Abstract
Bioactive gibberellins (GAs) affect many biological processes including germination, stem growth, transition to flowering, and fruit development. The location, timing, and level of bioactive GA are finely tuned to ensure that optimal growth and development occur. The balance between GA biosynthesis and deactivation is controlled by external factors such as light and by internal factors that include auxin. The role of auxin transport inhibitors (ATIs) and auxins on GA homeostasis in intact light-grown Arabidopsis thaliana (L.) Heynh. seedlings was investigated. Two ATIs, 1-N-naphthylthalamic acid (NPA) and 1-naphthoxyacetic acid (NOA) caused elevated expression of the GA biosynthetic enzyme AtGA20-oxidase1 (AtGA20ox1) in shoot but not in root tissues, and only at certain developmental stages. It was investigated whether enhanced AtGA20ox1 gene expression was a consequence of altered flow through the GA biosynthetic pathway, or was due to impaired GA signalling that can lead to enhanced AtGA20ox1 expression and accumulation of a DELLA protein, Repressor of ga1-3 (RGA). Both ATIs promoted accumulation of GFP-fused RGA in shoots and roots, and this increase was counteracted by the application of GA4. These results suggest that in ATI-treated seedlings the impediment to DELLA protein degradation may be a deficiency of bioactive GA at sites of GA response. It is proposed that the four different levels of AtGA20ox1 regulation observed here are imposed in a strict hierarchy: spatial (organ-, tissue-, cell-specific) > developmental > metabolic > auxin regulation. Thus results show that, in intact auxin- and auxin transport inhibitor-treated light-grown Arabidopsis seedlings, three other levels of regulation supersede the effects of auxin on AtGA20ox1.
Keywords: Auxin, auxin transport inhibitors, DELLA proteins, gibberellin 20-oxidase, gibberellin biosynthesis, RGA
Introduction
Hormone homeostasis is critical for normal plant growth and development. Homeostatic mechanisms involve regulation of hormone biosynthesis, deactivation, and transport. Bioactive members of the gibberellin (GA) group of phytohormones control many processes throughout the life cycle of a plant including seed germination, stem elongation, transition to flowering, and fruit development. The concentration of bioactive GA4 in Arabidopsis thaliana (L.) Heynh. is, in large part, regulated by flux through the GA biosynthetic pathway. The final rate-limiting steps in GA biosynthesis are catalysed by two small families of 2-oxoglutarate-dependent dioxygenases, the GA 20-oxidases (AtGA20ox1–5), and the GA 3-oxidases (AtGA3ox1–4) (reviewed by Hedden and Phillips, 2000; Olszewski et al., 2002; Sponsel and Hedden, 2004). Once synthesized the bioactive GA4 will transduce the signalling pathway and/or be deactivated by GA 2-oxidases [AtGA2ox1–3 (Thomas et al., 1999), 4–6 (Hedden and Phillips, 2000), 7–8 (Schomburg et al., 2003)], and other deactivating enzymes (Zhu et al., 2006; Varbanova et al., 2007). The balance between GA biosynthesis and deactivation is controlled by metabolic regulation involving both GA pool size and signalling, by other intrinsic factors such as other phytohormones, and by external factors including light and temperature (Yamaguchi and Kamiya, 2000; Garcia-Martinez and Gil, 2002).
Two aspects of metabolic regulation of the GA pathway are well-documented, namely negative feedback regulation of GA biosynthetic enzymes and positive feed-forward regulation of GA deactivating enzymes. In Arabidopsis, bioactive GA down-regulates the transcription of genes encoding at least three of its biosynthetic enzymes, AtGA20ox1, which is the main stem-expressed GA 20-oxidase, AtGA20ox2 and AtGA3ox1 (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Thomas et al., 1999). By contrast, bioactive GA up-regulates the expression of AtGA2ox1 and AtGA2ox2 that encode enzymes catalysing its irreversible deactivation (Thomas et al., 1999). Negative feedback and positive feed-forward regulation maintains the level of bioactive GA within physiological limits. In situations where the concentration of bioactive GA is extremely low, such as in the GA-deficient ga1 mutant or in plants treated with GA biosynthesis inhibitors, neither negative feedback nor positive feed-forward regulation is apparent, resulting in elevated transcripts of GA20-oxidases and GA3-oxidases, and reduced transcript levels of GA2-oxidases (Thomas et al., 1999).
The mechanism whereby biosynthesis and deactivation of GAs are regulated by bioactive GA is not understood, but it involves not only the pool size of bioactive GA, but the amount of GA signalling that occurs. The DELLA family of putative transcriptional regulators, for example, GA insensitive (GAI), Repressor of ga1-3 (RGA), and RGA-like (RGL1-3) in Arabidopsis repress GA signalling (for reviews see Sun and Gubler, 2004; Fleet and Sun, 2005). Bioactive GA, bound to one of its cognate receptors, facilitates proteolysis of DELLA proteins and allows GA-responsive genes to be expressed (reviewed by Ueguchi-Tanaka et al., 2007). In Arabidopsis, if DELLA proteolysis is prevented (as in the absence of bioactive GA, or as a consequence of gain of function mutations such as gai-1 or rga-Δ17) expression of AtGA20ox1 (Xu et al., 1995, 1999) and AtGA3ox1 (Dill et al., 2001; Dill and Sun, 2001; King et al., 2001) is elevated. Conversely, DELLA loss of function mutants such as ga1-3/rga-2 and ga1-3/gai-t6/rga-24 have reduced levels of AtGA3ox1 transcripts (Dill and Sun, 2001; Silverstone et al., 2001). Taken together these results suggest that DELLA proteins are positive regulators of GA 20-oxidation and GA 3-oxidation, and are negative regulators of GA 2-oxidation (Dill and Sun, 2001; King et al., 2001).
Negative feedback regulation of GA 20- and 3-oxidation has been well documented in other Angiosperms including Pisum sativum L. (Martin et al., 1996; Ross et al., 1999), Solanum tuberosum L. (Carrera et al., 1999), and Oryza sativa L. (Dai et al., 2007). Feed-forward regulation of GA deactivation has also been reported in pea (Elliott et al., 2001).
In addition to effects of GA on its own metabolic pathway by feedback or feed-forward loops, other hormones have documented effects on GA biosynthesis and deactivation. In Arabidopsis, AtGA20ox1 was shown to be up-regulated by indole-3-acetic acid (IAA) in microarray analysis of light-grown seedlings (Goda et al., 2004), whereas Frigerio et al. (2006), examining transcript levels of 13 GA oxidases by quantitative RT-PCR, showed that AtGA20ox1 and AtGA20ox2 transcript levels were increased after 24 h treatment with 1-naphthalene acetic acid (NAA), along with those of four GA 2-oxidases. In addition, auxins have been reported to be necessary for GA signalling in Arabidopsis roots since degradation of a DELLA protein is delayed in decapitated seedlings or those in which auxin transport or sensitivity is impaired (Fu and Harberd, 2003).
Other documented effects of auxins on expression of genes encoding GA-oxidases in pea (van Huizen et al., 1995, 1997; Ross, 1998; Ngo et al., 2002; O'Neill and Ross, 2002; Ozga et al., 2003), Nicotiana tabacum L. (Wolbang and Ross, 2001), and Hordeum vulgare L. (Wolbang et al., 2004) suggest that while auxin-regulation of GA biosynthesis and deactivation is widespread, the regulatory effects of auxin are often targeted to specific genes and integrated with other means of metabolic regulation in ways unique to each species.
