Abstract
Control of Salmonella enterica serovar Typhimurium (S. typhimurium) infection in the mouse model of typhoid fever is critically dependent on the natural resistance-associated macrophage protein 1, Nramp1. Here we examined the role of genetic polymorphisms in the human homologue, NRAMP1, in resistance to typhoid fever in southern Vietnam. Patients with blood-culture confirmed typhoid fever and healthy controls were genotyped for six polymorphic markers within and near NRAMP1 on chromosome 2q35. Four single base pair polymorphisms (274 C/T, 469+14 G/C, 1465-85 G/A, D543N), a (GT)n repeat in the promoter region of NRAMP1 and D2S1471, a microsatellite marker approximately 130 kb downstream of NRAMP1, were examined. The allelic and genotypic frequencies for each polymorphism were compared in cases and controls. No allelic association was identified between the NRAMP1 alleles and typhoid fever susceptibility. In addition, neither homozygotes or heterozygotes for any of the NRAMP1 variants were at increased risk of typhoid fever.
Introduction
The Nramp1 gene represents one of the most extensively characterised mouse host resistance loci. Initially it was observed that certain inbred mouse strains were innately susceptible to infection with the intracellular pathogens, S. typhimurium, Leishmania donovani and Mycobacterium bovis (bacille Calmette-Guerin) (for a recent review see [1]). This susceptibility was known to be under the control of a gene at the Ity/Bcg/Lsh locus on proximal chromosome 1, which was later positionally cloned and designated Nramp1 [2]. A single non-conservative amino acid substitution of glycine to aspartic acid at position 169 (G169D) within Nramp1 was found to be correlated with the susceptibility phenotype [2]. The causal relationship between Nramp1 and susceptibility to intracellular pathogens was confirmed by the construction of (i) a Nramp1 knockout mouse and (ii) a susceptible mouse (Nramp1D169) transgenic for the resistance allele (Nramp1G169). With S. typhimurium, mice containing the Nramp1 susceptibility allele are unable to control infection with a low number of organisms and die, whereas in resistant animals, S. typhimurium grow at a slower rate which results in eventual clearance of the organisms [3]. Salmonella enterica serovar Typhi (S. typhi), which is of the same species as S. typhimurium, is the causative agent of typhoid fever in humans. The fact that these two microorganisms are so closely related, and the extreme difference in phenotype observed in the mouse model of typhoid due to Nramp1, suggests that the human homologue of Nramp1, NRAMP1 is an obvious candidate when investigating host resistance genes to typhoid fever in human populations.
The macrophage is an integral cell in the defence against microbial agents and Nramp1 plays a significant role in the early innate phase of the macrophage-pathogen interaction. Nramp1 influences many functions important to macrophage activation including the up-regulation of chemokines (KC), cytokines [tumor necrosis factor α (TNF-α) and interleukin 1 β (IL1-β)], major histocompatibility complex (MHC) class II expression, respiratory burst activity, release of nitric oxide (NO), antigen processing and apoptosis [4]. The direct mechanism by which Nramp1 effects these macrophage functions is not clear. What is known is that Nramp1 encodes an integral membrane protein, which is localised to the late endosomal and lysosomal compartment of the macrophage. Nramp1 functions as a metal ion transporter modulating cellular iron levels which suggests that it may restrict the replication of intracellular pathogens by altering the phagolysosomal environment.
Despite the murine and human genes encoding NRAMP1 being highly conserved, the mutation (G169D) responsible for the murine susceptibility to intracellular pathogens is not present in the human homologue. However several other polymorphisms within human NRAMP1 have been identified [5, 6]. Here we genotyped six polymorphic markers within and near the NRAMP1 gene in typhoid fever patients and control individuals from Vietnam. In the southern provinces of Vietnam typhoid fever is endemic, with the current annual incidence rate in the Mekong Delta at 198/100,000 population [7] .
Subjects and Methods
Subjects
The samples genotyped were collected as part of larger epidemiological and treatment studies performed in Dong Thap and Dong Nai provinces Vietnam and have previously been described in Dunstan et al [8] and Luxemburger et al [9]. Blood was taken from patients with culture positive typhoid fever admitted to either Dong Thap Provincial Hospital (N=112) or Dong Nai Paediatric Centre (N=105). Healthy control samples were collected from the community in Dong Thap (N=77) and from patients undergoing minor surgery in Dong Nai (N=211). As age is a known risk factor for typhoid fever and some behaviour (such as eating outside the home) may be gender-related, samples were matched for age and sex. Typhoid cases and control individuals were also matched for their residential location. All typhoid patients and controls were unrelated and were of Vietnamese Kinh ethnicity.
