, Subbarao KV, Bressan RA. Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. The Plant Journal. 2003;35:574–587. doi: 10.1046/j.1365-313x.2003.01830.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Duby G, Purnelle B, Boutry M. Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. The Plant Cell. 2000;12:2129–2142. doi: 10.1105/tpc.12.11.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Cheng Y, Liu J, Chen X. HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis. Development. 2002;129:1569–1581. doi: 10.1242/dev.129.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. The Plant Cell. 2002;14(Suppl,):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant and Cell Physiology. 2006a;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. The Plant Journal. 2006b;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]