It has previously been shown that if auxin distribution is altered in Arabidopsis seedlings, as in a mutant of the BIG gene (encoding a calossin-like protein) and in wild-type seedlings treated with naphthylphthalamic acid (NPA), there is up-regulation of AtGA20ox1 (Desgagné-Penix et al., 2005). In this paper, the effects of ATIs and applied auxins on AtGA20ox1 expression in intact wild-type Arabidopsis seedlings are examined further to determine whether altered auxin status regulates AtGA20ox1 expression by affecting GA biosynthesis and/or signalling. It is shown that ATIs promote the accumulation of the GFP-fused DELLA protein, RGA, and that this accumulation can be counteracted by simultaneous application of GA4. These observations suggest that ATIs do not impair GA signalling. The results do suggest that ATIs directly or indirectly, lead to reduced levels of bioactive GA in normal sites of GA response. It is concluded, therefore, that the effects of ATIs on AtGA20ox1 expression are a consequence of metabolic regulation, and that in Arabidopsis seedlings metabolic regulation supersedes auxin regulation. These results also suggest that spatial (e.g. organ- tissue-, or cell-specific) and developmental regulation of AtGA20ox1 override both metabolic regulation and auxin-mediated regulation of GA pathways in intact auxin- and ATI-treated Arabidopsis seedlings.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana L. Heynh. Col-0 seeds were sterilized by incubation in freshly prepared 30% bleach plus 0.01% (v/v) Tween 20 for 10 min and then washed three times with sterile water. The surface-sterilized seeds were sown on standard A. thaliana salts (ATS) growth medium (Lincoln et al., 1990) supplemented with 30 mM sucrose and 0.8% agar and cold-treated for 2 d at 4 °C to synchronize germination before being placed under continuous cool-white fluorescent light (60 μmol m−2 s−1) at 25 °C. After 2 d, germinated seeds were transferred to liquid ATS medium supplemented with 30 mM sucrose with or without treatment as indicated in the figures. Seedlings were treated with 5 μM GA4, 12.5 μM 1-naphthoxyacetic acid (NOA), 0.1 μM indole 3-acetic acid (IAA), 0.1 μM 1-naphthalene acetic acid (NAA), all purchased from Sigma, 5 μM paclobutrazol (PAC, Allied Signal), 12.5 μM 1-N-naphthylthalamic acid (NPA, Chem Service), or mock treatment with ethanol 0.1% (v/v, final concentration) for 8 d, unless otherwise stated. The concentrations of hormones and other growth regulators were chosen after testing a range of concentrations and selecting those at which the test compounds gave a pronounced phenotypic effect after 8 d and no toxicity.
Reporter lines
Arabidopsis Col-0 AtGA20ox1::GUS reporter line (from Dr P Hedden, Rothamsted Research, UK) was constructed as a translational fusion comprising the promoter and transcribed region of AtGA20ox1 in-frame with the GUS reporter gene (Desgagné-Penix et al., 2005). Arabidopsis Col-0 DR5::GUS reporter line (from Dr T Guilfoyle, University of Missouri, Columbia) expresses the synthetic auxin response element DR5 fused to GUS (Ulmasov et al., 1997). Arabidopsis Col-0 pRGA::GFP::RGA reporter line (from Dr T-p Sun, Duke University) was used to monitor the presence of the protein repressor of GA signalling, RGA (Silverstone et al., 2001).
Reduced growth
Arabidopsis Col-O seeds were germinated (2 d) on nutrient agar and transferred to liquid media of different compositions; control: ATS minerals and 30 mM sucrose (full strength); 25% control: quarter-strength ATS minerals and 7.5 mM sucrose; 12.5% control: eighth-strength ATS minerals and 3.75 mM sucrose, and only 30 mM sucrose. The seedlings were treated for 8 d with or without 5 μM paclobutrazol or 12.5 μM NPA. Plants were grown in continuous light and measured at 10 d.
RNA extraction, cDNA synthesis, semi-quantitative RT-PCR, and northern blotting
Ten-day-old whole seedlings, shoot or roots, were frozen and ground using liquid nitrogen. Total RNA was extracted using the guanidium–phenol–chloroform method (Chomczynski and Sacchi, 1987). Five μg of total RNA were subjected to reverse transcription with Oligo dT18 using the RETROscript Kit (Ambion) according to the manufacturer's instructions. Semi-quantitative RT-PCR was performed to examine gene expression as described in Desgagné-Penix et al. (2005). The results are expressed as the ratio of the AtGA20ox1 transcript level compared to those of 18S rRNA.
GUS reporter lines analysis
Arabidopsis seeds containing AtGA20ox1::GUS and the DR5::GUS reporter construct were germinated and transferred as above. Ten-day-old seedlings were harvested. The GUS reporter activity analysis was conducted in two ways; (i) qualitative histochemical staining observations and (ii) quantitative fluorimetric measurements. GUS staining was performed as previously described in Desgagné-Penix et al. (2005). Briefly, qualitative analysis of GUS reporter activity was conducted by incubating the harvested seedlings in buffer containing the GUS substrate X-GLU (5-bromo-4-chloro3-indolyl-β-D-glucuronic acid) for 24 h (DR5::GUS seedlings) or 48 h (AtGA20ox1::GUS seedlings) at 37 °C (Jefferson et al., 1987). The GUS histochemical staining was visualized under a light microscope. For quantitative analysis of GUS reporter activity proteins of transgenic GUS seedlings were extracted, and quantified by Bradford assay. 32 μg of protein for each sample was incubated at 37 °C in GUS extraction buffer containing MUG (4-methylumbelliferone-β-D-glucuronide). Aliquots of 50 μl were removed at 30 min intervals for 2 h and added to 150 μl stop buffer (0.2 M sodium carbonate) in a 96-well microplate. The reporter activity was analysed using the Flx800 microplate fluorescence reader (Bio-Tek Instruments). Resulting fluorescence was measured and compared to a standard curve of 4-methylumbelliferone.
Confocal microscopy
Arabidopsis seeds containing the pRGA::GFP::RGA reporter construct were germinated and treated as described above. Five-day-old seedlings were harvested and the GFP-RGA distribution was visualized under a confocal microscope (Zeiss LSM 510 meta). Confocal images of seedlings were obtained with a constant set of microscopic and image intensity parameters with ×40 objective, at wavelength excitation at 488 nm and reading of the emission between 505–530 nm.
Quantitative GFP reporter line analysis
Arabidopsis seeds containing pRGA::GFP::RGA were germinated and transferred as above. Ten-day-old seedlings were harvested, frozen, and ground using liquid nitrogen. Proteins of transgenic GFP-RGA seedlings were extracted, and quantified by Bradford assay. For each sample, 640 μg of protein was analysed. The reporter fluorescence was analysed using the Fl×800 microplate fluorescence reader (Bio-Tek Instruments) by excitation at 485 nm and reading of the emission at 528 nm. Resulting fluorescence measured was compared to a standard curve of rGFP (Clontech labs).