NRAMP1 single nucleotide polymorphism (SNP) genotyping
Four SNPs within NRAMP1 (274 C/T, 469+14 G/C, 1465-85 G/A and D543N) were genotyped using the PCR primers, restriction enzymes and conditions described in Liu et al [6] .
D2S1471 microsatellite and (GT)n genotyping
D2S1471 and the repeat in the promoter region of NRAMP1 (designated [GT]n) were PCR amplified and the products generated were sized using an Applied Biosystems 377 automated sequencer. The fluorescent oligonucleotide primers and methods used have been published previously [5, 10] .
Statistical Analysis
Pearson’s chi-squared (χ2) test was used to test associations between disease phenotypes (typhoid cases or controls) and allele/genotype frequencies. Fisher’s exact test was used when an expected value in the contingency table was less than 5. For contingency tables larger than 2 × 2, which did not satisfy Cochran’s rule, the Fisher Freeman Halton test was used and the exact P value was reported. For analysis of D2S1471 in table 1 Pearson’s χ2 was used and the Monte Carlo estimate of the P value was reported. Linkage disequilibrium between loci within NRAMP1 in the Vietnamese control population was measured by Cramer’s association measure for contingency tables. When Wn=1, strong linkage disequilibrium is indicated; for Wn=0, there is no linkage disequilibrium. Overall allele frequencies for each polymorphic loci tested were compared using the likelihood-ratio test. The likelihood-ratio test was also used for testing haplotype associations with maximum-likelihood estimates for haplotype frequencies obtained by the expectation-maximisation (E-M) algorithm.
Table 1. Allelic and genotypic frequencies of NRAMP1 polymorphisms in typhoid patients and controls.
| NRAMP1 polymorphism | alleles/genotypesa | Typhoid Casesb,c | Controlsb,d | χ2(df) | P value |
|---|---|---|---|---|---|
| (GT)n | 1 (124 bp) | 4 (0.01) | 10 (0.02) | ||
| 2 (122 bp) | 46 (0.11) | 49 (0.08) | |||
| 3 (120 bp) | 378 (0.88) | 517 (0.90) | 2.491 (2) | 0.288 | |
| 1/2 | - | 1 (0.004) | |||
| 1/3 | 4 (0.02) | 9 (0.03) | |||
| 2/2 | 1 (0.005) | 4 (0.014) | |||
| 2/3 | 44 (0.205) | 40 (0.14) | |||
| 3/3 | 165 (0.77) | 234 (0.812) | - | 0.166e | |
| 274 C/T | T (allele 1) | 21 (0.09) | 16 (0.10) | ||
| C (allele 2) | 203 (0.91) | 138 (0.90) | 0.106 | 0.744 | |
| 1/1 (T/T) | 1 (0.01) | 1 (0.01) | |||
| 1/2 (T/C) | 19 (0.17) | 14 (0.18) | |||
| 2/2 (C/C) | 92 (0.82) | 62 (0.81) | - | 0.926e | |
| 469+14 G/C | G (allele 1) | 203 (0.91) | 136 (0.91) | ||
| C (allele 2) | 21 (0.09) | 14 (0.09) | 0.0002 | 0.989 | |
| 1/1 (G/G) | 92 (0.82) | 62 (0.83) | |||
| 1/2 (G/C) | 19 (0.17) | 12 (0.16) | |||
| 2/2 (C/C) | 1 (0.01) | 1 (0.01) | - | 0.947e | |
| 1465-85 G/A | A (allele 1) | 61 (0.27) | 40 (0.26) | ||
| G (allele 2) | 163 (0.73) | 114 (0.74) | 0.074 | 0.786 | |
| 1/1 (A/A) | 5 (0.04) | 8 (0.10) | |||
| 1/2 (A/G) | 51 (0.46) | 24 (0.31) | |||
| 2/2 (G/G) | 56 (0.50) | 45 (0.59) | 5.311 (2) | 0.070 | |
| D543N | Asn A (allele 1) | 33 (0.15) | 24 (0.16) | ||
| Asp G (allele 2) | 189 (0.85) | 130 (0.84) | 0.037 | 0.848 | |
| 1/1 (A/A) | - | 1 (0.01) | |||
| 1/2 (A/G) | 33 (0.30) | 22 (0.29) | |||
| 2/2 (G/G) | 78 (0.70) | 54 (0.70) | - | 0.603e | |
| D2S1471 | 1 [81] | 37 (0.086) | 44 (0.077) | ||
| 2 [83] | 3 (0.007) | 3 (0.005) | |||
| 3 [85] | 0 | 1 (0.002) | |||
| 4 [87] | 1 (0.002) | 1 (0.002) | |||
| 5 [89] | 40 (0.093) | 45 (0.079) | |||
| 6 [91] | 2 (0.005) | 1 (0.002) | |||
| 7 [93] | 3 (0.007) | 7 (0.012) | |||
| 8 [95] | 2 (0.005) | 2 (0.003) | |||