Results
The ATIs, NOA and NPA, cause an up-regulation of AtGA20ox1 expression in Arabidopsis seedlings
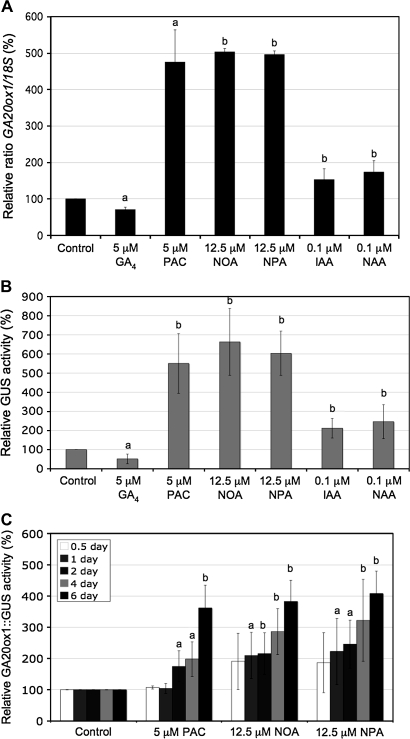
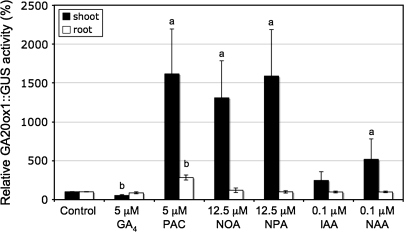
Previous work has shown that AtGA20ox1 expression was up-regulated in wild-type (Col-0) Arabidopsis seedlings growing on nutrient agar supplemented with the ATI, NPA, and in the tir3-1 allele of BIG, which has altered auxin transport (Desgagné-Penix et al., 2005). Subsequent work has used liquid culture that is a more tractable system with which to work, allowing additional treatments during the culture period. Also, in liquid culture, growth regulators have pronounced phenotypic effects at lower concentrations than those used by previous workers (Fu and Harberd, 2003; Frigerio et al., 2006). Changes in AtGA20ox1 transcript levels in 10-d-old Col-0 seedlings growing in nutrient solution containing NPA or other growth regulator treatments were measured. Semi-quantitative RT-PCR analysis extended our previous work (Fig. 1A). In 10-d-old light-grown Col-0 seedlings 5 μM GA4 caused a down-regulation, and paclobutrazol (PAC), an inhibitor of GA biosynthesis, caused up-regulation of AtGA20ox1 mRNA transcript levels. Treatment with NPA, which affects auxin efflux, and NOA which affects an influx component of the polar auxin transport, caused an increase of the AtGA20ox1 transcripts to a level comparable to the up-regulation seen by PAC treatment. Treatment with 0.1 μM IAA or its permeable analogue 1-NAA enhanced AtGA20ox1 expression, however, this increase was smaller than the one seen with the ATIs.
Fig. 1.
Molecular analysis of the AtGA20ox1 mRNA expression by RT-PCR analysis (A) and AtGA20ox1::GUS reporter by fluorimetric GUS assay (B) in response to GA4, PAC, ATIs or auxins treatment. Two-day-old Arabidopsis seedlings were transferred in liquid nutrient solution with or without GA4, PAC, ATIs or auxins for 8 d. (C) Time-course of AtGA20ox1::GUS activity in 10-d-old Arabidopsis seedlings in response to treatment with PAC, NOA or NPA for 0.5 d (white bars), 1 d (grey bars), 2 d (dark grey bars), 4 d (light grey bars), or 6 d (black bars). Values shown are means ±SD (n=3 different experiments) compared to respective control normalized to 100%. Different letters on the bars represent means that are statistically different relative to control using the Student t test where (a) P ≤0.05 and (b) P ≤0.01.
ATIs up-regulate the activity of the reporter construct AtGA20ox1::GUS
The β-glucoronidase (GUS) reporter gene fused to the AtGA20ox1 promoter has been used to monitor the expression of AtGA20ox1 (Hay et al., 2002; Chen et al., 2004). Quantification of the activity of the AtGA20ox1 promoter GUS gene fusion (Fig. 1B) corroborated our RT-PCR analysis (Fig. 1A). Gibberellin A4 treatment caused a significant decrease in AtGA20ox1::GUS reporter activity, and PAC, NOA, and NPA caused at least a 4-fold up-regulation of the AtGA20ox1::GUS activity. Treatment with IAA or NAA slightly increased AtGA20ox1::GUS activity but, similar to our RT-PCR results, this increase (1.5–2-fold), although significant, was much lower than the one mediated by the ATIs. Taken together, these data further suggest that altered auxin distribution plays a role in regulating the GA biosynthetic enzyme AtGA20ox1.
The timing of AtGA20ox1 up-regulation in the presence of PAC, NOA, and NPA was then examined (Fig. 1C). Seedlings were treated for varying durations from 0.5–8 d and all seedlings were extracted at 10 d. The increase in AtGA20ox1::GUS activity with ATI-treatment precedes that caused by PAC by at least one day. However, the characteristic phenotypic effects of each treatment are most apparent after 6–8 d of treatment. The expression of AtGA20ox1 continued to increase for the duration of the treatments, but values for 8 d were omitted from the figure as they were off scale.
Stunted growth does not induce AtGA20ox1 up-regulation
Paclobutrazol and ATIs affect the growth of Arabidopsis seedlings (Table 1) although different ATIs produce different phenotypes, presumably associated with how, and to what extent, they alter auxin transport. For example, NPA treatment reduced hypocotyl and primary root lengths and the number of lateral roots whereas NOA had no effect on hypocotyl and root elongation but reduced the number of lateral roots compared to control seedlings (Table 1). Gibberellins are required for both hypocotyl (Cowling and Harberd, 1999) and root (Fu and Harberd, 2003) elongation in light-grown Arabidopsis seedlings. In hypocotyls GAs are limiting so applied GA4 promoted growth (Table 1), but in roots of wild-type seedlings GAs are not limiting, so that applied GA4 did not promote root growth. In combination treatments, GA4 could partially reverse the effects of NPA on hypocotyl but not on root growth.
Table 1.
Measurement of 10-d-old Arabidopsis wild-type seedlings treated for 8 d with ATIs or auxins with or without GA4
| Control | +5 μM GA4 | |
| Control | ||
| Hypocotyl length (mm) | 2.5±0.6 | 3.9±0.5 |
| Root length (mm) | 34.3±5.3 | 31.7±2.9 |
| Lateral roots (no.) | 10.5±3.4 | 4.8±1.8 |
| 12.5 μM NOA | ||
| Hypocotyl length (mm) | 2.8±0.2 | 4.3±0.2 |
| Root length (mm) | 37.1±1.5 | 31.9±4.4 |
| Lateral roots (no.) | 4.2±2.3 | 3.6±1.2 |
| 12.5 μM NPA | ||
| Hypocotyl length (mm) | 1.2±0.4 | 2.1±0.3 |
| Root length (mm) | 10.7±1.2 | 8.3±2.7 |
| Lateral roots (no.) | 0.2±0.2 | 1.0±0.3 |
| 0.1 μM IAA | ||
| Hypocotyl length (mm) | 2.4±0.8 | 3.5±0.2 |
| Root length (mm) | 22.2±6.9 | 13.0±3.9 |
| Lateral roots (no.) | 9.7±0.9 | 5.0±1.4 |
| 0.1 μM NAA | ||
| Hypocotyl length (mm) | 1.8±0.3 | 2.6±0.8 |
| Root length (mm) | 16.9±2.2 | 21.5±1.4 |
| Lateral roots (no.) | 16.4±2.5 | 7.8±0.3 |
Values shown are means ±SD (=5 experiments; a minimum of 10 seedlings/treatment were measured for each experiment). Seeds were germinated on nutrient agar, and at 2 d were transferred onto fresh nutrient liquid solution with or without ATIs or auxins. Measurements were made 8 d later.
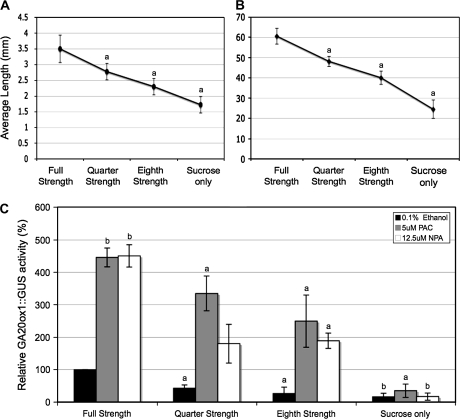
Many dwarf mutants show elevated levels of expression of AtGA20ox1 (Phillips et al., 1995; Xu et al., 1995). Since the up-regulation of AtGA20ox1 expression by NPA and NOA is accompanied by reduced hypocotyl and root growth and/or reduced lateral root number, it was investigated whether up-regulation of AtGA20ox1 is invariably coincident with reduced growth. Ten-day-old seedlings that had been grown in three nutrient-poor solutions showed sequentially less hypocotyl and root elongation during the culture period (Fig. 2A, B). The activity of AtGA20ox1::GUS was quantified in these seedlings (Fig. 2C), and was shown to decrease in parallel with reduced growth. Thus when growth is stunted because of nutrient deprivation AtGA20ox1 expression does not increase. Increase in AtGA20ox1::GUS activity could still be demonstrated in nutrient-deprived seedlings treated with PAC or NPA suggesting that it is altered GA or auxin status that determines the expression level of AtGA20ox1.
Fig. 2.
Measurements of hypocotyl (A) and root (B) lengths of 10-d-old Arabidopsis seedlings grown on serial dilutions of nutrient as specified in the Materials and methods section. (C) Fluorimetric GUS assay of GA20ox1::GUS activity of 10-d-old Arabidopsis seedlings in reduced growth conditions and treated for 8 d with 0.1% ethanol (black bars), 5 μM PAC (grey bars) or 12.5 μM NPA (white bars). Values shown are means ±SD (n=3). Letters on the graph represent means that are statistically different relative to control using the Student t test where (a) P ≤0.05 and (b) P ≤0.01.
Assessing auxin status of treated Arabidopsis seedlings
The DR5::GUS reporter gene (Ulmasov et al., 1997) has been widely used as a tool to monitor the distribution of auxin, as it has been suggested that the resulting GUS activity correlates with IAA distribution (Sabatini et al., 1999; Casimiro et al., 2001). Auxin status monitored in seedling extracts using the DR5::GUS reporter construct (DR5::GUS activity) increased gradually with increasing concentration of applied IAA or NAA (see Supplementary Fig. 1 at JXB online). The DR5::GUS activity was higher for NAA compared to IAA, most likely due to its difference in uptake. The lipophilic NAA is a permeable auxin, and it can enter the cells more easily to trigger DR5::GUS activity in comparison with IAA (Delbarre et al., 1996; Marchant et al., 1999). Furthermore, the increased DR5::GUS activity following treatment with IAA or NAA is not affected by GA4 treatment (see Supplementary Fig. 1 at JXB online).
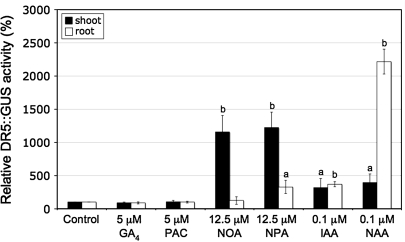
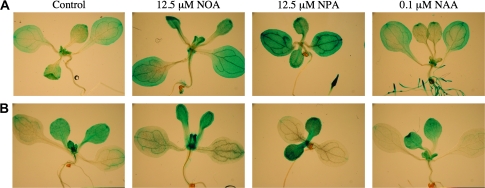
DR5::GUS reporter activity was quantified separately in shoots and roots of seedlings treated with growth regulators in an effort to determine auxin distribution. DR5::GUS reporter activity was unchanged in shoots or roots with GA4 or PAC, but was increased in shoots following ATI treatment (Fig. 3). There was also a small increase in roots of NPA-treated seedlings. Increases in the DR5::GUS reporter activity were seen in shoots and roots of IAA-treated seedlings, but these changes were small in comparison to the GUS activity in roots of NAA-treated seedlings. On histochemical examination, the DR5::GUS activity in untreated seedlings, although low, was clearly seen in the cotyledons and in the emerging and expanding leaves (Fig. 4A), where IAA is synthesized (Ljung et al., 2001). NOA treatment led to increased DR5::GUS activity in the cotyledons and leaves. NPA treatment also led to enhanced DR5::GUS activity in the cotyledons and leaves, and in the root tips, which are swollen. NAA treatment caused a small increase in DR5::GUS activity in the cotyledons and also a marked increase in both the primary and lateral roots, which are abundant in this treatment. IAA treatment caused a small increase in DR5::GUS activity, but it was not comparable to NAA treatment especially in the root (data not shown).
Fig. 3.
Fluorimetric GUS assay of DR5::GUS reporter construct of 10-d-old shoots (black bars) and roots (white bars) of Arabidopsis seedlings following 8 d treatment with GA4, PAC, ATIs or auxins to whole seedlings. Values shown are means ±SD (n=3 different experiments) compared to their respective control (shoot or root) normalized to 100%. Different letters on the bars represent means that are statistically different relative to control using the Student t test where (a) P ≤0.05 and (b) P ≤0.01.
Fig. 4.
Light microscopy pictures of histochemical staining of the (A) DR5::GUS or (B) AtGA20ox1::GUS activity in 10-d-old Arabidopsis seedlings following treatment with ATIs or auxin.
Assessing the relationship between DR5::GUS reporter activity and increased AtGA20ox1 in shoots and roots of ATI-treated seedlings
To determine the location of the AtGA20ox1 expression in treated seedlings, the activity was quantified (Fig. 5) and histochemical analysis was performed (Fig. 4B) of the reporter AtGA20ox1::GUS construct. In PAC-treated seedlings, the increase in AtGA20ox1::GUS activity takes place predominantly in the shoot (Fig. 5). ATIs and auxin treatment also increased AtGA20ox1::GUS activity in the shoot. No difference was noticeable in the roots of the ATI- or auxin-treated seedlings. The histochemical analysis of Arabidopsis seedlings containing the AtGA20ox1::GUS reporter show that the AtGA20ox1 promoter activity is localized mostly in the leaves of seedlings (Fig. 4B). Treatment with NOA and NPA resulted in increased staining in the leaves, whereas increased reporter activity in leaves of auxin-treated seedlings was less apparent.
Fig. 5.
Fluorimetric GUS assay of AtGA20ox1::GUS reporter construct following treatment with GA4, PAC, ATIs or auxins in shoots (black bars) and root (white bars) of 10-d-old Arabidopsis seedlings as described in Fig. 3. Values shown are means ±SD (n=3 different experiments) compared to their respective control (shoot or root) normalized to 100%. Different letters on the bars represent means that are statistically different relative to control using the Student t test where (a) P ≤0.05 and (b) P ≤0.01.
When comparing the activity of the two reporter genes, GUS::DR5 (Fig. 4A) and AtGA20ox1::GUS (Fig 4B), it is clear that there is enhanced AtGA20ox1::GUS activity in some of the places where DR5::GUS activity is high. Thus, in response to ATI treatment, enhanced DR5::GUS activity in leaves, but not in cotyledons, is associated with increased AtGA20ox1::GUS activity. The high DR5::GUS activity in roots of NAA-treated seedlings is not correlated with enhanced AtGA20ox1::GUS activity. Furthermore, it is also evident that elevated AtGA20ox1::GUS activity can occur independently of DR5::GUS activity. This is particularly evident in PAC-treated seedlings (Figs 3, 5).