| 9 [97] | 19 (0.044) | 32 (0.056) | |||
| 10 [100] | 46 (0.106) | 46 (0.081) | |||
| 11 [102] | 200 (0.463) | 272 (0.479) | |||
| 12 [104] | 45 (0.104) | 60 (0.106) | |||
| 13 [106] | 27 (0.062) | 39 (0.069) | |||
| 14 [108] | 4 (0.009) | 12 (0.021) | |||
| 15 [110] | 1 (0.002) | 3 (0.005) | |||
| 16 [112] | 1 (0.002) | 0 | |||
| 18 [116] | 1 (0.002) | 0 | 11.16 (16) | 0.849f |
[values] represent size of alleles approximated from mobility of PCR products on an ABI 377 DNA sequencer,
(values) represent allelic and genotypic frequencies,
cases sample sizes (GT)n N=214; 274C/T, 469+14G/C, 1465-85G/A N=112; D543N N=111; D2S1471 N=216,
control sample sizes (GT)n N=288; 274C/T, 1465-85G/A, D543N N=77; 469+14G/C N=75; D2S1471 N=284,
Fisher Freeman Halton test, exact P value,
Monte Carlo estimate for Pearson’s χ2 test
Results
Genotyping of polymorphisms in NRAMP1
The allelic and genotypic frequencies in typhoid patients and controls of five NRAMP1 polymorphisms, one within the promoter [(GT)n] and four SNPs within the NRAMP1 gene are shown in table 1. The allelic frequencies of the four intragenic SNPs in the Vietnamese controls were comparable to those in an Asian population reported by Lui et al [6]. The allelic frequencies of the five SNPs in typhoid cases were not significantly different to the frequencies in matched controls (P>0.05). In addition, genotypic frequencies of the four intragenic SNPs were not significantly different between cases and controls. The allele frequencies of the microsatellite marker D2S1471 are also presented in table 1. As the NRAMP1 (GT)n promoter polymorphism and the D2S1471 microsatellite are more polymorphic than the SNPs (3 and 18 alleles, respectively) a larger number of samples were genotyped. The larger sample set consisted of samples from Dong Thap and Dong Nai province. No allelic associations were detected with the D2S1471 microsatellite marker (χ2 11.16, df 16, P= 0.849).
Linkage disequilibrium and haplotyping
Linkage disequilibrium between loci within NRAMP1 in the Vietnamese control population was measured. Markers in the 5′ region of NRAMP1 were in strong linkage disequilibrium with each other [(GT)n and 274C/T (Wn=0.89, P=1.24e-26), (GT)n and 469+14G/C (Wn=0.96, P =9.47e-31), 274C/T and 469+14G/C (Wn=0.85, P =1.55e-25)]. Evidence for weaker linkage disequilibrium also occurs between markers within the 3′ region of NRAMP1 [1465-85G/A and D543N (Wn=0.63, P=4.1e-15)], with even weaker linkage disequilibrium between 5′ and 3′ markers [e.g. (GT)n and D543N (Wn=0.43, P=7.87e-07); 274C/T and D543N (Wn=0.14, P=0.07)]. Evidence for linkage disequilibrium between D2S1471 and either 5′ [e.g. (GT)n and D2S1471 (Wn=0.29, P=9.84e-26)] or 3′ [e.g. 1465-85G/A and D2S1471 (Wn=0.61, P=9.6e-7); D543N and D2S1471 (Wn=0.31, P=0.42)] NRAMP1 intragenic markers was variable depending on the marker, suggesting that there may be some extended haplotypes across this ∼130kb region. Based on these observations, we examined whether there was an association between particular 5′ versus 3′ NRAMP1 haplotypes and typhoid fever. Maximum-likelihood frequencies of the possible haplotypes of 2, 3, 5 or 6 markers across the NRAMP1 region were estimated using the E-M algorithm (data not shown). The χ2 data for maximum likelihood frequency comparisons between cases and controls for each individual marker, and for 5′, 3′, and combined 5′-to-3′ haplotypes, are shown in figure 1. No significant haplotype associations with disease were observed.