The AtGA20ox1 up-regulation caused by ATIs is not due to impaired GA signalling
The relationship between AtGA20ox1 expression and auxin status (as determined by DR5::GUS activity) is not simple. Treatment of seedlings with IAA and NAA does not lead to the very high levels of AtGA20ox1 expression observed in certain organs and tissues of NOA- and NPA-treated seedlings (Figs 5, 4B). This may be because the internal concentration of auxin in auxin-treated shoots does not reach the level of endogenous IAA that appears to accumulate in shoots after ATI treatment (Figs 3, 4A). Alternatively, NOA and NPA may be affecting AtGA20ox1 expression in more than one way.
Both GA-deficient and GA-insensitive mutants have elevated levels of AtGA20ox1 (Phillips et al., 1995; Xu et al., 1995, 1999). Bioactive GA activates its signalling pathway by removing specific GA signalling repressor proteins; for example RGA, GAI, and RGA-like, by facilitating their ubiquitin-mediated proteolysis (Dill et al., 2001; Fu et al., 2002; Gubler et al., 2002; Fleet and Sun, 2005; Ueguchi-Tanaka et al., 2007). Moreover, Fu and Harberd (2003), working with decapitated Arabidopsis seedlings or those with altered auxin distribution or response, reported that auxin derived from the shoot facilitates GA-induced proteolysis of RGA in roots. It was therefore determined if the observed effects of ATIs on AtGA20ox1 expression are a consequence of an effect of ATIs on GA signalling by monitoring the presence of RGA. RGA was visualized in the Arabidopsis transgenic line pRGA::GFP::RGA expressing the fusion protein GFP-RGA. In particular, it was necessary to determine RGA levels in those parts of the Arabidopsis seedlings in which DR5::GUS and AtGA20ox1::GUS activities are altered by growth regulators.
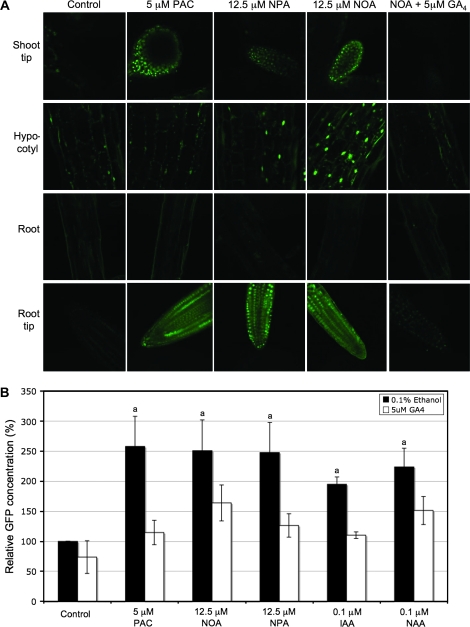
Three-day-old seedlings were transferred to liquid nutrient media containing PAC, NOA, or NPA for 48 h. In the case of the combination treatment, GA4 was added to the liquid nutrient media for the final 4 h. Confocal microscopic examination of component parts of 5-d-old seedlings showed that ATI-treatment increased (or stabilized) the RGA repressor in nuclei of cells in shoot tips, hypocotyls, and root tips in a similar manner to PAC treatment (Fig. 6A). Fluorimetric quantification of GFP-RGA (Fig. 6B) supported the confocal microscopy data, and confirmed that the effects of PAC, ATIs and auxins on the amount of GFP-RGA are reversed by GA4. Taken together, these results suggest that while PAC, ATIs, and auxins all lead to elevated or stabilized GFP-RGA, in none of the cases is this due to an irreversible impairment of GA signalling. Interestingly, the increased amount of GFP-RGA in PAC- or ATI-treated seedlings was not reversed or affected by concomitant application of 0.1 μM IAA or NAA (data not shown).
Fig. 6.
(A) Confocal microscopy images of GFP-RGA reporter accumulation in response to treatment with or without PAC or ATIs in 5-d-old pRGA::GFP::RGA Arabidopsis seedlings. Three-day-old Arabidopsis seedlings were treated for 48 h with or without PAC or ATIs. GFP-RGA fluorescence was monitored in shoot tips, hypocotyls, primary roots, and primary root tips. For combination treatment, GA4 was applied 4 h prior to acquiring the images. (B) Fluorimetric GFP assay of GFP-RGA reporter accumulation in 10-d-old pRGA::GFP::RGA Arabidopsis seedlings in response to 8 d treatment with PAC, ATIs or auxins alone (black bars) or with GA4 (white bars). Values shown are means ±SD (n=3 different experiments) compared to control normalized to 100%. Letters on the bars represent means that are statistically different relative to control using the Student t test where (a) P ≤0.01.
Since the effects of ATIs and exogenous auxins on growth are in some cases mediated by ethylene (Yu and Yang, 1979; Yoshii and Imaseki, 1981; Botella et al., 1992) the effects have been compared of ATIs on AtGA20ox1 expression in the presence or absence of aminoethoxyvinyl glycine (AVG) (10 μM) or sodium oxamate (50 μM), both of which are inhibitors of ethylene biosynthesis or silver nitrate (0.1 μM), an inhibitor of ethylene perception/response. The increased AtGA20ox1::GUS activity by ATIs was observed with or without AVG, sodium oxamate, or AgNO3, indicating that it is independent of ethylene (data not shown).
The up-regulation of AtGA20ox1 expression caused by ATIs or auxins can be reversed by bioactive GA
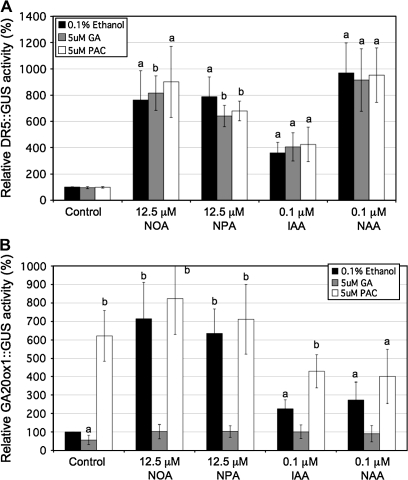
Additional experiments examined whether the AtGA20ox1 expression observed in NOA- and NPA-treated seedlings could be counteracted by simultaneous application of bioactive GA4 using the AtGA20ox1::GUS reporter line. DR5::GUS reporter construct activity was also monitored (Fig. 7). Neither GA4 or PAC, alone or in combination with the ATIs or auxins, affected the levels of DR5::GUS activity (Fig. 7A) suggesting that GA4 does not affect auxin status within these seedlings.
Fig. 7.
Fluorimetric GUS assay of DR5::GUS (A) and AtGA20ox1::GUS (B) reporter activity in response to various treatments with ATIs or auxins alone (black bars) or in combination with GA4 (grey bars) or PAC (white bars) in 10-d-old Arabidopsis seedlings. Values shown are means ±SD (n = at least 3 different experiments) compared to the 0.1% ethanol control normalized to 100%. Different letters on the bars represent means that are statistically different relative to control using the Student t test where (a) P ≤0.05 and (b) P ≤0.01.
The increased AtGA20ox1::GUS activity in ATI- and auxin-treated seedlings was not apparent when GA4 was co-applied with any of these growth regulators (Fig. 7B). Thus AtGA20ox1::GUS activity decreased in response to GA4 in ATI- and auxin-treated seedlings even though the auxin status remained unchanged (Fig. 7A). The effects of ATIs and PAC on AtGA20ox1::GUS activity were not additive.