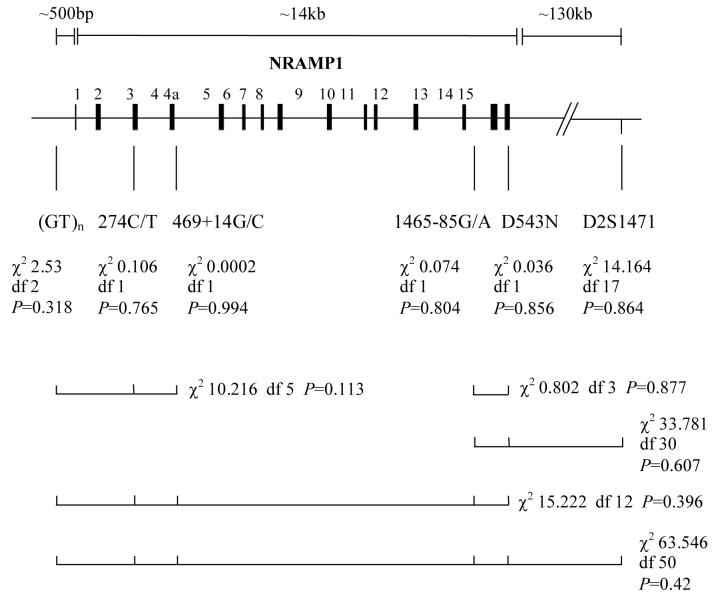
Figure 1.
Typhoid patients and controls were genotyped for six polymorphisms within and near NRAMP1. This schematic representation shows the location of the polymorphisms genotyped. Four intragenic NRAMP1 polymorhisms (274C/T, 469+14G/C, 1465-85G/A and D543N), one polymorphism in the promoter region of NRAMP1 ([GT]n) and the microsatellite marker D2S1471 were genotyped. The overall allele frequencies of the six individual markers in typhoid cases and controls were compared by the likelihood ratio test. Maximum likelihood frequencies of the possible haplotypes of the 5′ markers, the 3′ markers and all markers across the gene region were estimated using the E-M algorithm. Below the tests for the individual markers are bars that represent the various 2, 3, 5 and 6 loci haplotypes tested. The overall frequencies of the estimated haplotypes were compared in typhoid cases and controls. Empirical P values are stated.
Discussion
Genetic linkage/association has been identified between NRAMP1 alleles and infections caused by the intra-macrophage pathogens Mycobacterium tuberculosis [11] and Mycobacterium leprae [12]. However, in this sample of Vietnamese, no association was detected between any of six NRAMP1 polymorphisms and typhoid fever, a disease caused by infection with the intra-macrophage pathogen S. typhi. The sample we employed did have power to detect allelic association between HLA and acute typhoid fever in our earlier study [8]. Hence, one interpretation of our data is that NRAMP1 plays no role in susceptibility to typhoid fever. However, it is possible that a larger sample size might detect a weaker influence of NRAMP1 on disease. Other interpretations of our data are: (i) NRAMP1 plays no role in typhoid fever susceptibility in this population but may in other populations; and/or (ii) NRAMP1 is not associated with acute culture-confirmed typhoid fever, but may play a role in more serious sequelae of disease. The former is consistent with previous observations that NRAMP1 is associated/linked with leprosy in Vietnam, but not in French Polynesia or in Calcutta [12]. The effect of NRAMP1 may be more prominent on some ethnic genetic backgrounds than others, as would be expected if disease susceptibility were under polygenic control. This observation is also consistent with multifactorial disease control, where local environmental or microbial factors may modify the effect of specific susceptibility loci in different populations.
Bellamy et al [11] found a significant association between four intragenic NRAMP1 polymorphisms and susceptibility to tuberculosis in the Gambia. They reported that two polymorphic markers in the 5′ region of the gene [(CA)n and INT4 (termed 469+14G/C here)] were in strong linkage disequilibrium, as were two markers in the 3′ region (D543N and 3′UTR). However markers in the 5′ region were not in linkage disequilibrium with the markers in the 3′ region. Hence they concluded that separate polymorphisms in the 5′ and 3′ regions of the gene acted independently to control susceptibility to tuberculosis [11]. As in The Gambia, markers in the 5′ and 3′ regions of NRAMP1 were in strong linkage disequilibrium with each other in the Vietnamese, with some evidence for extended haplotypes across NRAMP1 and the ∼130kb region spanning the most 5′ to 3′ markers used in this study. Nevertheless, the potential increase in information content for regions of the NRAMP1 locus obtained by haplotype analysis did not provide any further evidence for an association between NRAMP1 and typhoid. Bellamy et al [11] also reported that heterozygosity of INT4 (5′ marker) and 3′UTR (3′ marker) had an additive effect and was associated with the highest risk of tuberculosis. Heterozygosity at 469+14G/C (INT4, 5′ marker), D543N (3′ marker), or combined heterozygosity, was not associated with typhoid fever in the Vietnamese (data not shown).
Recently we reported significant allelic associations between genes of the major histocompatibility complex (HLA-DR, HLA-DQ and TNFA) and typhoid fever in these samples from the southern provinces of Vietnam [8]. A previous study [13] suggested that the NRAMP1 (GT)n promoter polymorphism was associated with disease susceptibility in a subset of rheumatoid arthritis patients who did not possess certain HLA susceptibility alleles. We investigated this in our sample set by removing all individuals that were positive for HLA-DRB1*04 (typhoid resistance allele) or HLA-DRB1*0301 (typhoid susceptibility allele), and re-testing for an association between the NRAMP1 (GT)n promoter polymorphism, the microsatellite marker D2S1471 and typhoid fever. In the subset of typhoid patients (N=206) and controls (N=239) lacking HLA-DRB1*04 individuals, no association between typhoid and the NRAMP1 (GT)n promoter polymorphism (χ2 1.61, df 2, P= 0.48) or D2S1471 (χ2 15.29, df 17, P= 0.862) was detected. Likewise, in the subset of typhoid patients (N=180) and controls (N=262) lacking HLA-DRB1*0301 individuals, no association between typhoid and the NRAMP1 (GT)n promoter polymorphism (χ2 3.82, df 2, P=0.166) or D2S1471 (χ2 13.716, df 17, P=0.9) was detected. This suggests that HLA susceptibility or resistance alleles are not “masking” an association of NRAMP1 polymorphisms with typhoid fever in these subsets of patients.
NRAMP1 may not only contribute to the risk of infection, but also may modulate the risk of disease development. A role for NRAMP1 in determining disease severity following S. typhi infection could be tested by comparing patients with complicated (e.g. intestinal perforations) versus acute disease. This hypothesis is attractive given that, in addition to its effects on acute susceptibility to virulent S. typhimurium infection, Nramp1 has a strong influence on T helper 1:T helper 2 bias as demonstrated in Nramp1 wildtype versus mutant mice infected with less virulent S. typhimurium [14]. The hypothesis is also of interest given the recent observation [15] that NRAMP1 is linked to development of acquired antimycobacterial immune responses in Vietnam. Specifically, NRAMP1 was linked to the Mitsuda reaction, a skin reaction that measures a granulomatous response to intradermally injected heat killed leprosy bacilli. These findings are consistent with the suggestion that NRAMP1 may influence Th1/Th2 differentiation. Although unknown, it has been suggested that intestinal perforations due to typhoid fever are immune mediated. Hence, although we have not shown an association between NRAMP1 and acute typhoid fever in this region of Vietnam, it remains of interest to investigate the role of NRAMP1 in the development of the acquired T cell-mediated immunity in S. typhi infection.