Discussion
Gibberellin 20-oxidation is a rate-limiting step in GA biosynthesis in Arabidopsis and enhanced growth is seen in transgenic plants over-expressing AtGA20ox1 because of elevated levels of bioactive GA (Coles et al., 1998; Huang et al., 1998). However, increased AtGA20-oxidase mRNAs is not always associated with enhanced growth. A deficit of bioactive GA or perturbation of GA signalling leads to enhanced AtGA20ox1 expression because of metabolic regulation (Xu et al., 1995, 1999). In contrast to up-regulation caused by reduced GA response, positive GA responses down-regulate AtGA20ox mRNAs in GA-treated seedlings (Phillips et al., 1995; Xu et al., 1995). This negative feedback regulation of AtGA20ox1 requires active GA signalling, and RGA and other DELLA proteins that repress GA signalling must be removed by GA-induced proteolysis (Dill et al., 2001; Silverstone et al., 2001; Fu et al., 2002; Itoh et al., 2002; Sasaki et al., 2003) (Fig. 8).
Fig. 8.
Overview of AtGA20ox1 regulation in Arabidopsis seedlings. Organ-, tissue-, and cell-specific regulation supersedes developmental regulation of AtGA20ox, which encodes a multifunctional dioxygenase catalysing a rate-limiting step in the synthesis of bioactive GA4. GA4 can be deactivated by AtGA2ox or can bind to its receptor GID1a, b, c (low affinity) which triggers a conformational change to allow binding of DELLA proteins (RGA, GAI, RGLs). DELLA degradation via SCFSLY1/GAR2, through the 26S proteosome, allows GA signalling and response, including metabolic regulation to down-regulate AtGA20ox expression. Lastly, auxin regulates GA pathways by affecting DELLA stability in roots and promoting GA 20-oxidation in shoots. Numbers in parentheses indicate the following references: (1) Phillips et al., 1995; (2) Xu et al., 1995; (3) Thomas et al., 1999; (4) Desgagné-Penix et al., 2005; (5) Griffiths et al., 2006; (6) Nakajima et al., 2006; (7) Willige et al., 2007; (8) Iuchi et al., 2007; (9) Fu and Harberd, 2003; (10) Frigerio et al., 2006; (11) Pufky et al., 2003; (12) Goda et al., 2004; (13) Rieu et al., 2008.
In addition to metabolic regulation, intrinsic factors, such as other hormones, can affect the expression of genes encoding GA 20-oxidases. For example, 4 Cl-IAA, a native auxin in pea fruits promotes GA 20-oxidation in deseeded pea pericarp (van Huizen et al., 1995, 1997). Furthermore, Wolbang and Ross (2001) showed that decapitation of tobacco plants reduces the level of GA20 in the internodes below that point, an effect that is counteracted by the application of IAA. These studies differ from the present investigation, where the source(s) of native auxin was/were not surgically removed.
In the present work, gene expression analysis and transgenic reporter activity showed that in Arabidopsis seedlings two different ATIs, NOA and NPA, and two different auxins, IAA and NAA, enhance the expression of the AtGA20ox1 in the shoot of 10-d-old seedlings (Fig. 1). The changes in AtGA20ox1 expression mediated by either auxin were less than the up-regulation mediated by the ATIs (Fig. 1). However, it is possible that ATIs may be leading to localized accumulation of auxin concentrations in planta which are not attained by IAA or NAA treatment in our conditions.
All parts of 10-d-old seedlings can synthesis IAA, although young leaves (<0.5 mm in length) have the highest capacity (Ljung et al., 2001). The first developing leaves are the source of IAA for the emergence of lateral roots (Bhalerao et al., 2002) so the decreased number of lateral roots in ATI-treated seedlings (Table 1) is consistent with impaired shoot-to-root auxin transport. NOA and NPA affect auxin transport in different ways; NOA acts at the AUX1 influx carrier proteins (Parry et al., 2001; Ottenschlager et al., 2003; Yang et al., 2006) whereas NPA is known to block auxin transport by affecting efflux (Katekar and Geissler, 1980). Treatment with both led to intense DR5::GUS reporter activity in emerging and expanding leaves (Figs 3, 4A). The appearance of these seedlings (Table 1; Fig. 3) suggests that ATIs may be causing supra-optimal auxin accumulation in shoots. Strong reporter activity was also observed in the root caps of ATI-treated seedlings (Fig. 4A), consistent with the observation that, even though ATIs may be trapping auxin in the emerging and expanding leaves, root tips are additional sites of auxin synthesis (Ljung et al., 2005). NAA and IAA also led to enhanced DR5::GUS activity relative to untreated seedlings (Fig. 3), suggesting that both ATI and auxin-treatments lead to auxin concentrations in planta that exceed those seen in untreated wild-type plants.
In emerging and expanding leaves, increased AtGA20ox1 expression following treatment with ATIs correlates with high DR5::GUS activity, whereas in cotyledons increased DR5::GUS activity was not accompanied by AtGA20ox1 expression. In addition, elevated auxin response in NAA-treated roots (as shown by DR5::GUS activity and a multitude of lateral roots) was not accompanied by elevated AtGA20ox1 expression. Conversely, elevated AtGA20ox1 expression in shoots of PAC-treated seedlings was not accompanied by high DR5::GUS activity.
One explanation for our results is that several levels of regulation of AtGA20ox1, imposed in a strict hierarchical order, can be observed in intact wild-type Arabidopsis seedlings (Fig. 8). The first type of regulation would be organ-, tissue-, or cell-specific regulation. For example, in the present work, AtGA20ox1 expression is low in roots of 10-d-old Arabidopsis seedlings, in contrast to that in cotyledons and leaves (Fig. 4B). [With regard to the low level of expression of AtGA20ox1 in 10-d-old roots, our results are different from those of Rieu et al. (2008) who observed similar GA20ox1 transcripts levels in roots and shoots of 7-d-old seedlings.] The second would be developmental regulation. For example, AtGA20ox1 is strongly expressed in cotyledons of up to approximately 8 d (data not shown), but not in cotyledons of 10-d-old seedlings (Fig. 4B). Previous work has shown that several genes encoding GA metabolic enzymes are developmentally regulated (for example, Phillips et al., 1995; Silverstone et al., 1997; Mitchum et al., 2006). Indeed Rieu et al. (2008) have recently published developmental expression profiles for the five AtGA20ox genes throughout the Arabidopsis life cycle. The third level of regulation would be metabolic regulation, whereby the endogenous level of bioactive GA regulates the expression of its biosynthetic genes. For example, in 6-d-old seedlings PAC treatment up-regulates AtGA20ox1 in the cotyledons and leaves, whereas in 10-d-old seedlings, up-regulation in the cotyledons was not observed, suggesting that the developmental regulation overrides metabolic regulation. The fourth level of regulation of AtGA20ox1 expression is auxin-mediated. In emerging and expanding leaves, AtGA20ox1 expression occurred where DR5::GUS activity was high after treatment with ATIs. However, this up-regulation of AtGA20ox1 expression does not occur when ATIs and bioactive GA are applied concurrently (Fig. 7B), indicating that metabolic regulation overrides auxin regulation. The absence of AtGA20ox1 expression in roots of NAA-treated seedlings, despite high DR5-GUS activity, can also be explained by the lower overall level of expression in roots (organ-specific regulation) overriding the other types of regulation.