Acknowledgements
We thank the staff at the Centre for Tropical Diseases, Dong Thap Provincial hospital and Dong Nai Paediatric hospital for the clinical work associated with this study and we also extend our thanks to the Vietnamese individuals that took part in this study. This work was supported by The Wellcome Trust, UK.
Grant support: Wellcome Trust UK, Program grant number 035783.
Footnotes
Presented in part: Genetic Predisposition to Infectious Diseases, Inserm, November 1999, Aix-les-Bains, France
Informed consent was obtained from the individuals admitted into the study, which was approved by Dong Thap Hospital and the Health Services of Dong Thap Province. For studies in Dong Nai ethical approval was obtained by the Institutional Review Board of Dong Nai Paediatric Centre and the Ethical and Scientific Committee of the Centre for Tropical Diseases Ho Chi Minh, Vietnam
References
- 1.Bellamy R. The natural resistance-associated macrophage protein and susceptibility to intracellular pathogens. Microbes Infect. 1999;1:23–7. doi: 10.1016/s1286-4579(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 2.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–85. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 3.Hormaeche CE. Natural resistance to Salmonella typhimurium in different inbred mouse strains. Immunology. 1979;37:311–8. [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell JM. Structure and function of the natural-resistance-associated macrophage protein (Nramp1), a candidate protein for infectious and autoimmune disease susceptibility. Mol Med Today. 1996;2:205–11. doi: 10.1016/1357-4310(96)88773-9. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell JM, Barton CH, White JK, et al. Genomic organization and sequence of the human NRAMP gene: identification and mapping of a promoter region polymorphism. Mol Med. 1995;1:194–205. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Fujiwara TM, Buu NT, et al. Identification of polymorphisms and sequence variants in the human homologue of the mouse natural resistance-associated macrophage protein gene. Am J Hum Genet. 1995;56:845–53. [PMC free article] [PubMed] [Google Scholar]
- 7.Kossaczka Z, Lin FY, Ho VA, et al. Safety and immunogenicity of Vi conjugate vaccines for typhoid fever in adults, teenagers, and 2- to 4-year-Old children in Vietnam. Infect Immun. 1999;67:5806–10. doi: 10.1128/iai.67.11.5806-5810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunstan SJ, Stephens HA, Blackwell JM, et al. Genes of the Major Histocompatibility Complex are associated with Typhoid Fever. Journal of Infectious Diseases. 2001;183 doi: 10.1086/317940. In Press. [DOI] [PubMed] [Google Scholar]
- 9.Luxemburger C, Chau Minh Duc, Mai Ngoc Lanh, et al. Risk factors for typhoid fever in the Mekong Delta, southern Vietnam: A case-control study. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2000 doi: 10.1016/s0035-9203(01)90318-9. In Press. [DOI] [PubMed] [Google Scholar]
- 10.Sanjeevi CB, Miller EN, Dabadghao P, et al. Polymorphism at NRAMP1 and D2S1471 loci associated with Juvenile Rheumatoid Arthritis. Arthritis and Rheumatism. 2000:43. doi: 10.1002/1529-0131(200006)43:6<1397::AID-ANR25>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–4. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 12.Abel L, Sanchez FO, Oberti J, et al. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;177:133–45. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
- 13.John S, Marlow A, Hajeer A, Ollier W, Silman A, Worthington J. Linkage and association studies of the natural resistance associated macrophage protein 1 (NRAMP1) locus in rheumatoid arthritis. J Rheumatol. 1997;24:452–7. [PubMed] [Google Scholar]
- 14.Soo SS, Villarreal-Ramos B, Anjam Khan CM, Hormaeche CE, Blackwell JM. Genetic control of immune response to recombinant antigens carried by an attenuated Salmonella typhimurium vaccine strain: Nramp1 influences T-helper subset responses and protection against leishmanial challenge. Infect Immun. 1998;66:1910–7. doi: 10.1128/iai.66.5.1910-1917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcais A, Sanchez FO, Thuc NV, et al. Granulomatous reaction to intradermal injection of lepromin (Mitsuda reaction) is linked to the human NRAMP1 gene in Vietnamese leprosy sibships. J Infect Dis. 2000;181:302–8. doi: 10.1086/315174. [DOI] [PubMed] [Google Scholar]