As mentioned earlier, the expression of AtGA20ox1 is up-regulated by altered flux through the GA biosynthetic pathway or due to a block in the GA signalling pathway. The question arises through which of these means are the ATIs exerting their effect on AtGA20ox1 expression. The current model for GA signalling proposes that DELLA proteins such as RGA must be degraded in order for the GA response to be transduced (Fleet and Sun, 2005; and see Fig. 8). The binding of bioactive GA to its protein receptor is necessary for the proteolytic degradation of DELLA proteins (Griffiths et al., 2006; Nakajima et al., 2006; Iuchi et al., 2007; Willige et al., 2007). Treatment of Arabidopsis seedlings with PAC and ATIs leads to the stabilization of RGA in shoot tips, young leaves, and primary root tips (Fig. 6A). In the case of ATI-treated seedlings, these are the locations of high DR5::GUS staining, indicating they are sites of auxin response (Fig. 4A). This stabilization of RGA by ATIs is therefore unexpected if auxin facilitates RGA degradation as reported by Fu and Harberd (2003). While their work specifies shoot-derived auxin facilitates RGA degradation in root tips, we have no evidence of auxin promoting RGA degradation in tissues in which they appear to co-occur.
Results reported here suggest that high AtGA20ox1 expression in ATI- and auxin-treated seedlings is not due to auxin increasing flux through the GA biosynthetic pathway (in which case AtGA20ox1 expression would be high and RGA degraded). After ATI treatment, the high AtGA20ox1 expression and RGA persistence suggests that either there is a deficit of bioactive GA, or GA signalling is impaired. Concomitant treatment of ATIs and GA, or auxin and GA, led to RGA disappearance (Fig. 6A, B). Moreover, applied GA4 can, in part, overcome the effects of NPA on hypocotyl growth (Table 1). Thus the GA signal transduction pathway seems not to be impaired by ATIs, instead metabolic regulation appears to be the reason for RGA stabilization.
Frigerio et al. (2006) demonstrated that 50 μM NAA caused a spike in AtGA20ox1 expression 30 min after application, suggesting a direct regulation of AtGA20ox1 by auxin. On the other hand, a longer term effect of applied auxin on AtGA20ox1 expression, leading to metabolic regulation of GA biosynthesis and/or deactivation genes, could ultimately cause a reduction in the level of endogenous bioactive GA. This, in turn, would result in the accumulation of RGA and up-regulation of AtGA20ox1. While Frigerio et al. (2006) showed that, in 24 h treatments, auxin up-regulated AtGA20ox1 even in Arabidopsis mutants lacking RGA and GAI, these observations do not preclude the involvement of metabolic regulation in the longer term treatments described in this paper. While it is already known that enhanced AtGA3ox1 expression occurs with ATI (I Desgagné-Penix and VM Sponsel, unpublished results) and auxin treatment (Frigerio et al., 2006), further work will examine GA 2-oxidation in the experimental system described here. Using a concentration of NAA that is clearly supra-optimal (a 500-fold excess of NAA over that used in the current work), Frigerio et al. (2006) demonstrated that several members of the AtGA2ox family were up-regulated. Whether the much lower auxin concentrations used in the present work also enhance GA deactivation by 2-oxidation or other means (Zhu et al., 2006; Varbanova et al., 2007), contributing to a decrease in bioactive GA levels, will be examined. As noted earlier, it is also possible that the ATI treatments used in the present work led to the accumulation of supra-optimal IAA levels in treated shoots, so that the effects of both applied ATIs and auxins on 2-oxidation must be compared. It is also proposed to quantify endogenous GAs in ATI- treated and control seedlings by GC-MS to understand more fully the possible site(s) within the biosynthetic pathway where ATIs may be exerting an effect.
As key repressive factors of the GA signal transduction pathway, RGA and other DELLA proteins have been identified as probable sites of cross-talk between GAs and several other hormones (Achard et al., 2003; Vriezen et al., 2004). In some respects, the results reported can be discussed in the context of those for the interaction of ethylene and GAs in apical hooks of etiolated Arabidopsis seedlings (Achard et al., 2003; Vriezen et al., 2004). Analysis of ACC-treated etiolated Arabidopsis seedlings showed an increase in ent-copalyl diphosphate synthase (CPS; GA1) expression and accumulation of RGA (similar to our increased AtGA20ox1 and RGA accumulation induced by auxin or ATIs). They concluded that the effects of ethylene (Vriezen et al., 2004) [and auxin (Achard et al., 2003)] on apical hook structure are mediated via an effect on stabilizing RGA. However, it is possible that these effects are also mediated in part by lowering endogenous bioactive GA levels. Although an examination of GASA1::GUS activity suggested that the GA response was not reduced, a direct examination of GA levels would allow this question to be addressed definitively.
In conclusion, our results showed that perturbing auxin status in intact Arabidopsis seedlings leads to an up-regulation of the biosynthetic enzyme AtGA20ox1 mRNA levels and AtGA20ox1::GUS activity, accompanied by an increase or stabilization of RGA. It is proposed that these observations are consistent with the homeostatic regulation of GA levels. Moreover, developmental regulation, and organ-, tissue-, and cell-specific regulation override both auxin- and metabolic regulation, ensuring the appropriate temporal and spatial expression of AtGA20ox1. Since all our experiments were conducted in continuous white light, it is not possible at the present time to place light regulation within this hierarchy although, clearly, AtGA20ox1 is regulated by light of varying wavelengths (Achard et al., 2007; Zhao et al., 2007).
Just how these layers of regulation are imposed has yet to be defined. Chang and Sun (2002), after comprehensive promoter deletion analysis, were able to define positive and negative regulatory sequences for the expression of CPS, which catalyses the first committed step in GA biosynthesis. In addition, they defined a sequence necessary for the expression of CPS in developing seeds. To our knowledge, such a comprehensive analysis of the AtGA20ox1 promoter, or intragenic regulatory sequences, has yet to be conducted. Some auxin-regulated genes in Arabidopsis, though not AtGA20ox1, possess either an auxin response element TGTCTC or two copies of TGTC within 50 nucleotides of each other (Nemhauser et al., 2004). Thus the sequence element necessary and sufficient for auxin regulation of AtGA20ox1 has yet to be discovered. Similarly, although feedback regulation of AtGA3ox1, which catalyse the final metabolic step in the production of bioactive GA4 in Arabidopsis, appears to be mediated by an AT-hook DNA-binding protein, AGF1, no AT-hook binding sequence is observed in the AtGA20ox1 promoter region (Matsushita et al., 2007). Thus a considerable amount of further work is necessary to determine the nature of different cis-acting elements for AtGA20ox1, and how they are organized to allow multiple levels of regulation.
Supplementary data
Supplementary data are available at JXB online. Figure S1 shows DR5::GUS reporter activity in response to increasing concentration of IAA or NAA alone or in combination of 5 μM GA4 in 10-d-old Arabidopsis seedlings.
Supplementary Material
Acknowledgments
We thank Drs Peter Hedden, Tom Guilfoyle, and Tai-ping Sun for the reporter lines as specified in the Materials and methods. We also want to thank Dr Garry Sunter for training and access to the fluorimeter and Dr Colleen Witt for training and access to the confocal microscope. This work was supported by funding from the National Science Foundation (IBN-0080934 to VMS) and in part by the Research Centers at Minority Institutions Grant 5G12 RR013646-07 awarded to UTSA through the NCRR. IDP was supported by MBRS-RISE GM60655.
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP. DELLAs contribute to plant photomorphogenesis. Plant Physiology. 2007;143:1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. The Plant Cell. 2003;15:2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. The Plant Journal. 2002;29:325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- Botella JR, Schlangenhaufer CD, Arteca RN, Phillips AT. Identification and characterization of a full-length cDNA encoding for an auxin-induced 1-aminocyclopropane-1-carboxylate synthase from etiolated mung bean hypocotyl segments and expression of its mRNA in response to indole-3-acetic acid. Plant Molecular Biology. 1992;20:425–436. doi: 10.1007/BF00040602. [DOI] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S. Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiology. 1999;119:765–774. doi: 10.1104/pp.119.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, et al. Auxin transport promotes Arabidopsis lateral root initiation. The Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-w, Sun T-p. Characterization of cis-regulatory regions responsible for developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant Molecular Biology. 2002;49:579–589. doi: 10.1023/a:1015592122142. [DOI] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ. The tandem complex BEL and KNOX partners is required for repression of ga20ox1. The Plant Journal. 2004;38:276–284. doi: 10.1111/j.1365-313X.2004.02048.x. [DOI] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. The Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate–phenol–chloroform extraction. Analytical Biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. The Plant Journal. 1998;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- Cowling RJ, Harberd NP. Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. Journal of Experimental Botany. 1999;50:1351–1357. [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou D-X. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiology. 2007;144:121–133. doi: 10.1104/pp.107.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J. Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid and indole-3-acetic acid in suspension-cultured tobacco cells. Planta. 1996;198:532–541. doi: 10.1007/BF00262639. [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix I, Eakanunkul S, Coles JP, Phillips AL, Hedden P, Sponsel VM. The auxin transport inhibitor response 3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. The Plant Journal. 2005;41:231–242. doi: 10.1111/j.1365-313X.2004.02287.x. [DOI] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun T-p. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proceedings of the National Academy of Sciences, USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T-p. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Ross JJ, Smith JJ, Lester DR, Reid JB. Feed-forward regulation of gibberellin deactivation in pea. Journal of Plant Growth Regulation. 2001;20:87–94. [Google Scholar]
- Fleet CM, Sun T-p. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Current Opinion in Plant Biology. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Frigerio M, Aladi D, Perez-Gomez J, Garcia-Carcel L, Phillips AL, Hedden P, Blazquez MA. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiology. 2006;142:553–563. doi: 10.1104/pp.106.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-ali T, Hynes LW, Ougham H, Peng J, Harberd NP. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. The Plant Cell. 2002;14:3191–3200. doi: 10.1105/tpc.006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Gil J. Light regulation of gibberellin biosynthesis and mode of action. Journal of Plant Growth Regulation. 2002;20:354–368. doi: 10.1007/s003440010033. [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiology. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiology. 2002;129:191–200. doi: 10.1104/pp.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. The gibberellin pathway mediates KNOTTED1-type homeobox in plants with different body plans. Current Biology. 2002;12:1557–1565. doi: 10.1016/s0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiology. 1998;118:773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. The Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim Y-C, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. The Plant Journal. 2007;50:958–966. doi: 10.1111/j.1365-313X.2007.03098.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase, a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katekar GF, Geissler AE. Auxin transport inhibitors. IV. Evidence of a common mode of action for a proposed class of auxin transport inhibitors: the phytotropins. Plant Physiology. 1980;66:1190–1195. doi: 10.1104/pp.66.6.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. The Plant Cell. 1990;2:257–263. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. The Plant Journal. 2001;28:465–474. doi: 10.1046/j.1365-313x.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. The Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO Journal. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y. AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiology. 2007;15:1120–1130. doi: 10.1104/pp.106.093542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun T-p. Distinct and overlapping roles of two gibberellin 3-oxidase in Arabidopsis development. The Plant Journal. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, et al. Identification and characterization of Arabidopsis gibberellin receptors. The Plant Journal. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroids and auxin signaling in Arabidopsis. PLoS Biology. 2004;2:1460–1471. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo P, Ozga JA, Reinecke DM. Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp. Plant Molecular Biology. 2002;49:439–448. doi: 10.1023/a:1015522404586. [DOI] [PubMed] [Google Scholar]
- Olszewski NT, Sun T-p, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. The Plant Cell. 2002;14:S111–S130. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill D, Ross JJ. Auxin regulation of the gibberellin pathway in pea. Plant Physiology. 2002;130:1974–1982. doi: 10.1104/pp.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sanberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proceedings of the National Academy of Sciences, USA. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM. Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiology. 2003;131:1137–1146. doi: 10.1104/pp.102.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ. Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. The Plant Journal. 2001;25:399–406. doi: 10.1046/j.1365-313x.2001.00970.x. [DOI] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiology. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufky J, Qiu Y, Rao M, Hurban P, Jones A. The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Functional and Integrative Genomics. 2003;135:135–143. doi: 10.1007/s10142-003-0093-7. [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. The Plant Journal. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- Ross JJ. Effects of auxin transport inhibitors on gibberellins in pea. Journal of Plant Growth Regulation. 1998;17:141–146. [Google Scholar]
- Ross JJ, MacKenzie-Hose AK, Davies PJ, Lester DR, Twitchin B, Reid JB. Further evidence for feedback regulation of gibberellin biosynthesis in pea. Physiologia Plantarum. 1999;105:532–538. [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–189. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, C-w Chang, Krol E, Sun T-p. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. The Plant Journal. 1997;12:9–19. doi: 10.1046/j.1365-313x.1997.12010009.x. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun T-p. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. The Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. The Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel VM, Hedden P. Gibberellin biosynthesis and inactivation. In: Davies PJ, editor. Plant hormones: physiology, biochemistry and molecular biology. 2nd edn. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2004. pp. 66–97. [Google Scholar]
- Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annual Review of Plant Biology. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proceedings of the National Academy of Sciences, USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annual Review of Plant Biology. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiology. 1997;115:123–128. doi: 10.1104/pp.115.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN. Seed and 4-chloroindole-3-acetic acid regulation of gibberellin metabolism in pea pericarp. Plant Physiology. 1995;109:1213–1217. doi: 10.1104/pp.109.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova M, Yamaguchi S, Yang Y, et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. The Plant Cell. 2007;19:32–45. doi: 10.1105/tpc.106.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Achard P, Harberd NP, Van Der Straeten D. Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. The Plant Journal. 2004;37:505–516. doi: 10.1046/j.1365-313x.2003.01975.x. [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. The DELLA domain of GA INSENSITIVE mediated the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. The Plant Cell. 2007;19:1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ. Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiology. 2004;134:769–776. doi: 10.1104/pp.103.030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Ross JJ. Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta. 2001;214:153–157. doi: 10.1007/s004250100663. [DOI] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JA. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proceedings of the National Academy of Sciences, USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Gage DA, Zeevaart JA. Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. The Plant Cell. 1999;11:927–935. doi: 10.1105/tpc.11.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y. Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant and Cell Physiology. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammers UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Current Biology. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Yoshii H, Imaseki H. Biosynthesis of auxin-induced ethylene. Effects of indole-3-acetic acid, benzyladenine and abscisic acid on endogenous levels of 1-aminocyclopropane-1-carboxylic acid (ACC) and ACC synthase. Plant and Cell Physiology. 1981;22:369–379. [Google Scholar]
- Yu YB, Yang SF. Auxin induced ethylene production and its inhibition by aminoethoxyvinylglycine and cobalt ion. Plant Physiology. 1979;64:1074–1077. doi: 10.1104/pp.64.6.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu X, Foo E, et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiology. 2007;145:106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Yonghan X, et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. The Plant Cell. 2006;18:442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.